Evaluation of an Ultrafiltration-Based Procedure for Simultaneous Recovery of Diverse Microbes in Source Waters
Abstract
:1. Introduction
2. Materials and Methods
2.1. Water Samples
2.2. Water Quality Testing
2.3. Microorganisms and Microbial Assays
2.4. Ultrafilter Blocking
2.5. Ultrafiltration Setup
2.6. Ultrafiltration Procedure
2.7. Secondary Sample Processing Procedures
2.8. Data Analysis and Statistics
3. Results and Discussion
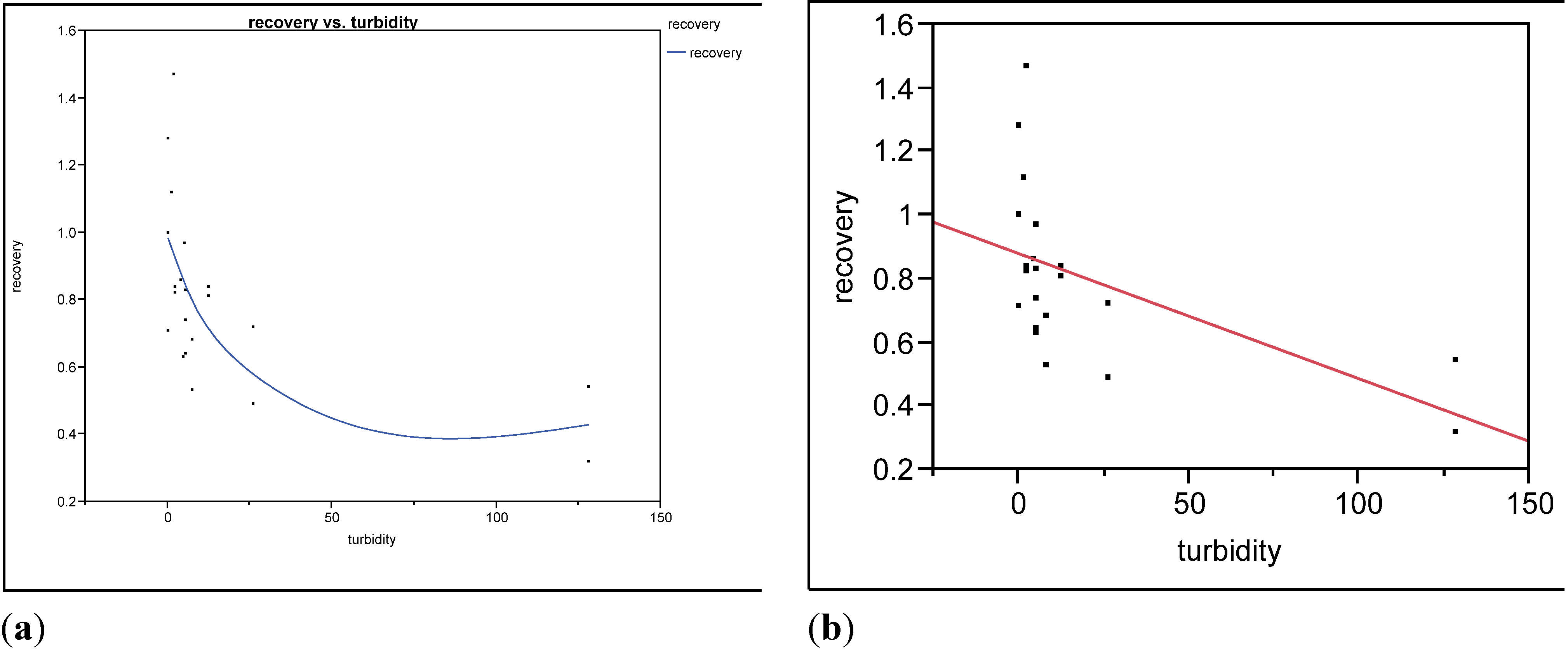
3.1. Water Quality
3.2. Microbial Recovery by ultrafiltration (UF)
| Site | pH | Turbidity (NTU) | Specific Conductance (µS/cm at 25 °C) | Hardness (mg/L as CaCO3) | Alkalinity (mg/L as CaCO3) | Total iron (mg/L Fe) | Total Suspended Solids (TSS) (mg/L as C) | Dissolved Organic Carbon (DOC) (mg/L as C) | Total Suspended Solids (TSS) (mg/L) |
|---|---|---|---|---|---|---|---|---|---|
| Chattahoochee river | 5.4 | 128 | 53.0 | 16 | 14 | 1.6 | 5.5 | 2.0 | 57 |
| 6.1 | 26.0 | 66.0 | 17 | 16 | 0.75 | 4.3 | 2.6 | NT * | |
| 6.9 | 5.25 | 61.0 | 16 | 16 | 0.24 | 1.8 | 1.4 | 7.2 | |
| Murphy Candler lake | 7.7 | 12.4 | 89.0 | 26 | 25 | 0.59 | 7.9 | 6.0 | 13 |
| 6.7 | 7.58 | 103 | 31 | 31 | 0.69 | 10 | 9.2 | 5.6 | |
| 7.6 | 4.09 | 119 | 28 | 27 | 0.82 | 12 | 13 | 4.0 | |
| Lawrenceville ground water | 8.2 | 0.097 | 233 | 120 | 124 | 0.08 | 7.0 | 7.3 | 0.3 |
| 8.3 | 0.113 | 235 | 110 | 96 | 0.09 | 10 | 9.9 | 0.15 | |
| 8.2 | 0.135 | 237 | 110 | 99 | 0.07 | 11 | 11 | <0.01 | |
| Jefferson City ground water | 8.1 | 2.11 | 693 | 290 | 250 | <0.02 | 28 | 20 | 2.4 |
| 7.8 | 1.89 | 572 | 290 | 240 | <0.02 | 28 | 28 | 1.7 | |
| 7.7 | 1.20 | 573 | 300 | 250 | 0.02 | 28 | 25 | 0.8 | |
| Allatoona lake | 9.2 | 5.25 | 70.5 | 22 | 24 | 0.08 | 7.7 | 0.90 | 6.5 |
| 9.0 | 4.98 | 73.1 | 21 | 22 | 0.07 | NT * | NT * | 7.2 | |
| 9.6 | 4.55 | 70.0 | 24 | 25 | 0.06 | NT * | NT * | 8.0 |
| Site | n | Average % Recovery Efficiency (SD) | |||||
|---|---|---|---|---|---|---|---|
| ΦX174 | MS2 | Echovirus 1 | E. coli | C. perfringens | C. parvum | ||
| Chattahoochee river | 6 * | 58 (16) | 91 (38) | 69 (9) | 98 (11) | 86 (15) | 78 (33) |
| Murphy Candler lake | 5 | 74 (14) | 65 (33) | 50 (15) | 85 (38) | 55 (10) | 70 (24) |
| Lawrenceville | 4 † | 100 (23) | 85 (23) | 130 (24) | ND ‡ | 73 (21) | 120 (37) |
| Jefferson City | 4 | 110 (31) | 77 (8) | 45 (27) | 87 (16) | 69 (10) | 120 (44) |
| Allatoona lake | 3 § | 81 (17) | 53 (19) | 53 (32) | 79 (12) | 100 (9) | 100 (11) |
| Cross-site avg. | 81 (26) | 76 (29) | 66 (33) | 88 (23) | 75 (20) | 95 (37) | |
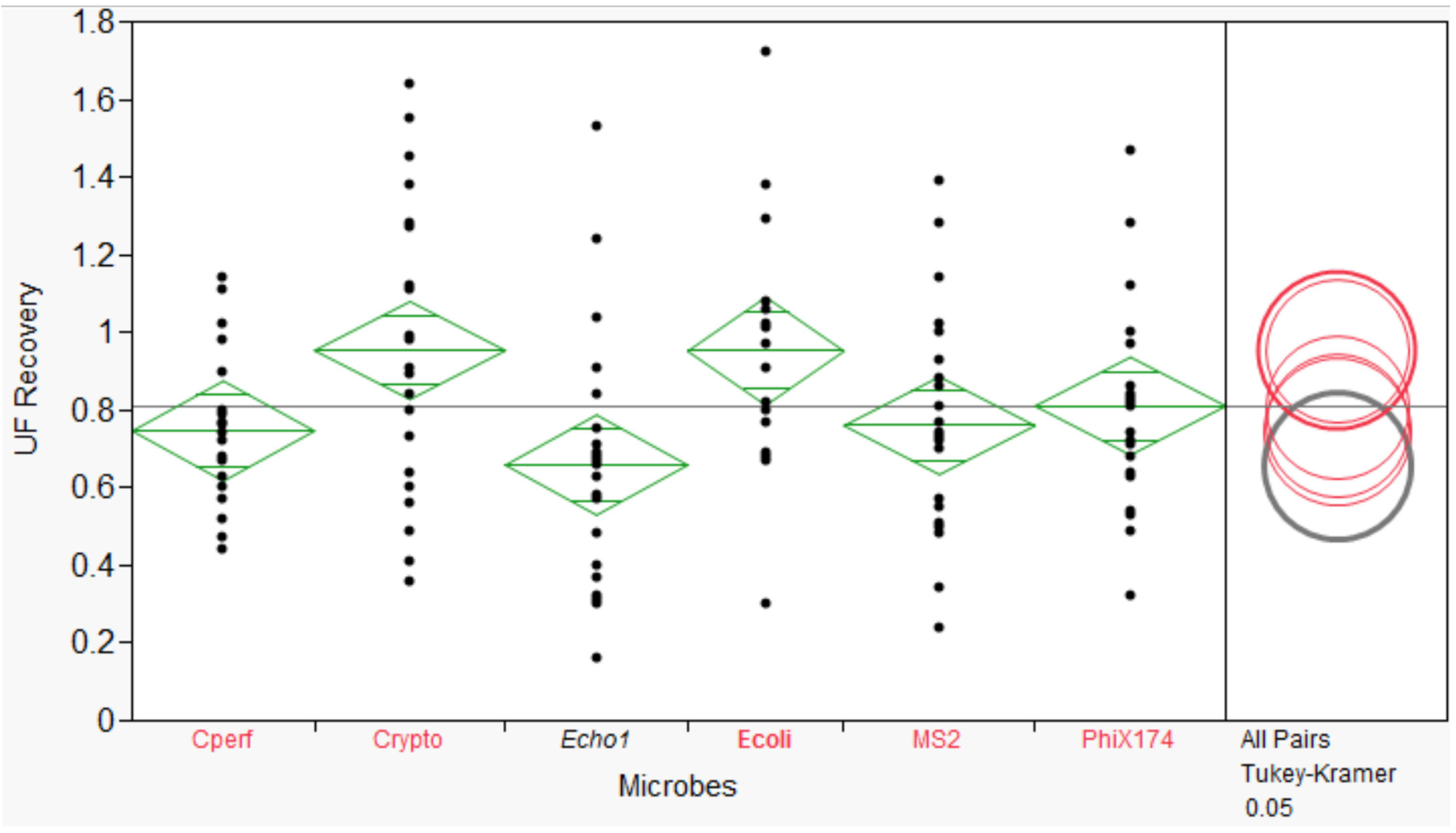
3.3. Microbial Recovery by Secondary Concentration
| Site | n | Average % Recovery Efficiency (SD) | ||||||
|---|---|---|---|---|---|---|---|---|
| ΦX174 | MS2 | Echovirus 1 | C. parvum | |||||
| Centricon | PEG | Centricon | PEG | Centricon | PEG | IMS | ||
| Chattahoochee river | 6 | 70 (27) | 88 (18) | 81 (16) | 69 (19) | 26 (28) | 52 (43) | 109 (56) |
| Murphy Candler | 5 | 67 (13) | 63 (14) | 81 (39) | 73 (12) | 30 (12) | 60 (41) | 87 (16) |
| Lawrenceville | 4 * | 80 (10) | 81 (11) | 93 (13) | 99 (28) | 56 (18) | 89 (22) | 85 (27) |
| Jefferson City | 4 * | 69 (6) | 72 (5) | 81 (8) | 75 (11) | 39 (19) | 220 (100) | 110 (47) |
| Lake Allatoona | 3 † | 64 (19) | 73 (20) | 55 (13) | 81 (9) | 31 (9) | 64 (21) | 48 |
| Cross-site avg. | 70 (17) | 76 (16) | 79 (23) | 78 (19) | 35 (21) | 92 (80) | 95 (40) | |
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Appendix
| Site | n | Overall % Recovery Efficiency (SD) | ||||||
|---|---|---|---|---|---|---|---|---|
| ΦX174 | MS2 | Echovirus 1 | C. parvum | |||||
| Centricon | PEG | Centricon | PEG | Centricon | PEG | IMS | ||
| Chattahoochee river | 6 | 41 (21) | 49 (14) | 69 (19) | 66 (36) | 17 (17) | 34 (28) | 82 (43) |
| Murphy Candler | 5 | 50 (14) | 46 (12) | 56 (33) | 44 (16) | 15 (7) | 32 (26) | 60 (21) |
| Lawrenceville | 4 * | 80 (24) | 83 (28) | 77 (13) | 80 (7) | 60 (13) | 100 (21) | 117 (43) |
| Jefferson City | 4 † | 73 (20) | 76 (21) | 62 (8) | 58 (9) | 15 (7) | 77 (21) | 109 (12) |
| Lake Allatoona | 3 ‡ | 53 (23) | 61 (29) | 27 (4) | 42 (13) | 15 (8) | 30 (9) | 47 |
| Cross-site avg. | 58 (24) | 61 (23) | 61 (24) | 59 (24) | 22 (19) | 51 (35) | 84 (38) | |
Conflicts of Interest
References
- Simmons, O.D.; Sobsey, M.D.; Heaney, C.D.; Schaefer, F.W.; Francy, D.S. Concentration and detection of cryptosporidium oocysts in surface water samples by method 1622 using ultrafiltration and capsule filtration. Appl. Environ. Microbiol. 2001, 67, 1123–1127. [Google Scholar] [CrossRef] [PubMed]
- Lambertini, E.; Spencer, S.K.; Bertz, P.D.; Loge, F.J.; Kieke, B.A.; Borchardt, M.A. Concentration of enteroviruses, adenoviruses, and noroviruses from drinking water by use of glass wool filters. Appl. Environ. Microbiol. 2008, 74, 2990–2996. [Google Scholar] [CrossRef] [PubMed]
- Francy, D.S.; Simmons, O.D., III; Ware, M.W.; Granger, E.J.; Sobsey, M.D.; Schaefer, F.W. Effects of seeding procedures and water quality on recovery of cryptosporidium oocysts from stream water by using U.S. Environmental agency method 1623. Appl. Environ. Microbiol. 2004, 70, 4118–4128. [Google Scholar] [CrossRef]
- Karim, M.R.; Rhodes, E.R.; Brinkman, N.; Wymer, L.; Fout, G.S. New electropositive filter for concentrating enteroviruses and noroviruses from large volumes of water. Appl. Environ. Microbiol. 2009, 75, 2393–2399. [Google Scholar] [CrossRef] [PubMed]
- Belfort, G.; Rotem, Y.; Katzenelson, E. Virus concentration using hollow fiber membranes. Water Res. 1974, 9, 79–85. [Google Scholar] [CrossRef]
- Belfort, G.; Rotem, Y.; Katzenelson, E. Virus concentration using hollow fiber membranes—II. Water Res. 1976, 10, 279–284. [Google Scholar] [CrossRef]
- Albinana-Gimenez, N.; Clemente-Casares, P.; Calgua, B.; Huguet, J.M.; Courtios, S.; Girones, R. Comparison of methods for concentrating human adenoviruses, polyomavirus JC and noroviruses in source waters and drinking water using quantitative PCR. J. Virol. Methods 2009, 158, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Bicknell, E.A.; Dziewulski, D.M.; Sturman, L.S.; Belfort, G. Concentration of seeded and naturally-occurring enteroviruses from waters of varying quality by hollow fiber ultrafiltration. Water Sci. Technol. 1985, 17, 47–62. [Google Scholar]
- Francy, D.S.; Bushon, R.N.; Brady, A.M.G.; Bertke, E.E.; Kephart, C.M.; Likirdopulos, C.A.; Mailot, B.E.; Schaefer, F.W., III; Lindquist, H.D.A. Comparison of traditional and molecular analytical methods for detecting biological agents in raw and drinking water following ultrafiltration. J. Appl. Microbiol. 2009, 107, 1479–1491. [Google Scholar] [CrossRef]
- Gibson, K.E.; Schwab, K.J. Tangential-flow ultrafiltration with integrated inhibition detection for recovery of surrogates and human pathogens from large-volume source water and finished drinking water. Appl. Environ. Microbiol. 2011, 77, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Hill, V.R.; Polaczyk, A.L.; Hahn, D.; Narayanan, J.; Cromeans, T.L.; Roberts, J.M.; Amburgey, J.E. Development of a rapid method for simultaneous recovery of diverse microbes in drinking water by ultrafiltration with sodium polyphosphate and surfactants. Appl. Environ. Microbiol. 2005, 71, 6878–6884. [Google Scholar] [CrossRef] [PubMed]
- Hill, V.R.; Kahler, A.M.; Jothikumar, N.; Johnson, T.B.; Hahn, D.; Cromeans, T.L. Multistate evaluation of an ultrafiltration-based procedure for simultaneous recovery of enteric microbes in 100-liter tap water samples. Appl. Environ. Microbiol. 2007, 73, 4218–4225. [Google Scholar] [CrossRef] [PubMed]
- Hill, V.R.; Mull, B.; Jothikumar, N.; Ferdinand, K.; Vinje, J. Detection of gi and gii noroviruses in ground water using ultrafiltration and taqman real-time RT-PCR. Food Environ. Virol. 2010, 2, 218–224. [Google Scholar] [CrossRef]
- Jansons, J.; Bucens, M.R. Virus detection in water by ultrafiltration. Water Res. 1986, 20, 1603–1606. [Google Scholar] [CrossRef]
- Jansons, J.; Bucens, M.R. Concentration of rotavirus by ultrafiltration. Water Res. 1986, 20, 79–83. [Google Scholar] [CrossRef]
- Kuhn, R.C.; Oshima, K.H. Evaluation and optimization of a reusable hollow fiber ultrafilter as a first step in concentrating cryptosporidium parvum oocysts from water. Water Res. 2001, 35, 2779–2783. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, R.C.; Oshima, K.H. Hollow-fiber ultrafiltration of cryptosporidium parvum oocysts from a wide variety of 10-l surface water samples. Can. J. Microbiol. 2002, 48, 542–549. [Google Scholar] [CrossRef] [PubMed]
- Morales-Morales, H.A.; Vidal, G.; Olszewski, J.; Rock, C.M.; Dasgupta, D.; Oshima, K.H.; Smith, G.B. Optimization of a reusable hollow-fiber ultrafilter for simultaneous concentration of enteric bacteria, protozoa, and viruses from water. Appl. Environ. Microbiol. 2003, 69, 4098–4102. [Google Scholar] [CrossRef] [PubMed]
- Mull, B.; Hill, V.R. Recovery and detection of escherichia coli o157: H7 in surface water, using ultrafiltration and real-time PCR. Appl. Environ. Microbiol. 2009, 75, 3593–3597. [Google Scholar] [CrossRef] [PubMed]
- Olszewski, J.; Winona, L.; Oshima, K.H. Comparison of two ultrafiltration systems for the concentration of seeded viruses from environmental waters. Can. J. Microbiol. 2005, 51, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Polaczyk, A.L.; Narayanan, J.; Cromeans, T.L.; Hahn, D.; Roberts, J.M.; Amburgey, J.E.; Hill, V.R. Ultrafiltration-based techniques for rapid and simultaneous concentration of multiple microbe classes from 100-L tap water samples. J. Microbiol. Methods 2008, 73, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Winona, L.J.; Ommani, A.W.; Olszewski, J.; Nuzzo, J.B.; Oshima, K.H. Efficient and predictable recovery of viruses from water by small scale ultrafiltration systems. Can. J. Microbiol. 2001, 47, 1033–1041. [Google Scholar] [CrossRef] [PubMed]
- Hill, V.R.; Polaczyk, A.L.; Kahler, A.M.; Cromeans, T.L.; Hahn, D.; Amburgey, J.E. Comparison of hollow-fiber ultrafiltration to the U.S. Environmental Protection Agency (USEPA) viradel technique and USEPA method 1623. J. Environ. Qual. 2009, 38, 822–825. [Google Scholar] [CrossRef] [PubMed]
- Wilson, I.G. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 1997, 63, 3741–3751. [Google Scholar] [PubMed]
- Kuhn, R.C.; Rock, C.M.; Oshima, K.H. Effects of ph and magnetic material on immunomagnetic separation of cryptosporidium oocysts from concentrated water samples. Appl. Environ. Microbiol. 2002, 68, 2066–2070. [Google Scholar] [CrossRef] [PubMed]
- Yakub, G.P.; Stadterman-Knauer, K.L. Evaluation of immunomagnetic separation for recovery of cryptosporidium parvum and giardia duodenalis from high-iron matrices. Appl. Environ. Microbiol. 2000, 66, 3628–3631. [Google Scholar] [CrossRef] [PubMed]
- Di Giovanni, G.D.; Hashemi, F.H.; Shaw, N.J.; Abrams, F.A.; LeChevallier, M.W.; Abbaszadegan, M. Detection of infections cryptosporidium parvum oocysts in surface and filter backwash water samples by immunomagnetic separation and integrated cell culture-pcr. Appl. Environ. Microbiol. 1999, 65, 3427–3432. [Google Scholar] [PubMed]
- American Public Health Association; American Water Works Association; Water Environment Federation. Standard Methods for the Examination of Water and Wastewater, 21st ed.; American Public Health Association: New York, NY, USA, 2005. [Google Scholar]
- Bisson, J.W.; Cabelli, V.J. Membrane filter enumeration method for clostridium perfringens. Appl. Environ. Microbiol. 1979, 37, 55–66. [Google Scholar] [PubMed]
- U.S. Environmental Protection Agency (USEPA). Method 1623: Cryptosporidium and Giardia in Water by Filtration/IMS/FA; EPA 815-R-05-002; Office of Water: Washington, DC, USA, 2005; p. 68.
- Leskinen, S.D.; Kearns, E.A.; Jones, W.L.; Miller, R.S.; Bevitas, C.R.; Kingsley, M.T.; Brigmon, R.L.; Lim, D.V. Automated dead-end ultrafiltration of large volume water samples to enable detection of low-level targets and reduce sample variability. J. Appl. Microbiol. 2012, 113, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Mull, B.; Hill, V.R. Recovery of diverse microbes in high turbidity surface water samples using dead-end ultrafiltration. J. Microbiol. Methods 2012, 91, 429–433. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.B.; Hill, V.R.; Hahn, D.; Johnson, T.B.; Pan, Y.; Jothikumar, N.; Moe, C.L. Hollow-fiber ultrafiltration for simultaneous recovery of viruses, bacteria and parasites from reclaimed water. J. Microbiol. Methods 2012, 88, 155–161. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kahler, A.M.; Johnson, T.B.; Hahn, D.; Narayanan, J.; Derado, G.; Hill, V.R. Evaluation of an Ultrafiltration-Based Procedure for Simultaneous Recovery of Diverse Microbes in Source Waters. Water 2015, 7, 1202-1216. https://doi.org/10.3390/w7031202
Kahler AM, Johnson TB, Hahn D, Narayanan J, Derado G, Hill VR. Evaluation of an Ultrafiltration-Based Procedure for Simultaneous Recovery of Diverse Microbes in Source Waters. Water. 2015; 7(3):1202-1216. https://doi.org/10.3390/w7031202
Chicago/Turabian StyleKahler, Amy M., Trisha B. Johnson, Donghyun Hahn, Jothikumar Narayanan, Gordana Derado, and Vincent R. Hill. 2015. "Evaluation of an Ultrafiltration-Based Procedure for Simultaneous Recovery of Diverse Microbes in Source Waters" Water 7, no. 3: 1202-1216. https://doi.org/10.3390/w7031202





