Review of the Application of Modern Cytogenetic Methods (FISH/GISH) to the Study of Reticulation (Polyploidy/Hybridisation)
Abstract
:1. Introduction
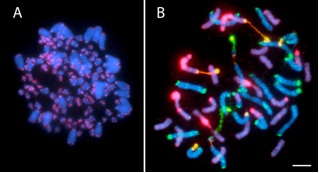
Applications of ISH
2. Recently Formed Allopolyploid Species
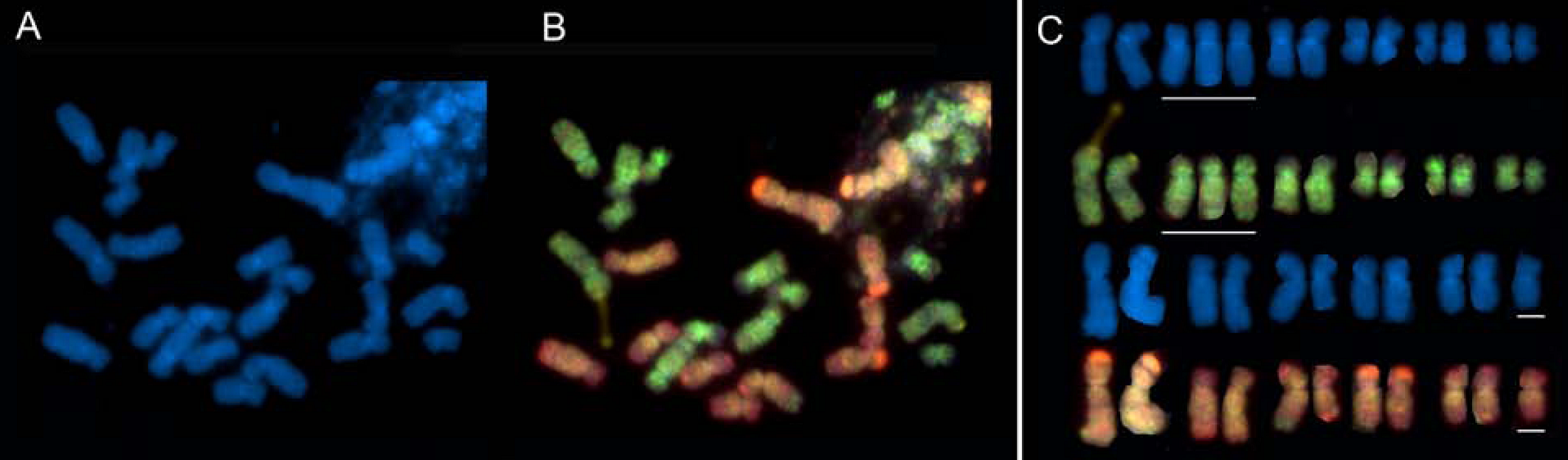
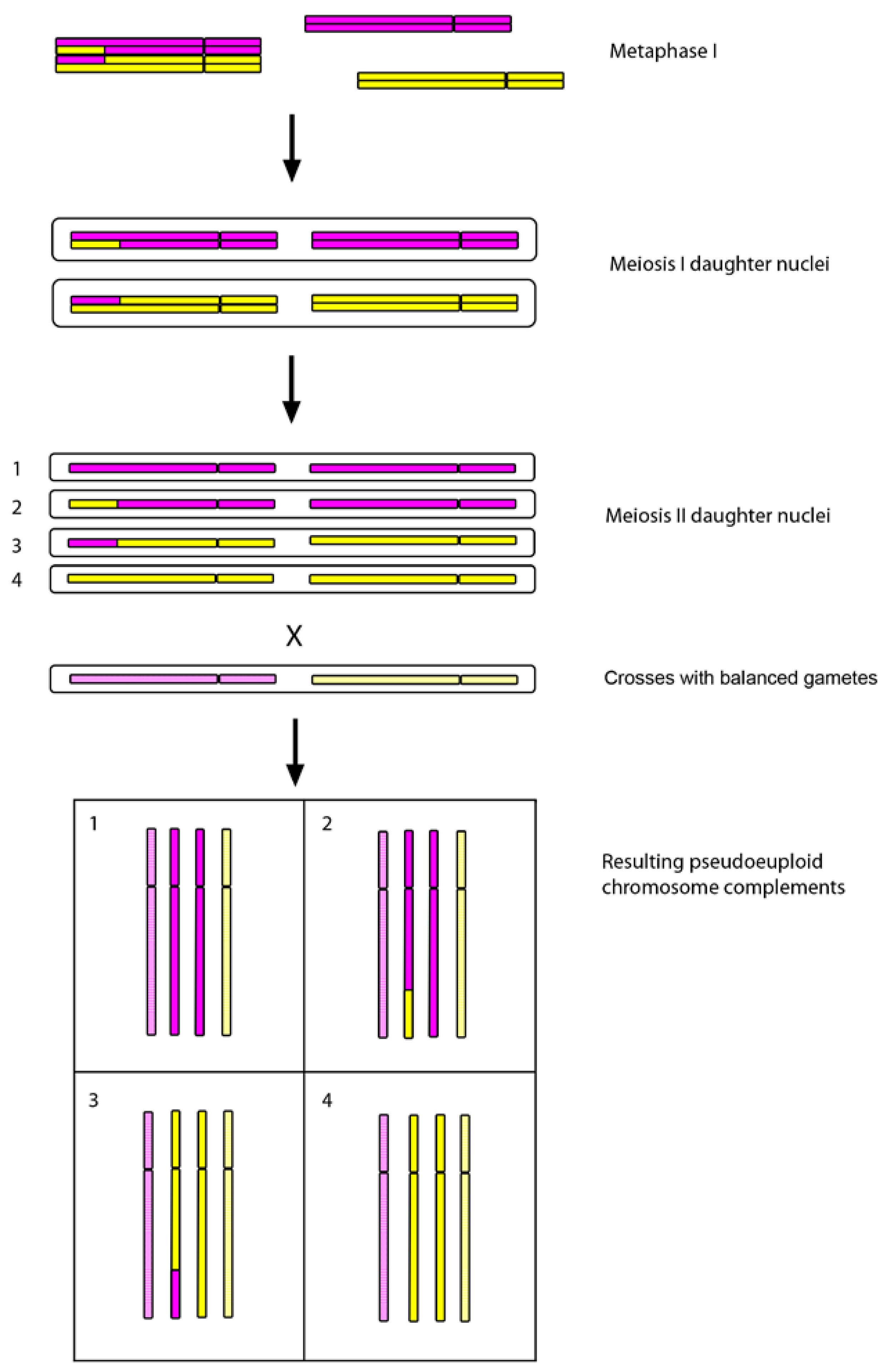
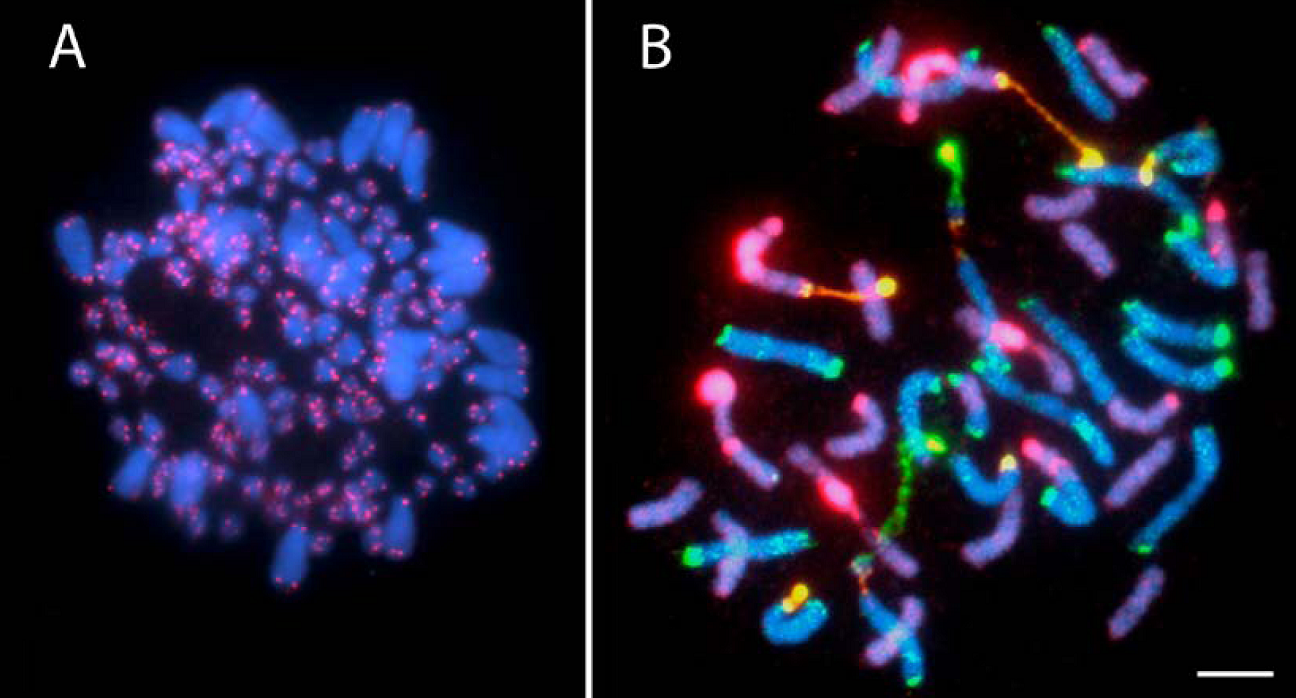
3. Autopolyploidy, Polyploid Species Complexes and Reticulate Evolution
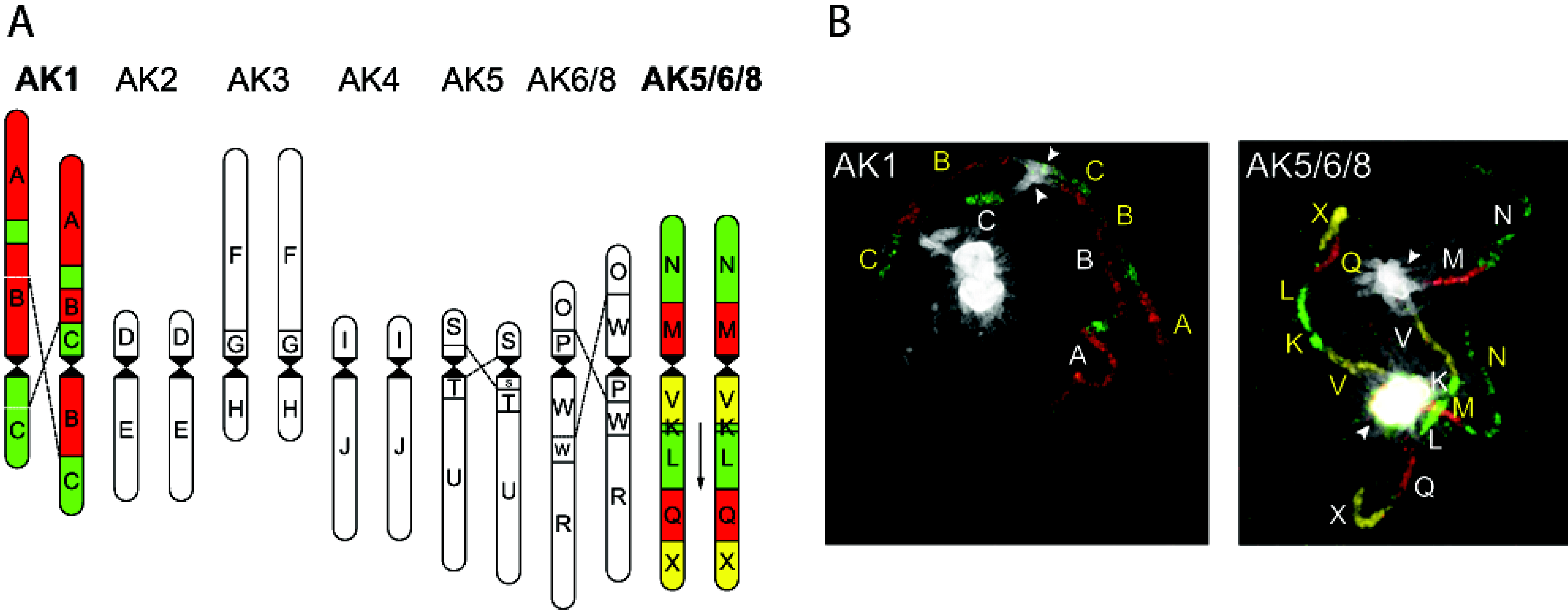
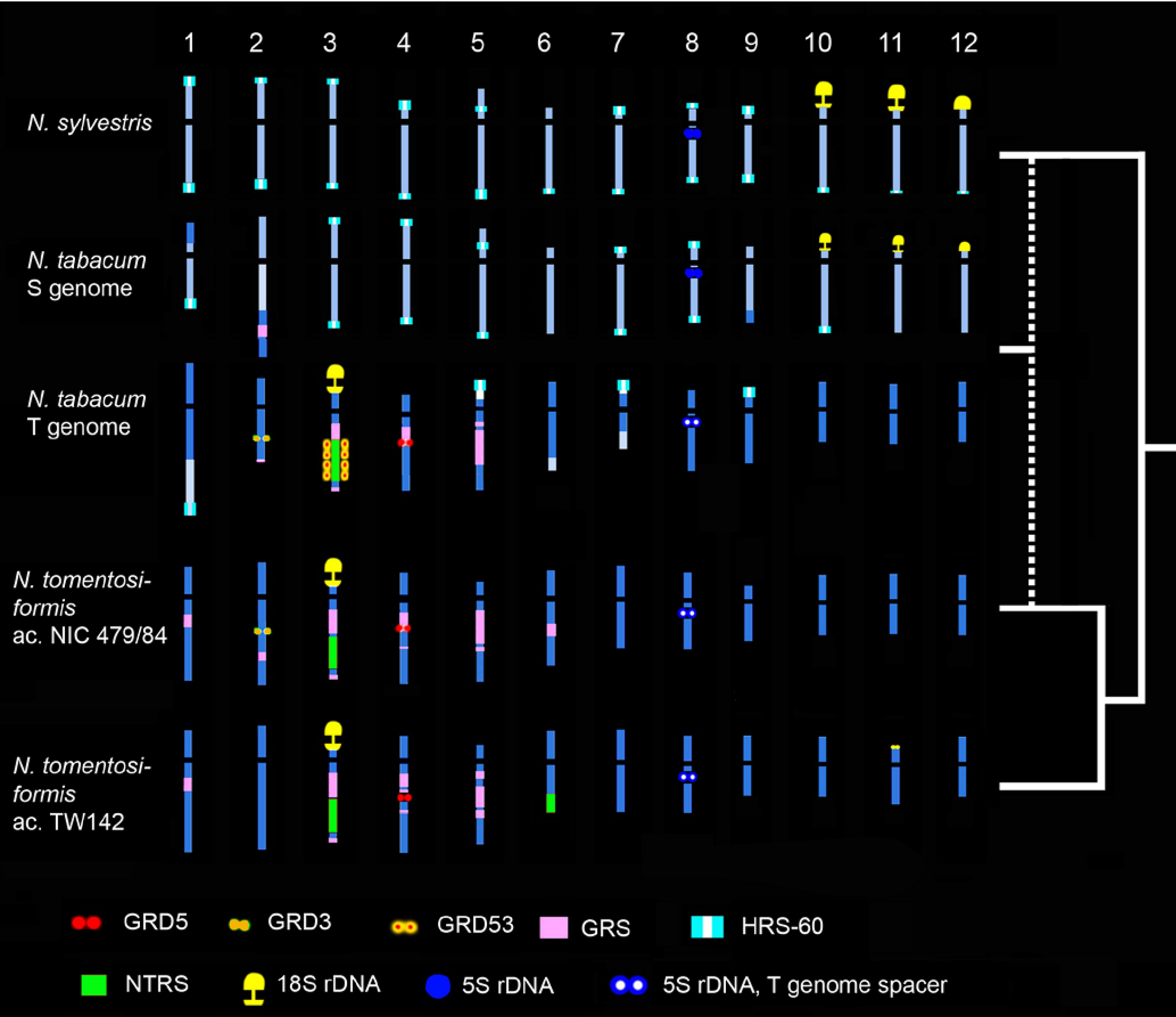
4. Interspecific Hybridisation and Introgression in Plant Breeding
5. Sequence Dynamics
6. Paleoallopolyploidy
7. Next-Generation Sequencing (NGS), FISH Studies and the Future
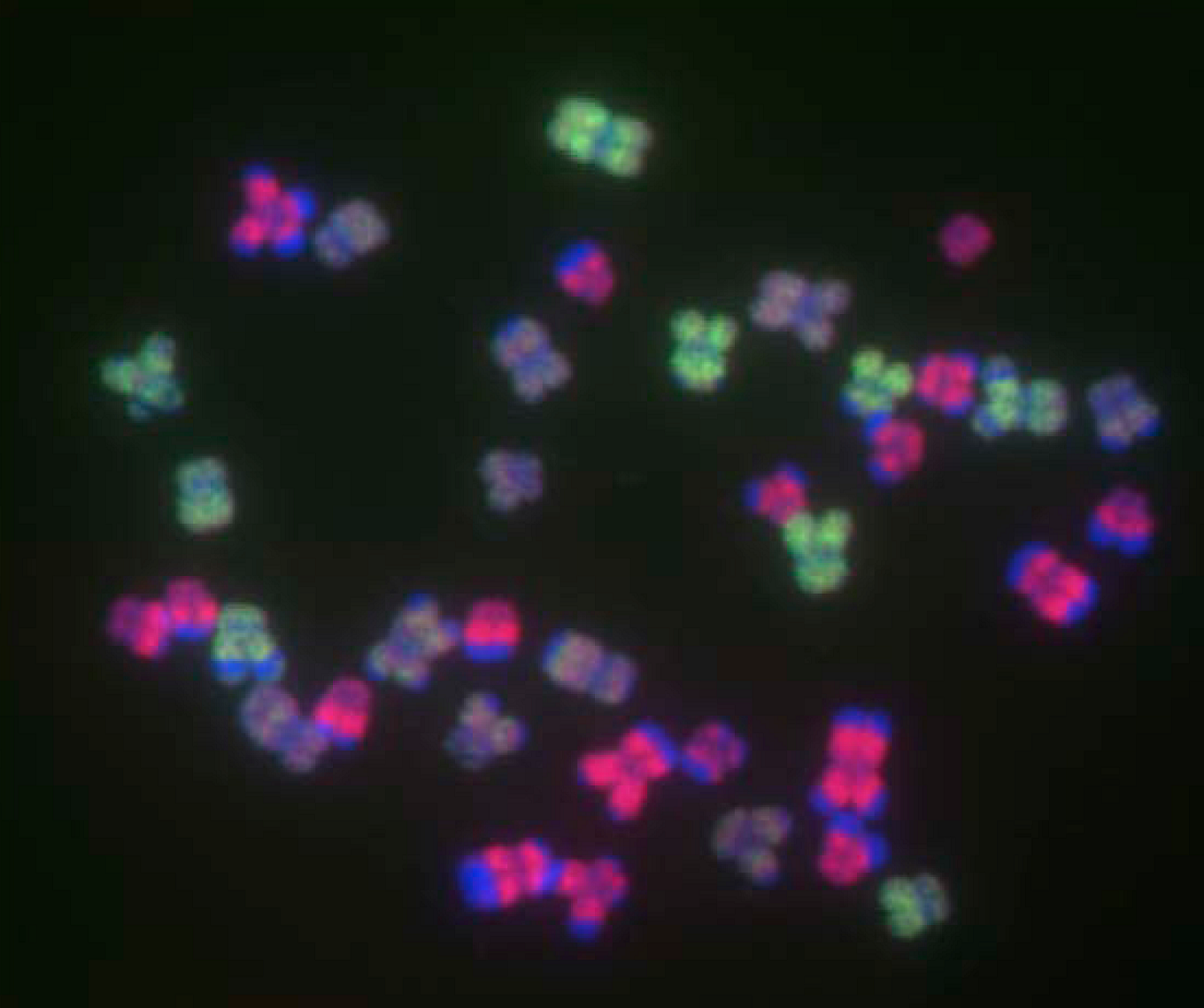
8. Conclusions
Acknowledgments
References
- Leitch, A.R. Higher levels of organization in the interphase nucleus of cycling and differentiated cells. Microbiol. Mol. Biol. Rev. 2000, 64, 138–152. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.Y.; Souckova-Skalicka, K.; Sarasan, V.; Clarkson, J.J.; Chase, M.W.; Kovarik, A.; Leitch, A.R. A genetic appraisal of a new synthetic Nicotiana tabacum (Solanaceae) and the Kostoff synthetic tobacco. Am. J. Bot. 2006, 93, 875–883. [Google Scholar] [CrossRef]
- Schwarzacher, T.; Leitch, A.R.; Bennett, M.D.; Heslop-Harrison, J.S. In situ localization of parental genomes in a wide hybrid. Ann. Bot. 1989, 64, 315–324. [Google Scholar]
- Le, H.; Armstrong, K.; Miki, B. Detection of rye DNA in wheat-rye hybrids and wheat translocation stocks using total genomic DNA as a probe. Plant Mol. Biol. Rep. 1989, 7, 150–158. [Google Scholar] [CrossRef]
- Fedak, G. Molecular aids for integration of alien chromatin through wide crosses. Genome 1999, 42, 584–591. [Google Scholar] [CrossRef]
- Jauhar, P.P.; Chibbar, R.N. Chromosome-mediated and direct gene transfers in wheat. Genome 1999, 42, 570–583. [Google Scholar] [CrossRef]
- Thomas, H.M.; Morgan, W.G.; Humphreys, M.W. Designing grasses with a future - combining the attributes of Lolium and Festuca. Euphytica 2003, 133, 19–26. [Google Scholar] [CrossRef]
- Raina, S.N.; Rani, V. GISH technology in plant genome research. Methods Cell Sci. 2001, 23, 83–104. [Google Scholar] [CrossRef] [PubMed]
- Bennett, S.T.; Kenton, A.Y.; Bennett, M.D. Genomic in situ hybridization reveals the allopolyploid nature of Milium montianum (Gramineae). Chromosoma 1992, 101, 420–424. [Google Scholar] [CrossRef]
- Markova, M.; Michu, E.; Vyskot, B.; Janousek, B.; Zluvova, J. An interspecific hybrid as a tool to study phylogenetic relationships in plants using the GISH technique. Chromosome Res. 2007, 15, 1051–1059. [Google Scholar] [CrossRef] [PubMed]
- Chase, M.W.; Knapp, S.; Cox, A.V.; Clarkson, J.J.; Butsko, Y.; Joseph, J.; Savolainen, V.; Parokonny, A.S. Molecular systematics, GISH and the origin of hybrid taxa in Nicotiana (Solanaceae). Ann. Bot.-London 2003, 92, 107–127. [Google Scholar] [CrossRef]
- Dierschke, T.; Mandakova, T.; Lysak, M.A.; Mummenhoff, K. A bicontinental origin of polyploid Australian/New Zealand Lepidium species (Brassicaceae)? Evidence from genomic in situ hybridization . Ann. Bot. 2009, 104, 681–688. [Google Scholar] [CrossRef] [PubMed]
- Guggisberg, A.; Baroux, C.; Grossniklaus, U.; Conti, E. Genomic origin and organization of the allopolyploid Primula egaliksensis investigated by in situ hybridization. Ann. Bot. 2008, 101, 919–927. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.S.; Ding, Z.Y.; Liu, W.; Pan, J.; Li, C.B.; Ge, S.; Zhang, D.M. Polyploid evolution in Oryza officinalis complex of the genus Oryza. BMC Evol. Biol. 2009, 9, 250. [Google Scholar] [CrossRef]
- Zhang, D.M.; Sang, T. Physical mapping of ribosomal RNA genes in peonies (Paeonia, Paeoniaceae) by fluorescent in situ hybridization: Implications for phylogeny and concerted evolution. Am. J. Bot. 1999, 86, 735–740. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.P.; Leitch, I.J.; Bennet, M.D.; Chase, M.W.; Leitch, A.R. Ribosomal DNA evolution and phylogeny in Aloe (Asphodelaceae). Am. J. Bot. 2000, 87, 1578–1583. [Google Scholar] [CrossRef] [PubMed]
- Ran, Y.D.; Hammett, K.R.W.; Murray, B.G. Phylogenetic analysis and karyotype evolution in the genus Clivia (Amaryllidaceae). Ann. Bot. 2001, 87, 823–830. [Google Scholar] [CrossRef]
- Clarkson, J.J.; Lim, K.Y.; Kovarik, A.; Chase, M.W.; Knapp, S.; Leitch, A.R. Long-term genome diploidization in allopolyploid Nicotiana section Repandae (Solanaceae). New Phytol. 2005, 168, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.Y.; Kovarik, A.; Matyasek, R.; Chase, M.W.; Clarkson, J.J.; Grandbastien, M.A.; Leitch, A.R. Sequence of events leading to near-complete genome turnover in allopolyploid Nicotiana within five million years. New Phytol. 2007, 175, 756–763. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.L.; Zhang, D.; Hong, D.Y.; Wang, X.R. Chromosomal localization of 5S and 18S-5.8S-25S ribosomal DNA sites in five Asian pines using fluorescence in situ hybridization . Theor. Appl. Genet. 2003, 106, 198–204. [Google Scholar] [PubMed]
- Jiang, J.M.; Gill, B.S. Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research. Genome 2006, 49, 1057–1068. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.M.; Gill, B.S.; Wang, G.L.; Ronald, P.C.; Ward, D.C. Metaphase and interphase fluorescence in-situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc. Natl. Acad. Sci. U. S. A. 1995, 92, 4487–4491. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Klein, P.E.; Klein, R.R.; Price, H.J.; Mullet, J.E.; Stelly, D.M. Chromosome identification and nomenclature of Sorghum bicolor. Genetics 2005, 169, 1169–1173. [Google Scholar] [CrossRef] [PubMed]
- Koumbaris, G.L.; Bass, H.W. A new single-locus cytogenetic mapping system for maize (Zea mays L.): overcoming FISH detection limits with marker-selected sorghum (S. propinquum L.) BAC clones. Plant J. 2003, 35, 647–659. [Google Scholar] [CrossRef] [PubMed]
- Howell, E.C.; Armstrong, S.J.; Barker, G.C.; Jones, G.H.; King, G.J.; Ryder, C.D.; Kearsey, M.J. Physical organization of the major duplication on Brassica oleracea chromosome O6 revealed through fluorescence in situ hybridization with Arabidopsis and Brassica BAC probes. Genome 2005, 48, 1093–1103. [Google Scholar] [CrossRef] [PubMed]
- Walling, J.G.; Shoemaker, R.; Young, N.; Mudge, J.; Jackson, S. Chromosome-level homeology in paleopolyploid soybean (Glycine max) revealed through integration of genetic and chromosome maps. Genetics 2006, 172, 1893–1900. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Guo, W.Z.; Zhang, T.Z. Detection and mapping of homologous and homoeologous segments in homoeologous groups of allotetraploid cotton by BAC-FISH . BMC Genomics 2007, 8. [Google Scholar]
- Wang, K.; Song, X.L.; Han, Z.G.; Guo, W.Z.; Yu, J.Z.; Sun, J.; Pan, J.J.; Kohel, R.J.; Zhang, T.Z. Complete assignment of the chromosomes of Gossypium hirsutum L. by translocation and fluorescence in situ hybridization mapping. Theor. Appl. Genet. 2006, 113, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Ming, R.; Hou, S.B.; Feng, Y.; Yu, Q.Y.; Dionne-Laporte, A.; Saw, J.H.; Senin, P.; Wang, W.; Ly, B.V.; Lewis, K.L.T.; et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus) . Nature 2008, 452, U991–U997. [Google Scholar] [CrossRef]
- Mandakova, T.; Lysak, M.A. Chromosomal phylogeny and karyotype evolution in x=7 crucifer species (Brassicaceae). Plant Cell 2008, 20, 2559–2570. [Google Scholar] [CrossRef] [PubMed]
- Lysak, M.A.; Berr, A.; Pecinka, A.; Schmidt, R.; McBreen, K.; Schubert, I. Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 5224–5229. [Google Scholar] [CrossRef] [PubMed]
- Lysak, M.A.; Koch, M.A.; Pecinka, A.; Schubert, I. Chromosome triplication found across the tribe Brassiceae. Genome Res. 2005, 15, 516–525. [Google Scholar] [CrossRef] [PubMed]
- Ziolkowski, P.A.; Kaczmarek, M.; Babula, D.; Sadowski, J. Genome evolution in Arabidopsis/Brassica: conservation and divergence of ancient rearranged segments and their breakpoints. Plant J. 2006, 47, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Lysak, M.A.; Cheung, K.; Kitschke, M.; Bures, P. Ancestral chromosomal blocks are triplicated in Brassiceae species with varying chromosome number and genome size. Plant Physiol. 2007, 145, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Ownbey, M. Natural hybridization and amphiploidy in the genus Tragopogon. Am. J. Bot. 1950, 37, 487–499. [Google Scholar] [CrossRef]
- Symonds, V.V.; Soltis, P.S.; Soltis, D.E. Dynamics of polyploid formation in Tragopogon (Asteracaeae): recurrent formation, gene flow, and population structure . Evolution 2010, in press. [Google Scholar]
- Soltis, P.S.; Plunkett, G.M.; Novak, S.J.; Soltis, D.E. Genetic variation in Tragopogon species - additional origins of the allotetraploids Tmirus and T. miscellus (Compositae). Am. J. Bot. 1995, 82, 1329–1341. [Google Scholar] [CrossRef]
- Soltis, P.S.; Soltis, D.E. Multiple origins of the allotetraploid Tragopogon mirus (Compositae) - rDNA evidence. Syst. Bot. 1991, 16, 407–413. [Google Scholar] [CrossRef]
- Ownbey, M.; McCollum, G.D. Cytoplasmic inheritance and reciprocal amphiploidy in Tragopogon. Am. J. Bot. 1953, 40, 788–796. [Google Scholar] [CrossRef]
- Lim, K.Y.; Soltis, D.E.; Soltis, P.S.; Tate, J.; Matyasek, R.; Srubarova, H.; Kovarik, A.; Pires, J.C.; Xiong, Z.Y.; Leitch, A.R. Rapid chromosome evolution in recently formed polyploids in Tragopogon (Asteraceae) . Plos One 2008, 3, e3353. [Google Scholar] [CrossRef] [PubMed]
- Szadkowski, E.; Eber, F.; Huteau, V.; Lode, M.; Huneau, C.; Belcram, H.; Coriton, O.; Manzanares-Dauleux, M.J.; Delourme, R.; King, G.J.; Chalhoub, B.; Jenczewski, E.; Chevre, A.M. The first meiosis of resynthesized Brassica napus, a genome blender. New Phytol. 2010, 186, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.A.; Joshi, P.; Soltis, K.A.; Soltis, P.S.; Soltis, D.E. On the road to diploidization? Homoeolog loss in independently formed populations of the allopolyploid Tragopogon miscellus (Asteraceae) . BMC Plant Biol. 2009, 9, 80. [Google Scholar] [CrossRef] [PubMed]
- Buggs, R.J.A.; Doust, A.N.; Tate, J.A.; Koh, J.; Soltis, K.; Feltus, F.A.; Paterson, A.H.; Soltis, P.S.; Soltis, D.E. Gene loss and silencing in Tragopogon miscellus (Asteraceae): comparison of natural and synthetic allotetraploids. Heredity 2009, 103, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.A.; Ni, Z.F.; Scheen, A.C.; Koh, J.; Gilbert, C.A.; Lefkowitz, D.; Chen, Z.J.; Soltis, P.S.; Soltis, D.E. Evolution and expression of homeologous loci in Tragopogon miscellus (Asteraceae), a recent and reciprocally formed allopolyploid. Genetics 2006, 173, 1599–1611. [Google Scholar] [CrossRef] [PubMed]
- Buggs, R.J.A.; Chamala, S.; Wu, W.; Gao, L.; May, G.D.; Schnable, P.S.; Soltis, D.E.; Soltis, P.S.; Barbazuk, W.B. Characterization of duplicate gene evolution in the recent natural allopolyploid Tragopogon miscellus by next-generation sequencing and Sequenom iPLEX MassARRAY genotyping. Mol. Ecol. 2010, 19, 132–146. [Google Scholar] [CrossRef] [PubMed]
- Koh, J.; Soltis, P.S.; Soltis, D.E. Homeolog loss and expression changes in natural populations of the recently and repeatedly formed allotetraploid Tragopogon mirus (Asteraceae). BMC Genomics 2010, 11, 97–112. [Google Scholar] [CrossRef] [PubMed]
- Marchant, C.J. Corrected chromosome numbers for Spartina x townsendii and its parent species. Nature 1963, 199, 929. [Google Scholar] [CrossRef]
- Ainouche, M.L.; Baumel, A.; Salmon, A. Spartina anglica C.E. Hubbard: a natural model system for analysing early evolutionary changes that affect allopolyploid genomes. Biol. J. Linn. Soc. 2004, 82, 475–484. [Google Scholar] [CrossRef]
- Hubbard, J.C.E. Spartina marshes in southern England. 6. Pattern of invasion in Poole Harbor. J. Ecol. 1965, 53, 799–813. [Google Scholar] [CrossRef]
- Ayres, D.R.; Strong, D.R. Origin and genetic diversity of Spartina anglica (Poaceae) using nuclear DNA markers. Am. J. Bot. 2001, 88, 1863–1867. [Google Scholar] [CrossRef]
- Raybould, A.F.; Gray, A.J.; Lawrence, M.J.; Marshall, D.F. The evolution of Spartina anglica Hubbard, C.E. (Gramineae) - Genetic variation and status of the parental species in Britain. Biol. J. Linn. Soc. 1991, 44, 369–380. [Google Scholar] [CrossRef]
- Renny-Byfield, S.; Ainouche, M.; Leitch, I.J.; Lim, K.Y.; Comber, S.C.L.; Leitch, A.R. Flow cytometry and GISH reveal mixed ploidy populations and Spartina nonaploids with genomes of Salterniflora and S. maritima origin. Ann. Bot. 2010, 105, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Clapham, A.R.; Tutin, T.G.; Warburg, E.F. Flora of the British Isles, 2nd ed; Cambridge University Press: Cambridge; New York, NY, USA, 1962. [Google Scholar]
- Ramsey, J.; Schemske, D.W. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 1998, 29, 467–501. [Google Scholar] [CrossRef]
- Richards, A.J. Eutriploid Facultative Agamospermy in Taraxacum. New Phytol. 1970, 69, 761–774. [Google Scholar] [CrossRef]
- Tel-Zur, N.; Abbo, S.; Mizrahi, Y. Cytogenetics of semi-fertile triploid and aneuploid intergeneric vine cacti hybrids. J. Hered. 2005, 96, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Upcott, M.; Philp, J. The genetic structure of Tulipa - IV balance, selection and fertility. J. Genet. 1939, 38, 91–123. [Google Scholar] [CrossRef]
- Puizina, J.; Papes, D. Further cytogenetic analyses of the Croatian triploid shallot "Ljutika" (Allium cepa var. viviparum, Alliaceae) and its comparison with the Indian triploid "Pran". Plant Syst. Evol. 1997, 208, 11–23. [Google Scholar] [CrossRef]
- Carputo, D. Cytological and breeding behavior of pentaploids derived from 3x x 4x crosses in potato. Theor. Appl. Genet. 2003, 106, 883–888. [Google Scholar] [PubMed]
- Henry, I.M.; Dilkes, B.P.; Tyagi, A.P.; Lin, H.Y.; Comai, L. Dosage and parent-of-origin effects shaping aneuploid swarms in A. thaliana. Heredity 2009, 103, 458–468. [Google Scholar] [CrossRef] [PubMed]
- Laverty, T.; Vorsa, N. Fertility of aneuploids between the 5x and 6x levels in blueberry - the potential for gene-transfer from 4x to 6x Levels. J. Am. Soc. Hort. Sci. 1991, 116, 330–335. [Google Scholar]
- Stebbins, G.L. Chromosome evolution in higher plants; 1971; J. W. Arrowsmith Ltd.: Bristol, UK. [Google Scholar]
- Hasterok, R.; Draper, J.; Jenkins, G. Laying the cytotaxonomic foundations of a new model grass, Brachypodium distachyon (L.) Beauv . Chromosome Res. 2004, 12, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Schranz, M.E.; Lysak, M.A.; Mitchell-Olds, T. The ABC's of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 2006, 11, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Dobes, C.H.; Mitchell-Olds, T.; Koch, M.A. Extensive chloroplast haplotype variation indicates Pleistocene hybridization and radiation of North American Arabis drummondii, A. x divaricarpa, and A. holboellii (Brassicaceae). Mol. Ecol. 2004, 13, 349–370. [Google Scholar] [CrossRef] [PubMed]
- Dobes, C.; Mitchell-Olds, T.; Koch, M.A. Intraspecific diversification in North American Boechera stricta (=Arabis drummondii), Boechera x divaricarpa, and Boechera holboellii (Brassicaceae) inferred from nuclear and chloroplast molecular markers - An integrative approach. Am. J. Bot. 2004, 91, 2087–2101. [Google Scholar] [CrossRef]
- Kohler, C.; Scheid, O.M.; Erilova, A. The impact of the triploid block on the origin and evolution of polyploid plants. Trends Genet. 2010, 26, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Ehlenfeldt, M.K.; Ortiz, R. Evidence on the nature and origins of endosperm dosage requirements in Solanum and other angiosperm genera. Sexual Plant Reproduction 1995, 8, 189–196. [Google Scholar] [CrossRef]
- Kantama, L.; Sharbel, T.F.; Schranz, M.E.; Mitchell-Olds, T.; de Vries, S.; de Jong, H. Diploid apomicts of the Boechera holboellii complex display large-scale chromosome substitutions and aberrant chromosomes. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 14026–14031. [Google Scholar] [CrossRef] [PubMed]
- Speeckaert, F.; Jacobs, M. Study of the divergence of moderately repetitive sequences in Nicotiana species and in protoclones of Nicotiana plumbaginifolia Viviani. Theor. Appl. Genet. 1988, 75, 746–750. [Google Scholar] [CrossRef]
- Kenton, A.; Khashoggi, A.; Parokonny, A.; Bennett, M.D.; Lichtenstein, C. Chromosomal location of endogenous geminivirus-related DNA sequences in Nicotiana tabacum L. Chromosome Res. 1995, 3, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.Y.; Matyasek, R.; Lichtenstein, C.P.; Leitch, A.R. Molecular cytogenetic analyses and phylogenetic studies in the Nicotiana section Tomentosae. Chromosoma 2000, 109, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Murad, L.; Lim, K.Y.; Christopodulou, V.; Matyasek, R.; Lichtenstein, C.P.; Kovarik, A.; Leitch, A.R. The origin of tobacco's T genome is traced to a particular lineage within Nicotiana tomentosiformis (Solanaceae). Am. J. Bot. 2002, 89, 921–928. [Google Scholar] [CrossRef]
- Murad, L.; Bielawski, J.P.; Matyasek, R.; Kovarik, A.; Nichols, R.A.; Leitch, A.R.; Lichtenstein, C.P. The origin and evolution of geminivirus-related DNA sequences in Nicotiana. Heredity 2004, 92, 352–358. [Google Scholar] [CrossRef] [PubMed]
- Schubert, I.; Fransz, P.F.; Fuchs, J.; de Jong, J.H. Chromosome painting in plants. Methods Cell Sci. 2001, 23, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.M.; Gill, B.S. Nonisotopic in-situ hybridization and plant genome mapping - the first 10 years. Genome 1994, 37, 717–725. [Google Scholar] [PubMed]
- Lysak, M.A.; Lexer, C. Towards the era of comparative evolutionary genomics in Brassicaceae. Plant Syst. Evol. 2006, 259, 175–198. [Google Scholar] [CrossRef]
- Ali, H.B.M.; Lysak, M.A.; Schubert, I. Genomic in situ hybridization in plants with small genomes is feasible and elucidates the chromosomal parentage in interspecific Arabidopsis hybrids. Genome 2004, 47, 954–960. [Google Scholar] [CrossRef] [PubMed]
- Säll, T.; Jakobsson, M.; Lind-Halldén, C.; C., H. Chloroplast DNA indicates a single origin of the allotetraploid Arabidopsis suecica. J. Evol. Biol. 2003, 16, 1019–1029. [Google Scholar] [CrossRef] [PubMed]
- O'Kane, S.L.; Schaal, B.A.; AlShehbaz, I.A. The origins of Arabidopsis suecica (Brassicaceae) as indicated by nuclear rDNA sequences. Syst. Bot. 1996, 21, 559–566. [Google Scholar] [CrossRef]
- Jakobsson, M.; Hagenblad, J.; Tavare, S.; Sall, T.; Hallden, C.; Lind-Hallden, C.; Nordborg, M. A unique recent origin of the allotetraploid species Arabidopsissuecica: Evidence from nuclear DNA markers. Mol. Biol. Evol. 2006, 23, 1217–1231. [Google Scholar] [CrossRef] [PubMed]
- Madlung, A.; Tyagi, A.P.; Watson, B.; Jiang, H.M.; Kagochi, T.; Doerge, R.W.; Martienssen, R.; Comai, L. Genomic changes in synthetic Arabidopsis polyploids. Plant J. 2005, 41, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Wright, K.M.; Pires, J.C.; Madlung, A. Mitotic instability in resynthesized and natural polyploids of the genus Arabidopsis (Brassicaceae). Am. J. Bot. 2009, 96, 1656–1664. [Google Scholar] [CrossRef]
- Gill, B.S. Nucleo-cytoplasmic interaction (NCI) hypothesis of genome evolution and speciation in polyploid plants. Nuclear and organellar genomes of wheat species . In Proceedings of the Kihara Memorial International Symposium on Cytoplasmic Engineering in Wheat; Sasakuma, T., Kinoshita, T., Eds.; Yokohama, Japan, 1991; pp. 48–53. [Google Scholar]
- Leitch, I.J.; Bennett, M.D. Polyploidy in angiosperms. Trends Plant Sci. 1997, 2, 470–476. [Google Scholar] [CrossRef]
- Kenton, A.; Parokonny, A.S.; Gleba, Y.Y.; Bennett, M.D. Characterization of the Nicotiana tabacum L genome by molecular cytogenetics. Mol. Gen. Genet. 1993, 240, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.Y.; Kovarik, A.; Matyasek, R.; Bezdek, M.; Lichtenstein, C.P.; Leitch, A.R. Gene conversion of ribosomal DNA in Nicotiana tabacum is associated with undermethylated, decondensed and probably active gene units. Chromosoma 2000, 109, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Jellen, E.N.; Gill, B.S.; Cox, T.S. Genomic in-situ hybridization differentiates between A/D-genome and C-genome chromatin and detects intergenomic translocations in polyploid oat species (genus Avena). Genome 1994, 37, 613–618. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.F.; Armstrong, K. Genomic in situ hybridization in Avena Sativa. Genome 1994, 37, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Leggett, J.M.; Thomas, H.M.; Meredith, M.R.; Humphreys, M.W.; Morgan, W.G.; Thomas, H.; King, I.P. Intergenomic translocations and the genomic composition of Avena maroccana Gdgr. revealed by FISH. Chromosome Res. 1994, 2, 163–164. [Google Scholar] [CrossRef]
- Yang, Q.; Hanson, L.; Bennett, M.D.; Leitch, I.J. Genome structure and evolution in the allohexaploid weed Avena fatua L (Poaceae). Genome 1999, 42, 512–518. [Google Scholar] [CrossRef] [PubMed]
- Linc, G.; Friebe, B.R.; Kynast, R.G.; Molnar-Lang, M.; Koszegi, B.; Sutka, J.; Gill, B.S. Molecular cytogenetic analysis of Aegilops cylindrica Host. Genome 1999, 42, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Linares, C.; Ferrer, E.; Fominaya, A. Discrimination of the closely related A and D genomes of the hexaploid oat Avena sativa L. Proc. Natl. Acad. Sci. U. S. A. 1998, 95, 12450–12455. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.L. Evolution through genetic exchange; 2006; Oxford University Press: Oxford. [Google Scholar]
- Anderson, E. Introgressive hybridization; J. Wiley: New York, NY, USA, 1949. [Google Scholar]
- Baack, E.J.; Rieseberg, L.H. A genomic view of introgression and hybrid speciation. Curr. Opin. Genet. Dev. 2007, 17, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Rieseberg, L.H.; Wendel, J.F. Introgression and its consequences in plants . In Hybrid zones and the evolutionary process; Harrison, R.G。, Ed.; Oxford University Press: New York, NY, USA, 1993. [Google Scholar]
- Orr, H.A. The population genetics of speciation - the evolution of hybrid incompatibilities. Genetics 1995, 139, 1805–1813. [Google Scholar] [PubMed]
- Lexer, C.; Widmer, A. The genic view of plant speciation: recent progress and emerging questions. Philos. Trans. R. Soc. Lond., Ser. B: Biol. Sci. 2008, 363, 3023–3036. [Google Scholar] [CrossRef]
- Lowry, D.B.; Modliszewski, J.L.; Wright, K.M.; Wu, C.A.; Willis, J.H. The strength and genetic basis of reproductive isolating barriers in flowering plants. Philos. Trans. R. Soc. Lond., Ser. B: Biol. Sci. 2008, 363, 3009–3021. [Google Scholar] [CrossRef]
- Rieseberg, L.H.; Whitton, J.; Gardner, K. Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics 1999, 152, 713–727. [Google Scholar] [PubMed]
- Rieseberg, L.H. Chromosomal rearrangements and speciation. Trends Ecol. Evol. 2001, 16, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Blakeslee, A.F.; Avery, A.G. Methods of inducing doubling of chromosomes in plants - By treatment with colchicine. J. Hered. 1937, 28, 393–411. [Google Scholar]
- Smith, H.H. The induction of polyploidy in Nicotiana species and species hybrids. J. Hered. 1939, 30, 291–306. [Google Scholar]
- Sears, E.R. Amphidiploids in the Triticinae induced by colchicine. J. Hered. 1939, 30, 38–43. [Google Scholar]
- Endo, T.R. Induction of chromosomal structural changes by a chromosome of Aegilops cylindrica L in common wheat. J. Hered. 1988, 79, 366–370. [Google Scholar]
- Friebe, B.; Kynast, R.G.; Gill, B.S. Gametocidal factor-induced structural rearrangements in rye chromosomes added to common wheat. Chromosome Res. 2000, 8, 501–511. [Google Scholar] [CrossRef]
- Ashida, T.; Nasuda, S.; Sat, K.; Endo, T.R. Dissection of barley chromosome 5H in common wheat. Genes Genet. Syst. 2007, 82, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, M.; Fukushima, T.; Nasuda, S.; Masoudi-Nejad, A.; Ishikawa, G.; Nakamura, T.; Endo, T.R. Dissection of rye chromosome 1R in common wheat. Genes Genet. Syst. 2008, 83, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Sakai, K.; Nasuda, S.; Sato, K.; Endo, T.R. Dissection of barley chromosome 3H in common wheat and a comparison of 3H physical and genetic maps. Genes Genet. Syst. 2009, 84, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Riley, R.; Chapman, V. Genetic control of the cytologically diploid behaviour of hexaploid wheat. Nature 1958, 182, 713–715. [Google Scholar] [CrossRef]
- Riley, R.; Chapman, V.; Kimber, G. Genetic control of chromosome pairing in intergeneric hybrids with wheat. Nature 1959, 183, 1244–1246. [Google Scholar] [CrossRef] [PubMed]
- Mestiri, I.; Chague, V.; Tanguy, A.M.; Huneau, C.; Huteau, V.; Belcram, H.; Coriton, O.; Chalhoub, B.; Jahier, J. Newly synthesized wheat allohexaploids display progenitor-dependent meiotic stability and aneuploidy but structural genomic additivity. New Phytol. 2010, 186, 86–101. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Moran, E.; Benavente, E.; Orellana, J. Analysis of karyotypic stability of homoeologous-pairing (ph) mutants in allopolyploid wheats. Chromosoma 2001, 110, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Pasakinskiene, I.; Jones, N. A decade of "chromosome painting" in Lolium and Festuca. Cytogenet. Genome Res. 2005, 109, 393–399. [Google Scholar]
- Zwierzykowski, Z.; Kosmala, A.; Zwierzykowska, E.; Jones, N.; Joks, W.; Bocianowski, J. Genome balance in six successive generations of the allotetraploid Festuca pratensis x Lolium perenne. Theor. Appl. Genet. 2006, 113, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Chetelat, R.T. GISH analysis of meiotic chromosome pairing in Solanum lycopersicoides introgression lines of cultivated tomato. Genome 2007, 50, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Chetelat, R.T.; Meglic, V.; Cisneros, P. A genetic map of tomato based on BC1 Lycopersicon esculentum x Solanum lycopersicoides reveals overall synteny but suppressed recombination between these homeologous genomes. Genetics 2000, 154, 857–867. [Google Scholar] [PubMed]
- Gaeta, R.T.; Pires, J.C.; Iniguez-Luy, F.; Leon, E.; Osborn, T.C. Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 2007, 19, 3403–3417. [Google Scholar] [CrossRef] [PubMed]
- Song, K.M.; Lu, P.; Tang, K.L.; Osborn, T.C. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl. Acad. Sci. U. S. A. 1995, 92, 7719–7723. [Google Scholar] [CrossRef] [PubMed]
- Jenczewski, E.; Alix, K. From diploids to allopolyploids: The emergence of efficient pairing control genes in plants. Crit. Rev. Plant Sci. 2004, 23, 21–45. [Google Scholar] [CrossRef]
- Le Comber, S.C.; Ainouche, M.L.; Kovarik, A.; Leitch, A.R. Making a functional diploid: from polysomic to disomic inheritance. New Phytol. 2010, 186, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Salmon, A.; Flagel, L.; Ying, B.; Udall, J.A.; Wendel, J.F. Homoeologous nonreciprocal recombination in polyploid cotton. New Phytol. 2010, 186, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Ungerer, M.C.; Strakosh, S.C.; Zhen, Y. Genome expansion in three hybrid sunflower species is associated with retrotransposon proliferation . Curr. Biol. 2006, 16, R872–R873. [Google Scholar] [CrossRef] [PubMed]
- Ungerer, M.C.; Strakosh, S.C.; Stimpson, K.M. Proliferation of Ty3/gypsy-like retrotransposons in hybrid sunflower taxa inferred from phylogenetic data. BMC Biol. 2009, 7, 40. [Google Scholar] [CrossRef] [PubMed]
- Lai, J.S.; Ma, J.X.; Swigonova, Z.; Ramakrishna, W.; Linton, E.; Llaca, V.; Tanyolac, B.; Park, Y.J.; Jeong, Y.; Bennetzen, J.L.; Messing, J. Gene loss and movement in the maize genome. Genome Res. 2004, 14, 1924–1931. [Google Scholar] [CrossRef] [PubMed]
- Ozkan, H.; Levy, A.A.; Feldman, M. Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell 2001, 13, 1735–1747. [Google Scholar] [CrossRef] [PubMed]
- Shaked, H.; Kashkush, K.; Ozkan, H.; Feldman, M.; Levy, A.A. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 2001, 13, 1749–1759. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Vega, J.M.; Segal, G.; Abbo, S.; Rodova, H.; Feldman, M. Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops. I. Changes in low-copy noncoding DNA sequences. Genome 1998, 41, 272–277. [Google Scholar] [CrossRef]
- Han, F.P.; Fedak, G.; Guo, W.L.; Liu, B. Rapid and repeatable elimination of a parental genome-specific DNA repeat (pGcIR-1a) in newly synthesized wheat allopolyploids. Genetics 2005, 170, 1239–1245. [Google Scholar] [CrossRef] [PubMed]
- Ozkan, H.; Tuna, M.; Arumuganathan, K. Nonadditive changes in genome size during allopolyploidization in the wheat (Aegilops-Triticum) group. J. Hered. 2003, 94, 260–264. [Google Scholar] [CrossRef] [PubMed]
- Skalicka, K.; Lim, K.Y.; Matyasek, R.; Matzke, M.; Leitch, A.R.; Kovarik, A. Preferential elimination of repeated DNA sequences from the paternal, Nicotianatomentosiformis genome donor of a synthetic, allotetraploid tobacco. New Phytol. 2005, 166, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Han, F.P.; Fedak, G.; Ouellet, T.; Liu, B. Rapid genomic changes in interspecific and intergeneric hybrids and allopolyploids of Triticeae. Genome 2003, 46, 716–723. [Google Scholar] [CrossRef] [PubMed]
- Baumel, A.; Ainouche, M.L.; Levasseur, J.E. Molecular investigations in populations of Spartina anglica C.E. Hubbard (Poaceae) invading coastal Brittany (France). Mol. Ecol. 2001, 10, 1689–1701. [Google Scholar] [CrossRef] [PubMed]
- Baumel, A.; Ainouche, M.; Kalendar, R.; Schulman, A.H. Retrotransposons and genomic stability in populations of the young allopolyploid species Spartina anglica C.E. Hubbard (Poaceae). Mol. Biol. Evol. 2002, 19, 1218–1227. [Google Scholar] [PubMed]
- Salmon, A.; Ainouche, M.L.; Wendel, J.F. Genetic and epigenetic consequences of recent hybridization and polyploidy in Spartina (Poaceae). Mol. Ecol. 2005, 14, 1163–1175. [Google Scholar] [CrossRef] [PubMed]
- Parisod, C.; Salmon, A.; Zerjal, T.; Tenaillon, M.; Grandbastien, M.A.; Ainouche, M. Rapid structural and epigenetic reorganization near transposable elements in hybrid and allopolyploid genomes in Spartina. New Phytol. 2009, 184, 1003–1015. [Google Scholar] [CrossRef] [PubMed]
- McClintock, B. The significance of responses of the genome to challenge. Science 1984, 226, 792–801. [Google Scholar] [PubMed]
- Liu, B.; Wendel, J.F. Epigenetic phenomena and the evolution of plant allopolyploids. Mol. Phylogen. Evol. 2003, 29, 365–379. [Google Scholar] [CrossRef]
- Kashkush, K.; Feldman, M.; Levy, A.A. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat Genet 2003, 33, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Wendel, J.F. Retrotransposon activation followed by rapid repression in introgressed rice plants. Genome 2000, 43, 874–880. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.Y.; Kovarik, A.; Matyasek, R.; Chase, M.W.; Knapp, S.; McCarthy, E.; Clarkson, J.J.; Leitch, A.R. Comparative genomics and repetitive sequence divergence in the species of diploid Nicotiana section Alatae. Plant J. 2006, 48, 907–919. [Google Scholar] [CrossRef] [PubMed]
- Rose, M.R.; Doolittle, W.F. Molecular biological mechanisms of speciation. Science 1983, 220, 157–162. [Google Scholar] [PubMed]
- Melayah, D.; Lim, K.Y.; Bonnivard, E.; Chalhoub, B.; De Borne, F.D.; Mhiri, C.; Leitch, A.R.; Grandbastien, M.A. Distribution of the Tnt1 retrotransposon family in the amphidiploid tobacco (Nicotianatabacum) and its wild Nicotiana relatives. Biol. J. Linn. Soc. 2004, 82, 639–649. [Google Scholar] [CrossRef]
- Petit, M.; Guidat, C.; Daniel, J.; Denis, E.; Montoriol, E.; Bui, Q.T.; Lim, K.Y.; Kovarik, A.; Leitch, A.R.; Grandbastien, M.A.; Mhiri, C. Mobilization of retrotransposons in synthetic allotetraploid tobacco. New Phytol. 2010, 186, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Senchina, D.S.; Alvarez, I.; Cronn, R.C.; Liu, B.; Rong, J.K.; Noyes, R.D.; Paterson, A.H.; Wing, R.A.; Wilkins, T.A.; Wendel, J.F. Rate variation among nuclear genes and the age of polyploidy in Gossypium. Mol. Biol. Evol. 2003, 20, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Hanson, R.E.; Islam-Faridi, M.N.; Crane, C.F.; Zwick, M.S.; Czeschin, D.G.; Wendel, J.F.; McKnight, T.D.; Price, H.J.; Stelly, D.M. Ty1-copia-retrotransposon behavior in a polyploid cotton. Chromosome Res. 2000, 8, 73–76. [Google Scholar] [CrossRef]
- Hu, G.; Hawkins, J.S.; Grover, C.E.; Wendel, J.F. The history and disposition of transposable elements in polyploid Gossypium. Genome in press.
- Nagaki, K.; Walling, J.; Hirsch, C.; Jiang, J.; Murata, M. Structure and Evolution of Plant Centromeres . In Centromere; Ugarkovic, D., Ed.; Springer-Verlag: Berlin, Germany, 2009. [Google Scholar]
- Kamm, A.; Galasso, I.; Schmidt, T.; Heslop-Harrison, J.S. Analysis of a repetitive DNA family from Arabidopsis arenosa and relationships between Arabidopsis species. Plant Mol. Biol. 1995, 27, 853–862. [Google Scholar] [CrossRef] [PubMed]
- Kawabe, A.; Nasuda, S. Structure and genomic organization of centromeric repeats in Arabidopsis species. Mol. Genet. Genomics 2005, 272, 593–602. [Google Scholar] [CrossRef] [PubMed]
- Kawabe, A.; Nasuda, S. Polymorphic chromosomal specificity of centromere satellite families in Arabidopsis halleri ssp. gemmifera. Genetica 2006, 126, 335–342. [Google Scholar] [CrossRef]
- Belyayev, A.; Raskina, O.; Nevo, E. Evolutionary dynamics and chromosomal distribution of repetitive sequences on chromosomes of Aegilops speltoides revealed by genomic in situ hybridization. Heredity 2001, 86, 738–742. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, X.M.; Cheng, Z.K.; Mucller, L.; Giovannoni, J.; Tanksley, S.D. Euchromatin and pericentromeric heterochromatin: Comparative composition in the tomato genome. Genetics 2006, 172, 2529–2540. [Google Scholar] [CrossRef] [PubMed]
- Saunders, V.A.; Houben, A. The pericentromeric heterochromatin of the grass Zingeria biebersteiniana (2n=4) is composed of Zbcen1-type tandem repeats that are intermingled with accumulated dispersedly organized sequences. Genome 2001, 44, 955–961. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Bao, Z.; Temnykh, S.; Cheng, Z.; Jiang, J.; Wing, R.A.; McCouch, S.R.; Wessler, S.R. Dasheng: A recently amplified nonautonomous long terminal repeat element that is a major component of pericentromeric regions in rice. Genetics 2002, 161, 1293–1305. [Google Scholar] [PubMed]
- Liu, Z.; Yue, W.; Li, D.Y.; Wang, R.R.C.; Kong, X.Y.; Lu, K.; Wang, G.X.; Dong, Y.S.; Jin, W.W.; Zhang, X.Y. Structure and dynamics of retrotransposons at wheat centromeres and pericentromeres. Chromosoma 2008, 117, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Blanc, G.; Barakat, A.; Guyot, R.; Cooke, R.; Delseny, I. Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 2000, 12, 1093–1101. [Google Scholar] [CrossRef] [PubMed]
- Soltis, D.E.; Albert, V.A.; Leebens-Mack, J.; Bell, C.D.; Paterson, A.H.; Zheng, C.F.; Sankoff, D.; dePamphilis, C.W.; Wall, P.K.; Soltis, P.S. Polyploidy and angiosperm diversification. Am. J. Bot. 2009, 96, 336–348. [Google Scholar] [CrossRef]
- Vision, T.J.; Brown, D.G.; Tanksley, S.D. The origins of genomic duplications in Arabidopsis. Science 2000, 290, 2114–2117. [Google Scholar] [CrossRef] [PubMed]
- Bowers, J.E.; Chapman, B.A.; Rong, J.K.; Paterson, A.H. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 2003, 422, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.Y.; Wall, P.K.; Leebens-Mack, J.H.; Lindsay, B.G.; Soltis, D.E.; Doyle, J.J.; Soltis, P.S.; Carlson, J.E.; Arumuganathan, K.; Barakat, A.; Albert, V.A.; Ma, H.; dePamphilis, C.W. Widespread genome duplications throughout the history of flowering plants. Genome Res. 2006, 16, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Blanc, G.; Wolfe, K.H. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 2004, 16, 1667–1678. [Google Scholar] [CrossRef] [PubMed]
- Salse, J.; Bolot, S.; Throude, M.; Jouffe, V.; Piegu, B.; Quraishi, U.M.; Calcagno, T.; Cooke, R.; Delseny, M.; Feuillet, C. Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. Plant Cell 2008, 20, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Genome sequencing and analysis of the model grass Brachypodium distachyon. Vogel, J.P.; Garvin, D.F.; Mockler, T.C.; Schmutz, J.; Rokhsar, D.; Bevan, M.W.; Barry, K.; Lucas, S.; Harmon-Smith, M.; Lail, K.; et al. Nature 2010, 463, 763–768. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Wang, J.; Lin, W.; Li, S.G.; Li, H.; Zhou, J.; Ni, P.X.; Dong, W.; Hu, S.N.; Zeng, C.Q.; et al. The Genomes of Oryza sativa: A history of duplications . PLoS Biol. 2005, 3 , 266–281. [Google Scholar] [CrossRef] [Green Version]
- Gaut, B.S.; Doebley, J.F. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. U. S. A. 1997, 94, 6809–6814. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Coe, E.; Nelson, W.; Bharti, A.K.; Engler, F.; Butler, E.; Kim, H.; Goicoechea, J.L.; Chen, M.; Lee, S.; Fuks, G.; Sanchez-Villeda, H.; Schroeder, S.; Fang, Z.; McMullen, M.; Davis, G.; Bowers, J.E.; Paterson, A.H.; Schaeffer, M.; Gardiner, J.; Cone, K.; Messing, J.; Soderlund, C.; Wing, R.A. Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet. 2007, 3, 1254–1263. [Google Scholar] [CrossRef]
- Rhoades, M.M. Meiosis in Maize. J. Hered. 1950, 41, 58–67. [Google Scholar]
- Gonzalez, G.; Comas, C.; Confalonieri, V.; Naranjo, C.A.; Poggio, L. Genomic affinities between maize and Zea perennis using classical and molecular cytogenetic methods (GISH-FISH). Chromosome Res. 2006, 14, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Gill, N.; Findley, S.; Walling, J.G.; Hans, C.; Ma, J.X.; Doyle, J.; Stacey, G.; Jackson, S.A. Molecular and chromosomal evidence for allopolyploidy in soybean. Plant Physiol. 2009, 151, 1167–1174. [Google Scholar] [CrossRef] [PubMed]
- Schlueter, J.A.; Dixon, P.; Granger, C.; Grant, D.; Clark, L.; Doyle, J.J.; Shoemaker, R.C. Mining EST databases to resolve evolutionary events in major crop species. Genome 2004, 47, 868–876. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.X.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.J.; Thelen, J.J.; Cheng, J.L.; et al. Genome sequence of the palaeopolyploid soybean . Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Gomez, M.I.; Islam-Faridi, M.N.; Zwick, M.S.; Jr., D.G.C.; Hart, G.E.; Wing, R.A.; Stelly, D.M.; Price, H.J. Tetraploid nature of Sorgum bicolor (L.) Moench . J. Hered. 1998, 89, 188–190. [Google Scholar] [CrossRef]
- Miller, J.T.; Jackson, S.A.; Nasuda, S.; Gill, B.S.; Wing, R.A.; Jiang, J. Cloning and characterization of a centromere-specific repetitive DNA element from Sorghum bicolor. Theor. Appl. Genet. 1998, 96, 832–839. [Google Scholar] [CrossRef]
- Zwick, M.S.; Islam-Faridi, M.N.; Zhang, H.B.; Hodnett, G.L.; Gomez, M.I.; Kim, J.S.; Price, H.J.; Stelly, D.M. Distribution and sequence analysis of the centromere-associated repetitive element CEN38 of Sorghum bicolor (Poaceae). Am. J. Bot. 2000, 87, 1757–1764. [Google Scholar] [CrossRef] [PubMed]
- Paterson, A.H.; Bowers, J.E.; Chapman, B.A. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 9903–9908. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.M.; Nasuda, S.; Dong, F.G.; Scherrer, C.W.; Woo, S.S.; Wing, R.A.; Gill, B.S.; Ward, D.C. A conserved repetitive DNA element located in the centromeres of cereal chromosomes. Proc. Natl. Acad. Sci. U. S. A. 1996, 93, 14210–14213. [Google Scholar] [CrossRef] [PubMed]
- Jurka, J.; Kapitonov, V.V.; Pavlicek, A.; Klonowski, P.; Kohany, O.; Walichiewicz, J. Repbase update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005, 110, 462–467. [Google Scholar]
- Benson, G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, K.; Varala, K.; Hudson, M.E. Global repeat discovery and estimation of genomic copy number in a large, complex genome using a high-throughput 454 sequence survey. BMC Genomics 2007, 8, 132. [Google Scholar] [CrossRef]
- Macas, J.; Neumann, P.; Navratilova, A. Repetitive DNA in the pea (Pisum sativum L.) genome: comprehensive characterization using 454 sequencing and comparison to soybean and Medicago truncatula. BMC Genomics 2007, 8, 427. [Google Scholar] [CrossRef]
- Wicker, T.; Taudien, S.; Houben, A.; Keller, B.; Graner, A.; Platzer, M.; Stein, N. A whole-genome snapshot of 454 sequences exposes the composition of the barley genome and provides evidence for parallel evolution of genome size in wheat and barley. Plant J. 2009, 59, 712–722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, A.; Albert, P.S.; Vega, J.M.; Birchler, J.A. Sensitive fluorescence in situ hybridization signal detection in maize using directly labeled probes produced by high concentration DNA polymerase nick translation. Biotech Histochem 2006, 81, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Kato, A.; Lamb, J.C.; Birchler, J.A. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 13554–13559. [Google Scholar] [CrossRef] [PubMed]
- Lamb, J.C.; Birchler, J.A. Retroelement genome painting: Cytological visualization of retroelement expansions in the genera Zea and Tripsacum. Genetics 2006, 173, 1007–1021. [Google Scholar] [CrossRef] [PubMed]
- Soltis, D.E.; Soltis, P.S.; Endress, P.K.; Chase, M.W. Phylogeny and evolution of the angiosperms; 2005; Sinauer: Sunderland, MA, USA. [Google Scholar]
- Soltis, D.E.; Bell, C.D.; Kim, S.; Soltis, P.S. Origin and early evolution of angiosperms. Ann. N. Y. Acad. Sci. 2008, 1133, 3–25. [Google Scholar] [CrossRef] [PubMed]
- Stokstad, K. Will Malthus continue to be wrong. Science 2005, 309, 102–102. [Google Scholar] [CrossRef] [PubMed]
- Costanza, R.; dArge, R.; deGroot, R.; Farber, S.; Grasso, M.; Hannon, B.; Limburg, K.; Naeem, S.; ONeill, R.V.; Paruelo, J.; Raskin, R.G.; Sutton, P.; vandenBelt, M. The value of the world's ecosystem services and natural capital. Nature 1997, 387, 253–260. [Google Scholar] [CrossRef]
© 2010 by the authors; licensee MDPI, Basel, Switzerland This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Share and Cite
Chester, M.; Leitch, A.R.; Soltis, P.S.; Soltis, D.E. Review of the Application of Modern Cytogenetic Methods (FISH/GISH) to the Study of Reticulation (Polyploidy/Hybridisation). Genes 2010, 1, 166-192. https://doi.org/10.3390/genes1020166
Chester M, Leitch AR, Soltis PS, Soltis DE. Review of the Application of Modern Cytogenetic Methods (FISH/GISH) to the Study of Reticulation (Polyploidy/Hybridisation). Genes. 2010; 1(2):166-192. https://doi.org/10.3390/genes1020166
Chicago/Turabian StyleChester, Michael, Andrew R. Leitch, Pamela S. Soltis, and Douglas E. Soltis. 2010. "Review of the Application of Modern Cytogenetic Methods (FISH/GISH) to the Study of Reticulation (Polyploidy/Hybridisation)" Genes 1, no. 2: 166-192. https://doi.org/10.3390/genes1020166