1. Introduction
Soil is a fundamental and irreplaceable natural resource, providing the essential connection between the land, air and water resources that allow life on Earth. As an example, it maintains the life of microorganisms that are involved in biogeochemical cycles, nutrient recycling and carbon storage. Soil also sustains agricultural production, which, together with the oceans, is the most important source of food for animals and humans. However, soil resources are threatened due to degradation and desertification. According to the Food and Agriculture Organization [
1], soil degradation is a “process which lowers the current and/or potential capability of soil to produce goods and services”. Thus, soil degradation is a threat that national and international organizations are trying to hinder and reverse.
The flat land area located between the Andes Mountains, known as the Altiplano, shared between Peru and Bolivia, is facing a rapid process of soil erosion, due to increasing drought, climate change, population growth and altered farming practices. This process leads to desertification, which results in soils with low vegetable cover and very poor organic matter content. These problems have caused drastic decreases in the production of crops, reducing the already low incomes of the farmers. In some cases, the farmers have used agrochemicals to mitigate soil fertility problems, with the potential of negative effects on the environment. In parallel, there is an increasing demand in international markets for organic products, such as the quinoa (
Chenopodium quinoa). Quinoa is a native pseudograin from the Altiplano, where local people domesticated it since ancestral times. Despite its ancestral condition, quinoa has lately become particular popular, due to its gluten-free and high nutritional value properties (with an average of 14.8% protein and an exceptional balance between oil, protein and starch, [
2]). During the last 10 years, Bolivian quinoa exportation increased 26 times in value and nine times in volume [
3]. This is the reason why quinoa, the most economically attractive crop from this area, is urging Andean growers to intensify its production beyond sustainability.
Organic production is a national priority in countries like Bolivia and has been mainly driven by the international demand for organic products for exportation. This motivates the agricultural sector, which seeks to produce food respecting the environment and the health of farmers. For this reason, it is necessary to develop adequate technologies to support organic production and according to local requirements. These technologies include bioproducts based on living microorganisms that are used as biofertilizers and biocontrol agents for pests and diseases, depending on the microbial species with which they have been made. Bioproducts are important tools to help reduce the harmful effects of chemical fertilizers and pesticides in the environment and to produce food free of contaminants and at a lower cost, in this way helping the economy of small farmers. Bioproducts can also help to improve the quality of the soils in the Altiplano by retaining microbial populations, which will help to mitigate soil degradation and enable farmers to produce organic crops, including quinoa, broad beans and potato, among others).
In this work, we focused on the isolation of native microorganisms associated with quinoa at the Bolivian Altiplano and studied their nutrient recycling potential and beneficial effects on plant growth. The best microbial isolates were used to formulate bioproducts for organic production, which were distributed among local farmers. We intentionally sought to work with native microorganisms to reduce the disturbance of the Altiplano soil ecosystem and to keep the technology as simple as possible, in order to provide cost-effective technologies for Andean farmers.
2. Results and Discussion
2.1. Microbial Isolates
Given the need for ecological fertilizers and biopesticides for organic agriculture, our investigation has been directed towards the development of new biotechnologies, to contribute to the clean and healthy production of quinoa and to help mitigate soil degradation. In the last few years, there has been a growing interest in plant growth-promoting bacteria (PGPBs) and fungi (PGPFs), because they can recycle nutrients, promote plant growth and, in some cases, prevent the infection of plant tissue by pathogens. Such microorganisms can be symbiotic or free living [
4,
5]. To reach this goal, we explored the native microorganisms associated with quinoa plants, isolating and identifying a series of bacterial groups. Results indicate that one hundred and four different bacterial isolates were obtained from internal plant tissue (endophytic bacteria), of which fifty five are from roots, eight from stems and forty one from leaves. Of the fungi group, only rhizospheric individuals were isolated. Bacteria and fungi were molecularly identified (see below), consisting of the following:
Rhizosphere microorganisms: Fungi individuals were isolated that belong to the following genera or species: Trichoderma harzianum, T. asperellum, T. koningiopsis, Beauveria bassiana, B. brongniartii and Metarhizium sp.
Endophytic microorganisms: Bacterial species isolated from leaves: Bacillus amyloliquefaciens, B. tequilensis, B. subtilis, B. pumilus, B. licheniformis, B. horikoshii, B. atrophaeus, B. thuringiensis and Paenibacillus sp. Bacterial species isolated from stems and roots: B. aryabhattai, B. subtilis, B. horikoshii, B. megaterium, Pseudomonas sp., Paenibacillus sp. and P. odorifer.
2.3. Greenhouse Testing
Bacterial isolates that showed positive results for the three tests performed under
in vitro conditions were used to inoculate plants under greenhouse conditions in order to confirm their beneficial properties in an
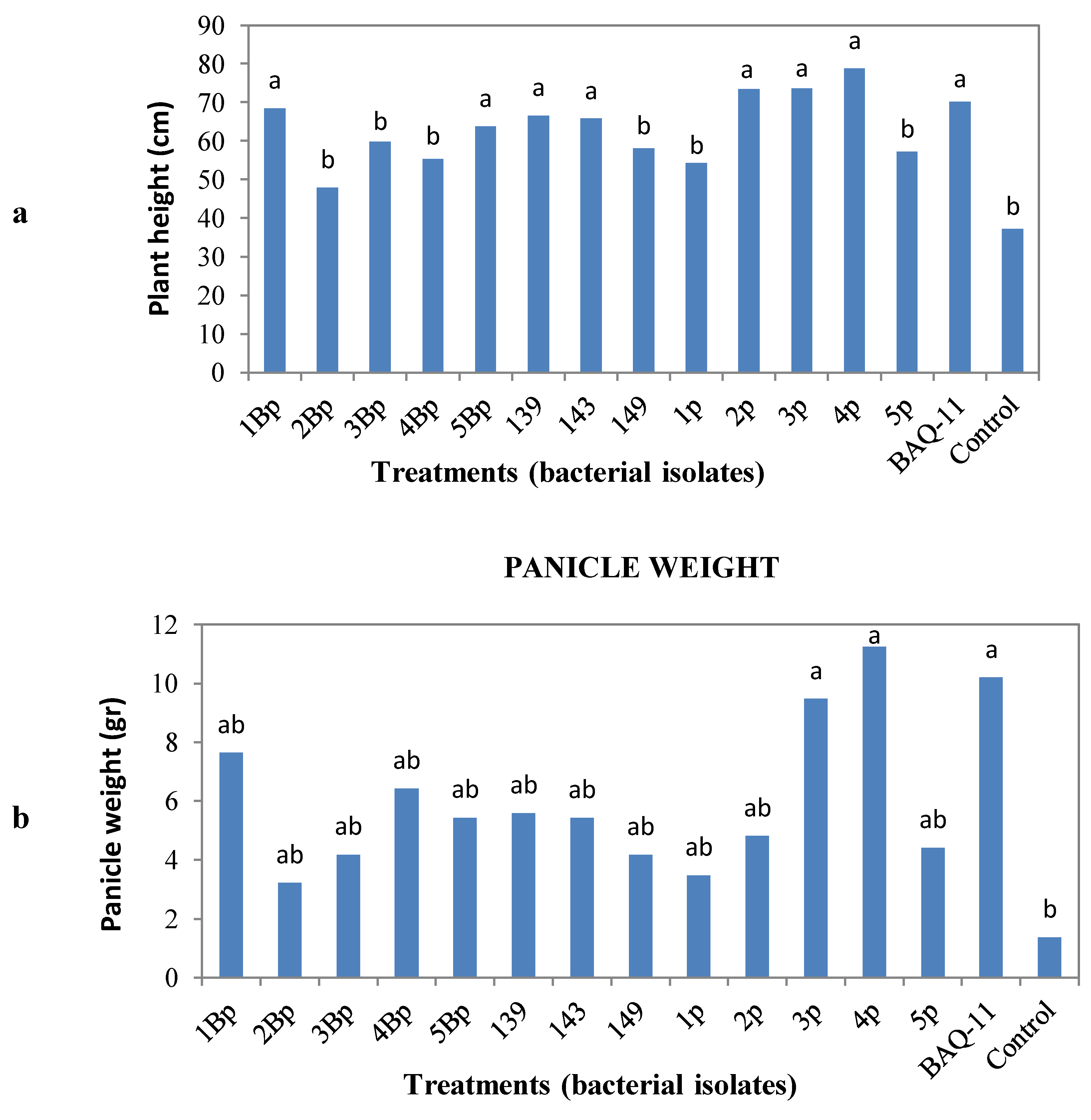
in vivo assessment. After a post-inoculation period of eight weeks, quinoa plants were harvested and measured for plant height, panicle weight and root weight, volume and length. Statistical analysis showed that the treatments with bacterial isolates did not influence the root weight, volume or length, but increased plant height and panicle weight (
Figure 1). The bacterial isolates that increased plant height were 1Bp and 5Bp (
B. pumilus, P-solubilization bacteria), 139 and 143 (
B. simplex, N-fixing bacteria), 2p, 3p and 4p (
Paenibacillus sp
., N-fixing bacteria) and BAQ-11 (
B. subtilis, N-fixing bacteria) and for panicle weight were 3p and 4p (
Paenibacillus sp.) and BAQ-11 (
B. subtilis) (
Figure 1). Thus, some bacterial isolates that increased panicle weight also increased plant height. All other isolates showed the same behavior on plant height and panicle weight compared to the uninoculated control.
2.6. Field Trials: Response of Quinoa to Treatments with Bioproducts
It is important to screen the response of quinoa plants to microbial inoculation under field conditions since in vitro screenings for PGPBs do not always reflect the reality of production on the field (i.e., microbial performance in situ is different from in vitro). Thus, the bioproducts were tested under organic production of quinoa by local Altiplano farmers for plant growth performance and disease control responses. The bioproducts, called Biobacillus (composed of B. subtilis strain BAQ-11) and Tricotop (containing T. koningiopsis y T. harzianum), were tested.
Table 1 shows the results indicating that Biobacillus or Tricotop increased quinoa yield compared to the control without microorganism amendment. Interestingly, using both products, the highest yields were obtained. This led to the idea of creating a third bioproduct, called Tricobal, which is formulated with
B. subtilis,
T. koningiopsis and
T. harzianum.
Table 1.
Comparison of the average yield (kg/ha) of quinoa with the use of bioproducts.
Table 1.
Comparison of the average yield (kg/ha) of quinoa with the use of bioproducts.
| Treatment | Average yield (kg/ha) * |
|---|
| Control (no bioproduct) | 1,051.67 c |
| Biobacillus | 1,206.67 b |
| Tricotop | 1,346.67 b |
| Tricotop + Biobacillus | 1,516.67 a |
We evaluated Tricobal over two quinoa production seasons (two different years). Tricobal was soil-applied to farmers’ fields at the time of planting. We measured the plant growth, leaf area, panicle size and average grain yields.
Table 2 shows that the differences between treatments with bioproduct (Tricobal) and the controls were significant. Plants treated with Tricobal looked overall healthier and more vigorous than untreated plants (
Figure 3f). This result demonstrates that Tricobal is a potential tool for farmers to improve the crop productivity of quinoa.
Table 2.
Bioproduct amendment effect on the development and yield of quinoa.
Table 2.
Bioproduct amendment effect on the development and yield of quinoa.
| Year | Treatment | Plant size (cm) m | Leaf area (%) m | Panicle length (cm) m | Yield (qq/ha) m,n |
|---|
| 1 (2009) | Tricobal | 125 a | 70 a | 40 a | 20 a |
| Control (no bioproduct) | 80 b | 50 b | 25 b | 11 b |
| 2 (2010) | Tricobal | 150 a | 80 a | 2 a | 25 a |
| Control (no bioproduct) | 110 b | 55 b | 30 b | 12 b |
3. Experimental Section
3.1. Microorganisms Isolation
To explore the microbe biodiversity of the Altiplano soil associated with quinoa plants, twenty organic quinoa crop fields were sampled from each Altiplano region: southern (S 19°41'0'', W 66°34'0"), central (S 18°3'0", W 66°59'0") and northern (S 17°4'0", W 68°9'0"), close to Uyuni, Quillacas, Salinas de Garci Mendoza, Jalsuri, Lacaya and Cohana villages, which corresponds to the districts of Potosí, Oruro and La Paz, respectively. Soil samples were obtained by discarding the top 5 cm and collecting the next 20 cm. Plant root samples were taken 15 cm from the tip, stems from the section above the roots and leaves from the upper segment of the plant. All samples were transported to the laboratory under cold temperature for microorganism isolation.
Microorganisms were isolated on culture plates using specific culture media to separate bacteria (TSA) and fungi (potato dextrose agar (PDA)). For rhizosphere microorganism isolation, 10 g of roots with soil were washed with 90 mL of 0.85% NaCl and gentle agitation. Soil residues were vigorously shaken and then plated on respective medium, making dilutions until 10−9. In order to isolate endophytic bacteria, small pieces of leaves (about 1 cm2), stems and roots (1 cm-long) were cut and submerged in 15 mL of potassium phosphate buffer (0.1 M, pH 7.0) for 3 h with gently shaking. After this step, plant samples were directly treated with 70% ethanol for 1 min and then with 1.2% sodium hypochlorite for 15 min for superficial sterilization. After rinsing (four times with sterile 0.85% NaCl solution), the tissue fragments were smashed in 30 mL of sterile 0.85% NaCl solution. Serial dilutions were spread on TSA medium and incubated at 28 °C for 72 h. To isolate only endospore-forming bacteria, dilutions were subject to heat (75 °C for 15 min) before plating.
Bacterial colonies were studied at the morphological (color, shape, growing speed, etc.), biochemical (catalase, potassium hydroxide and oxidase) and cellular (Gram stain, flagella and spore presence/absence) level. Taxonomic identification of each bacterial and fungal isolate was performed by following the method described below.
3.2. Establishing the Functional Activity of Microbial Isolates
We attempted to determine the nitrogen (N) fixing, phosphorus (P) solubilizing and growth-promoting activities of bacterial isolates by means of the following methods:
To select bacteria able to fix atmospheric nitrogen, the leaves and roots were tightly washed and then treated with ethanol 70%. Five rinses were done with sterile distilled water to remove ethanol. Under sterile conditions, small fragments (0.5 cm) were cut and deposited on N-free Rennie media (2 g/L agar). Plant tissue fragments were incubated for 6 days at 30 °C without shaking. After incubation, a veil and then a film were grown, which constitutes an inoculum that was later diluted and plated on the same and fresh medium. After an incubation period of 48 h at 30 °C, isolated colonies were obtained. Since the growth medium contains tiny amounts of nitrogen, all bacteria that were able to grow on it could be nitrogen fixers. To confirm this supposition, we determined the identity of bacteria by means of the molecular techniques detailed below.
To select phosphorus-solubilizing bacteria, isolates were culture on NBRIP (National Botanical Research Institute's phosphate) medium [
13] at 37 °C for 2 days. After this period, a clear halo was recorded as positive, which means that the phosphate was solubilized.
To select growth-promoting bacteria, we focused on finding isolates that produce IAA. Bacteria were cultured in tryptone soy broth (TSB) supplemented with 5 mM of
l-tryptophan at 28 °C for 7 days. After the incubation period, cultures were centrifuged (3802×
g for 5 min), and an aliquot of 75 µL of the supernatant was mixed with 225 µL of Salkowski reagent for 30 min under dark conditions. Reddish colored supernatant was read as a positive signal [
14].
3.3. Taxonomic Identification of Isolates
To determine or confirm the taxonomic affiliation of the selected microorganisms, we extracted genomic DNA from each isolate following the protocol described by Wilson [
15], for bacteria, and Melo and collaborators [
16], for fungi. We have amplified by PCR a small genomic region that is conserved among all bacteria using universal primers (
Table 3), and then, the amplified fragments were sequenced. For PCR, Phusion high fidelity DNA polymerase (manufactured by NEB, Ipswich, MA, USA) and the following conditions were used: each 25-µL amplification reaction mixture comprised ~10 ng chromosomal DNA, 1× HF (high-fidelity) buffer, 0.4 µM forward and reverse primers, 0.2 mM deoxynucleotide triphosphates (NEB), 1× Q reagent (Qiagen, Germantown, MD, USA) and 0.5 U Phusion DNA Polymerase. Reaction conditions for all the primers were as follows: initial denaturation at 95 °C for 10 min, 35 cycles of denaturation at 95 °C for 40 s, primer annealing at 55 °C for 40 s for bacteria and 52–59 °C for 1 min for fungi and extension at 72 °C for 1 min, followed by a final extension step of 72 °C for 5 min. The amplification product was then purified using Qiaquick columns (Qiagen) following the manufacturer’s protocol before being used in a sequencing reaction. DNA sequencing was performed at the DNA sequencing facility, the University of Chicago, Chicago, IL, USA, following the standard Sanger sequencing protocol. Resulting sequences were edited and trimmed with the BioEdit program [
17] and then used to find the most similar sequences in the database using the BLAST (Basic Local Alignment Search Tool) program [
18]. Only matches reaching 99%–100% of identity and covering most of the sequence (>90%) were considered to assign a taxonomic name to the isolates. For analysis of
Trichoderma sequences, TrichoBLAST and TrichOKEY programs [
19] were also used to analyze DNA sequences from intergenic regions of ribosomal genes and internal segments of the
tef1 gene [
20,
21].
Table 3.
Primers used in this work for the identification of microbial isolates; the same primers were used for amplification and sequencing.
Table 3.
Primers used in this work for the identification of microbial isolates; the same primers were used for amplification and sequencing.
| Microorganism | Primer name | Gene marker a | Reference |
|---|
| Bacteria | 27F | SSU rRNA | [22] |
| | 1492R | SSU rRNA | [22] |
| Trichoderma | ITS1F | SSU rRNA | [23] |
| | ITS5 | SSU rRNA | [24] |
| | ITS4 | LSU rRNA | [24] |
| | LR1 | LSU rRNA | [24] |
| | SR6R | SSU rRNA | [24] |
| | EF1-728F | tef1 gene | [25] |
| | TEF1LLErev | tef1 gene | [26] |
3.4. Plant Inoculation with Beneficial Bacteria under Greenhouse Conditions
Bacterial isolates that showed functional activity on N-fixing, P-solubilizing and IAA production, as assayed in culture plates, were selected for further analyses on their performance in planta. Each pot containing 1 kg of sterile soil (silt, sand, organic matter mixed in equal parts) was inoculated with 0.5 mL of a bacterial suspension (1 × 108 cfu/mL). Then, three quinoa seeds were sown per pot.
After two months, plants were harvested, and the following parameters were measured: plant height (from the neck to the tip of the panicle, in centimeters); panicle weight (in grams); root weight (discarding neck portion, in grams); root volume (calculating the difference in volume of water displaced in a beaker); and root length of each root from bottom to tip (in centimeters). The assay was intended to follow a complete randomized blocks design, and data were analyzed by the Duncan applied test (p = 0.05) with four repetitions per treatment.
3.5. Large-Scale Multiplication of Bacterial Isolates
For bacteria-based bioproducts, a starter inoculum was prepared by multiplying
B. subtilis strain BAQ-11 in TSB medium and incubating them for 3 days at room temperature with agitation until reaching a bacterial concentration of 1 × 10
8 cfu/mL. Different liquid culture media were tested to define which media was able to produce the highest number of colony forming units per milliliter. We used five different broths made of: (1) tryptic soy broth (commercial culture medium, control); (2) soybean protein (a subproduct from the edible oil industry); (3) potato (
Solanum tuberosum L. ssp
. andigenum cv. Waych’a); (4) a mix of rice grains and soybean protein (1:1 proportion); (5) rice grains; and (6) a mix of potato and soybean protein (6:1 proportion). These broths were prepared by boiling the product in water for 1 h and then were autoclaved for sterilization. The inoculated broths were incubated in a shaker at 100 rpm and 35 °C for ten days. After incubation, staining was performed with malachite green to verify bacterial spore formation [
27]. To determine the number of spores/milliliter, three dilutions were plated on PDA medium for each treatment, using three repetitions of each dilution and 20 observations per repetition. Analysis of variance with four repetitions was employed to statistically validate the differences among treatments.
The best culture medium as assayed above was used for large-scale multiplication. Inoculum grown in TSB medium was employed to establish a larger culture (80 L). Forced sterile air was supplied without agitation, since air itself generates turbulence in the liquid. The culture was incubated at 22–25 °C for 10 days to reach a cell density of 1 × 109 spores/mL. This culture was used to formulate bioproducts (see below).
3.6. Large-Scale Multiplication of Fungi Isolates
The starter inoculum of Trichoderma species was obtained by growing single spores of the fungus on PDA medium. To determine the best substrate for large-scale and fast multiplication of T. harzianum, the starter inoculum was mixed with different sterile substrates: (1) rice grain; (2) clay + sugarcane molasses; (3) calcium carbonate + sugarcane molasses; (4) rice husk; (5) rice grain + rice husk (proportion 1:1); (6) rice grain + rice husk (proportion 1:2.33); and (7) rice husk + sugarcane molasses. Each substrate was incubated for 9 days at 28 °C, and at the end of this period, the number of spores per gram of substrate was measured using the Neubauer chamber under microscope as evidence for fungus growth. Spore calculation was obtained by adding the number of spores of five small squares (0.0025 mm2) and multiplying it by 5000. A variance analysis applied to four repetitions of each treatment helped to statistically define the best substrate for the fungus growth.
The best culture medium as assayed according to the previous experiment was used for large-scale multiplication. The PDA-medium-based starter inoculum was employed to establish a larger culture in sterile rice grains and incubated for 15 days at 28 °C. In this way, a larger fungi biomass was achieved, which, in turn, was employed to inoculate the selected substrate (rice mixed with rice husk (1:1)) for large-scale multiplication. After a period of 15 days, the multiplication medium containing Trichoderma spores was dried on trays, which stresses the fungus, generating more spores. Spores were separated from the multiplication medium by sieving and then were used to formulate bioproducts, as described below.
3.8. Plant Inoculation with Bioproducts under Open Field Conditions
At the time of planting (late November), about 7 kg/ha of decomposed sheep manure were deposited as the basal dressing, and then, four quinoa seeds from the “Real Blanca” variety were sown per spot on Altiplano soil, which is mostly sandy (Quillacas area, Oruro district). Bioproducts (Tricotop, Biobacillus or Tricobal) were applied to the soil at doses of 1 kg/ha each. Later and at the branching stage, Biobacillus was also used at 1 kg/ha as foliage fertilizer for all assays, except the control. After two months, quinoa plants were harvested, and the response variables evaluated were plant size, leaf area, panicle length and yield. We assayed the bioproducts on 20 ha (40,000 plants/ha) and evaluated another 20 ha as the control without bioproducts. For the statistical analysis, we used a completely random block design with a split plot layout, where the main plot was the organic matter and small plots were: control (no bioproduct), Biobacillus, Tricotop, Biobacillus + Tricotop or Tricobal. Then, the means were compared by the Duncan applied test (p = 0.05).