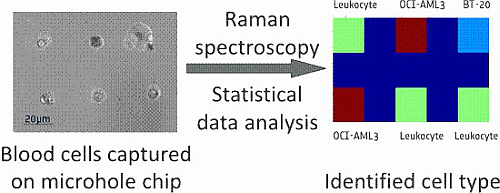
Raman-Spectroscopy Based Cell Identification on a Microhole Array Chip
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Sample Preparation
2.2. Microhole Array Chip
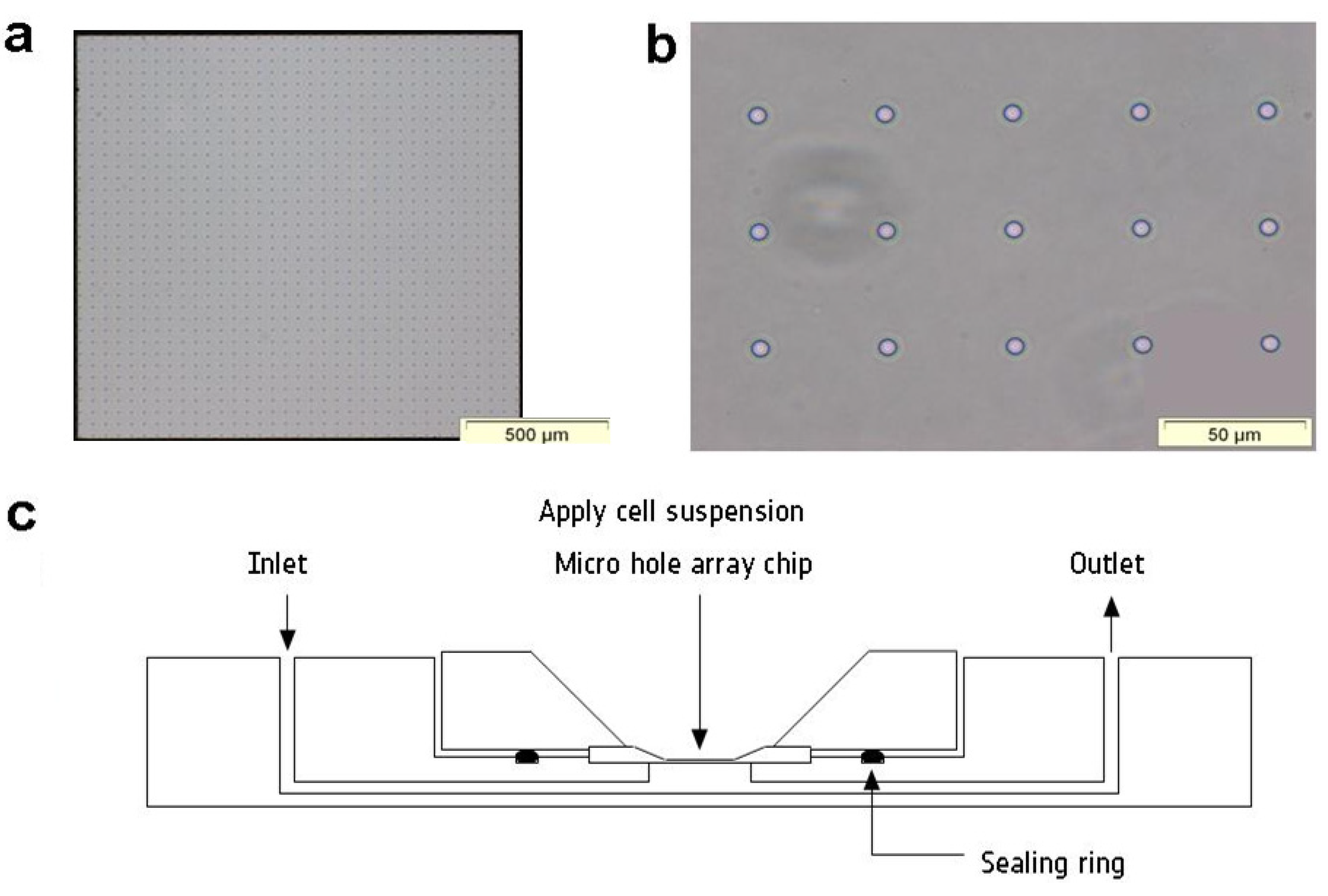
2.3. Raman Measurements
2.4. Fluorescence Analysis
2.5. Statistical Analysis
3. Results and Discussion
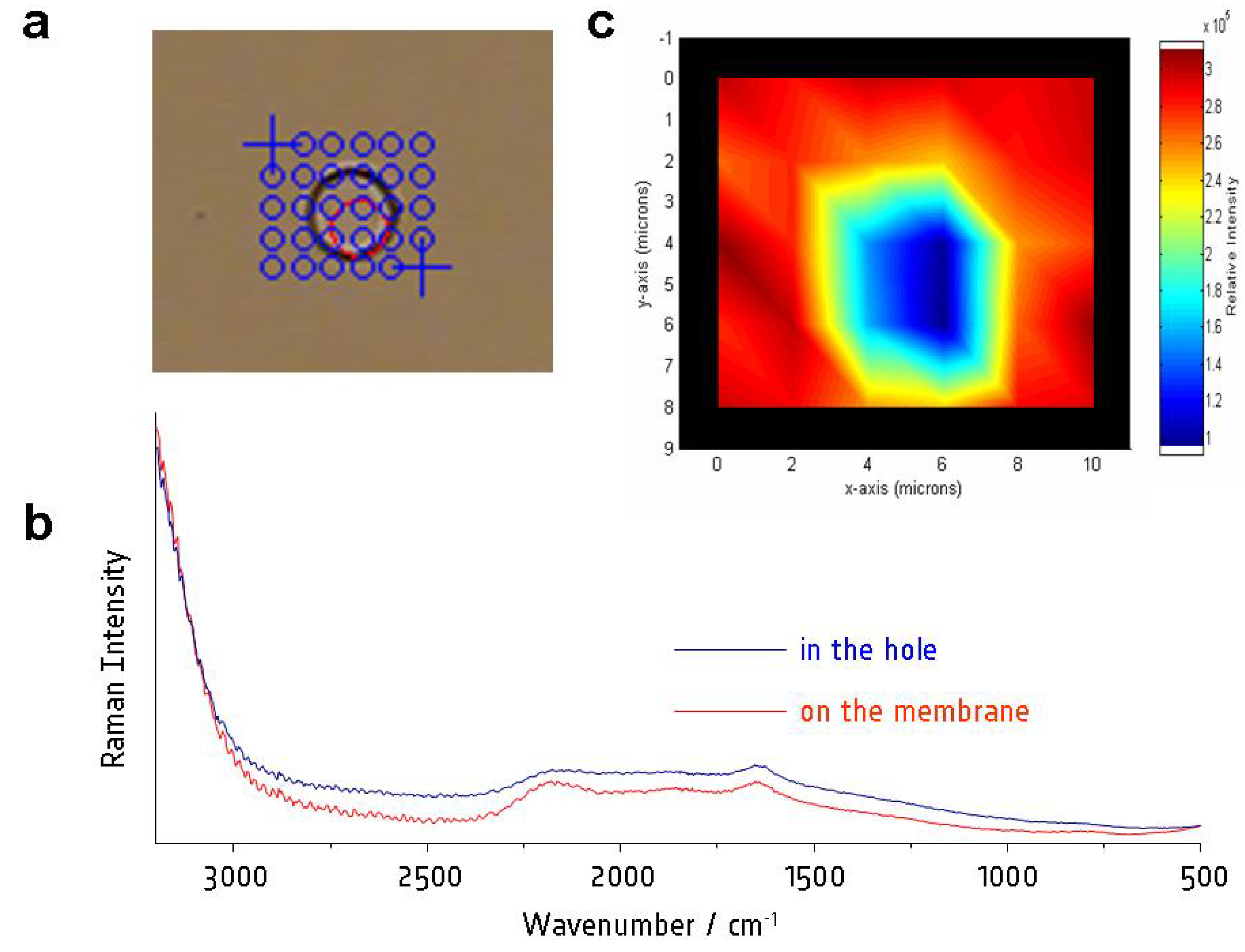
3.1. Characterization of the Microhole Chip by Raman Spectroscopy
3.2. Capturing Cells on the Microhole Chip

3.3. Raman Spectroscopy of Captured Cells in Solution
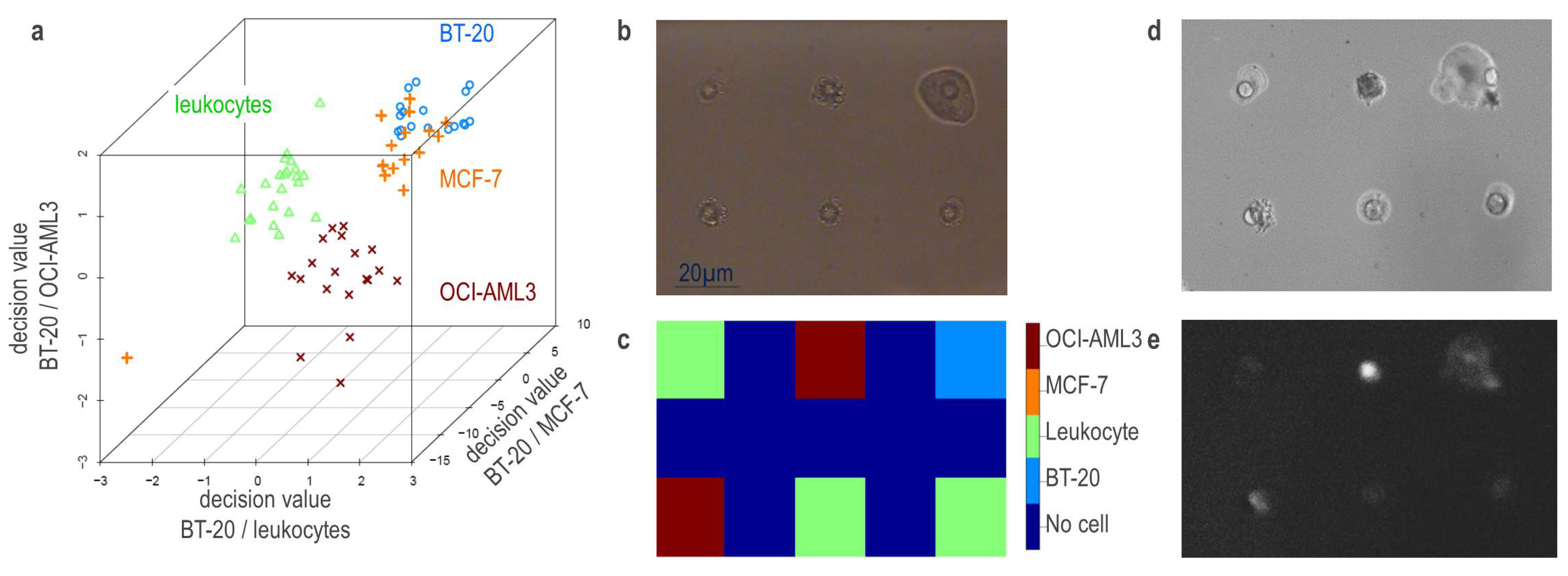
3.4. Raman Based Classification Model of Captured Cells
| Predicted labels | |||||
|---|---|---|---|---|---|
| Leukocytes | OCI-AML3 | BT-20 | MCF-7 | ||
| True labels | Leukocytes | 22 | 0 | 0 | 0 |
| OCI-AML3 | 0 | 19 | 0 | 0 | |
| BT-20 | 0 | 0 | 12 | 6 | |
| MCF-7 | 0 | 1 | 5 | 10 | |
| Predicted labels | |||
|---|---|---|---|
| Healthy cell | Tumor cell | ||
| True labels | Healthy cell | 22 (100%) | 0 (0%) |
| Tumor cell | 0 (0%) | 53 (100%) | |
3.5. Identification of Captured Cells in Mixtures
4. Conclusions and Outlook
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Yu, M.; Stott, S.; Toner, M.; Maheswaran, S.; Haber, D.A. Circulating tumor cells: Approaches to isolation and characterization. J. Cell Biol. 2011, 192, 373–382. [Google Scholar] [CrossRef]
- Pantel, K.; Alix-Panabieres, C. Real-time Liquid Biopsy in Cancer Patients: Fact or Fiction? Cancer Res. 2013, 73, 6384–6388. [Google Scholar] [CrossRef]
- Gorges, T.M.; Pantel, K. Circulating tumor cells as therapy-related biomarkers in cancer patients. Cancer Immunol. Immunother. 2013, 62, 931–939. [Google Scholar] [CrossRef]
- Riethdorf, S.; Fritsche, H.; Mueller, V.; Rau, T.; Schindibeck, C.; Rack, B.; Janni, W.; Coith, C.; Beck, K.; Jaenicke, F.; et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: A validation study of the CellSearch system. Clin. Cancer Res. 2007, 13, 920–928. [Google Scholar] [CrossRef]
- Yoon, H.J.; Kozminsky, M.; Nagrath, S. Emerging role of nanomaterials in circulating tumor cell isolation and analysis. ACS Nano 2014, 8, 1995–2017. [Google Scholar] [CrossRef]
- Attard, G.; Swermenhuis, J.F.; Olmos, D.; Reid, A.H.M.; Vickers, E.; A’Hern, R.; Levink, R.; Coumans, F.; Moreira, J.; Riisnaes, R.; et al. Characterization of ERG, AR and PTEN Gene Status in Circulating Tumor Cells from Patients with Castration-Resistant Prostate Cancer. Cancer Res. 2009, 69, 2912–2918. [Google Scholar] [CrossRef]
- Stott, S.L.; Hsu, C.-H.; Tsukrov, D.I.; Yu, M.; Miyamoto, D.T.; Waltman, B.A.; Rothenberg, S.M.; Shah, A.M.; Smas, M.E.; Korir, G.K.; et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl. Acad. Sci. USA 2010, 107, 18392–18397. [Google Scholar] [CrossRef]
- Neugebauer, U.; Clement, J.H.; Bocklitz, T.; Krafft, C.; Popp, J. Identification and differentiation of single cells from peripheral blood by Raman spectroscopic imaging. J. Biophotonics 2010, 3, 579–587. [Google Scholar] [CrossRef]
- Neugebauer, U.; Bocklitz, T.; Clement, J.H.; Krafft, C.; Popp, J. Towards detection and identification of circulating tumor cells using Raman spectroscopy. Analyst 2010, 135, 3178–3182. [Google Scholar]
- Chan, J.W. Recent advances in laser tweezers Raman spectroscopy (LTRS) for label-free analysis of single cells. J. Biophotonics 2013, 6, 36–48. [Google Scholar] [CrossRef]
- Dochow, S.; Krafft, C.; Neugebauer, U.; Bocklitz, T.; Henkel, T.; Mayer, G.; Albert, J.; Popp, J. Tumor cell identification by means of Raman spectroscopy in combination with optical traps and microfluidic environments. Lab Chip 2011, 11, 1484–1490. [Google Scholar] [CrossRef]
- Dochow, S.; Beleites, C.; Henkel, T.; Mayer, G.; Albert, J.; Clement, J.; Krafft, C.; Popp, J. Quartz microfluidic chip for tumor cell identification by Raman spectroscopy in combination with optical traps. Anal. Bioanal. Chem. 2013, 405, 2743–2746. [Google Scholar] [CrossRef]
- Zheng, G.A. Chip-scale microscopy imaging. J. Biophotonics 2012, 5, 639–649. [Google Scholar] [CrossRef]
- Kurz, C.M.; Buth, H.; Sossalla, A.; Vermeersch, V.; Toncheva, V.; Dubruel, P.; Schacht, E.; Thielecke, H. Chip-based impedance measurement on single cells for monitoring sub-toxic effects on cell membranes. Biosens. Bioelectron. 2011, 26, 3405–3412. [Google Scholar] [CrossRef]
- Matsunaga, T.; Hosokawa, M.; Arakaki, A.; Taguchi, T.; Mori, T.; Tanaka, T.; Takeyama, H. High-efficiency single-cell entrapment and fluorescence in situ hybridization analysis using a poly(dimethylsiloxane) microfluidic device integrated with a black poly(ethylene terephthalate) micromesh. Anal. Chem. 2008, 80, 5139–5145. [Google Scholar] [CrossRef]
- Hosokawa, M.; Arakaki, A.; Takahashi, M.; Mori, T.; Takeyama, H.; Matsunaga, T. High-density microcavity array for cell detection: Single-cell analysis of hematopoietic stem cells in peripheral blood mononuclear cells. Anal. Chem. 2009, 81, 5308–5313. [Google Scholar]
- Cho, S.; Thielecke, H. Micro hole-based cell chip with impedance spectroscopy. Biosens. Bioelectron. 2007, 22, 1764–1768. [Google Scholar] [CrossRef]
- Kurz, C.M.; Maurer, A.; Thees, K.; Schillberg, S.; Velten, T.; Thielecke, H. Impedance-controlled cell entrapment using microhole-array chips allows the isolation and identification of single, highly productive cells. Sens. Actuators B Chem. 2011, 158, 345–352. [Google Scholar] [CrossRef]
- Pachmann, K.; Clement, J.H.; Schneider, C.P.; Willen, B.; Camara, O.; Pachmann, U.; Hoffken, K. Standardized quantification of circulating peripheral tumor cells from lung and breast cancer. Clin. Chem. Lab. Med. 2005, 43, 617–627. [Google Scholar]
- Schwalbe, M.; Pachmann, K.; Hoffken, K.; Clement, J.H. Improvement of the separation of tumor cells from peripheral blood cells using magnetic nanoparticles. J. Phys. Condens. Matter 2006, 18, S2865–S2876. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2007. [Google Scholar]
- Ryan, C.G.; Clayton, E.; Griffin, W.L.; Sie, S.H.; Cousens, D.R. SNIP, a statistics-sensitive background treatment for the quantitative analysis of pixe spectra in geosience aplications. Nucl. Instrum. Methods Phys. Res. Sect. B 1988, 34, 396–402. [Google Scholar] [CrossRef]
- Chang, C.-C.; Lin, C.-J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011, 2, 1–27. [Google Scholar] [CrossRef]
- Giorgis, F.; Pirri, C.F. Characterization and Physical Properties of Non-Crystalline and Nanostructured Silicon-Based Alloys. In Slicon-Based Materials and Devices; Singh, N.H., Ed.; Acedemic Press: San Diego, CA, USA, 2001; pp. 187–236. [Google Scholar]
- Dochow, S.; Bergner, N.; Krafft, C.; Clement, J.; Mazilu, M.; Balagopal, B.; Marchington, R.; Dholakia, K.; Popp, J. Classification of Raman spectra of single cells with autofluorescence suppression by wavelength modulated excitation. Anal. Methods 2013, 5, 4608–4614. [Google Scholar] [CrossRef]
- Beleites, C.; Neugebauer, U.; Bocklitz, T.; Krafft, C.; Popp, J. Sample size planning for classification models. Anal. Chim. Acta 2013, 760, 25–33. [Google Scholar] [CrossRef]
- Suhalim, J.L.; Boik, J.C.; Tromberg, B.J.; Potma, E.O. The need for speed. J. Biophotonics 2012, 5, 387–395. [Google Scholar] [CrossRef]
- Esseling, M.; Kemper, B.; Antkowiak, M.; Stevenson, D.J.; Chaudet, L.; Neil, M.A.A.; French, P.W.; von Bally, G.; Dholakia, K.; Denz, C. Multimodal biophotonic workstation for live cell analysis. J. Biophotonics 2012, 5, 9–13. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Neugebauer, U.; Kurz, C.; Bocklitz, T.; Berger, T.; Velten, T.; Clement, J.H.; Krafft, C.; Popp, J. Raman-Spectroscopy Based Cell Identification on a Microhole Array Chip. Micromachines 2014, 5, 204-215. https://doi.org/10.3390/mi5020204
Neugebauer U, Kurz C, Bocklitz T, Berger T, Velten T, Clement JH, Krafft C, Popp J. Raman-Spectroscopy Based Cell Identification on a Microhole Array Chip. Micromachines. 2014; 5(2):204-215. https://doi.org/10.3390/mi5020204
Chicago/Turabian StyleNeugebauer, Ute, Christian Kurz, Thomas Bocklitz, Tina Berger, Thomas Velten, Joachim H. Clement, Christoph Krafft, and Jürgen Popp. 2014. "Raman-Spectroscopy Based Cell Identification on a Microhole Array Chip" Micromachines 5, no. 2: 204-215. https://doi.org/10.3390/mi5020204
APA StyleNeugebauer, U., Kurz, C., Bocklitz, T., Berger, T., Velten, T., Clement, J. H., Krafft, C., & Popp, J. (2014). Raman-Spectroscopy Based Cell Identification on a Microhole Array Chip. Micromachines, 5(2), 204-215. https://doi.org/10.3390/mi5020204