Talaromyces marneffei Genomic, Transcriptomic, Proteomic and Metabolomic Studies Reveal Mechanisms for Environmental Adaptations and Virulence
Abstract
:1. Introduction
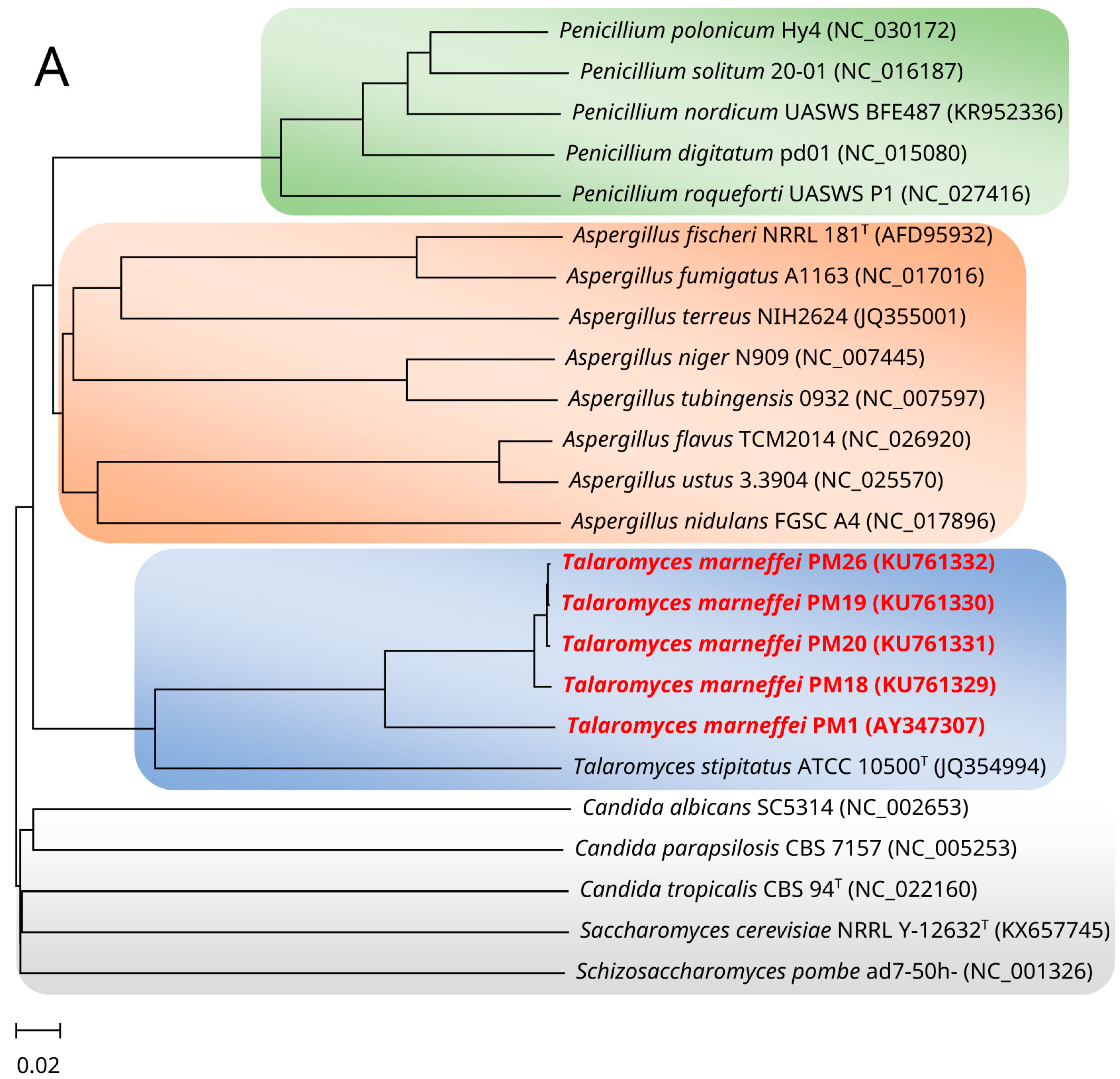
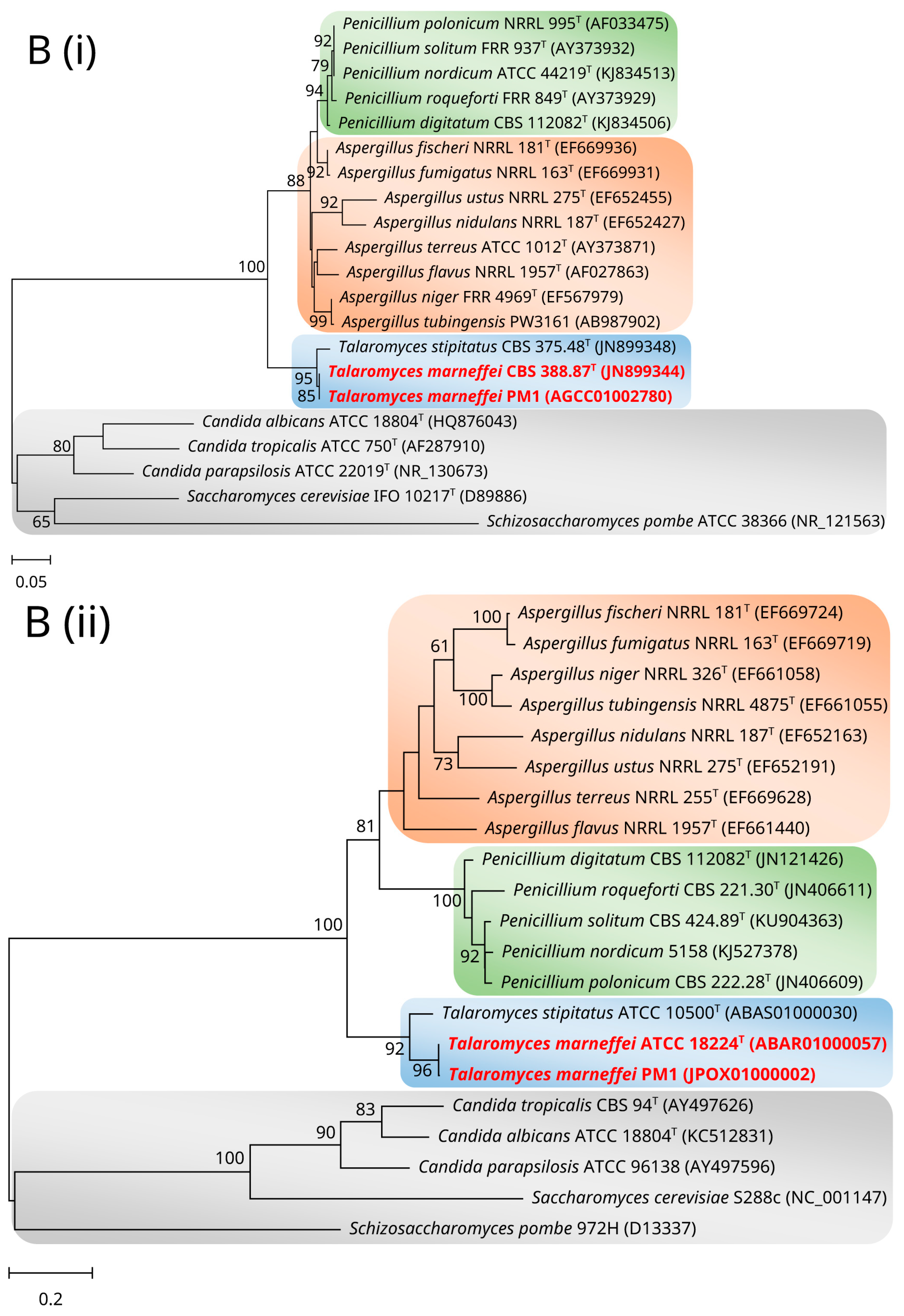
2. Mitochondrial Genome and Phylogeny
3. Sexual Stage
4. MP1 and Its Homologs
5. Virulence Properties
6. Multilocus Sequence Typing
7. Polyketide Synthases and Pigments
8. microRNA
9. Transcriptome Profiling
10. Proteome Profiling
11. Concluding Remarks
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Capponi, M.; Sureau, P.; Segretain, G. Pénicilliose de Rhizomys sinensis. Bull. Soc. Pathol. Exot. 1956, 49, 418–421. [Google Scholar]
- Deng, Z.; Yun, M.; Ajello, L. Human penicilliosis marneffei and its relation to the bamboo rat (Rhizomys pruinosus). J. Med. Vet. Mycol. 1986, 24, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Ajello, L.; Padhye, A.A.; Sukroongreung, S.; Nilakul, C.H.; Tantimavanic, S. Occurrence of Penicillium marneffei infections among wild bamboo rats in Thailand. Mycopathologia 1995, 131, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Chariyalertsak, S.; Vanittanakom, P.; Nelson, K.E.; Sirisanthana, T.; Vanittanakom, N. Rhizomys sumatrensis and Cannomys badius, new natural animal hosts of Penicillium marneffei. J. Med. Vet. Mycol. 1996, 34, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Samson, R.A.; Yilmaz, N.; Houbraken, J.; Spierenburg, H.; Seifert, K.A.; Peterson, S.W.; Varga, J.; Frisvad, J.C. Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Stud. Mycol. 2011, 70, 159–183. [Google Scholar] [CrossRef] [PubMed]
- Vanittanakom, N.; Cooper, C.R.; Fisher, M.C.; Sirisanthana, T. Penicillium marneffei infection and recent advances in the epidemiology and molecular biology aspects. Clin. Microbiol. Rev. 2006, 19, 95–110. [Google Scholar] [CrossRef] [PubMed]
- Nittayananta, W. Penicilliosis marneffei: Another AIDS defining illness in Southeast Asia. Oral Dis. 1999, 5, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Ribas, J.L.; Dean, W.G.; Connor, D.H. Infections caused by Penicillium marneffei in China and Southeast Asia: Review of eighteen published cases and report of four more chinese cases. Rev. Infect. Dis. 1988, 10, 640–652. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.W.; Chan, T.S.Y.; Gill, H.; Lam, F.Y.F.; Trendell-Smith, N.J.; Sridhar, S.; Tse, H.; Lau, S.K.P.; Hung, I.F.N.; Yuen, K.-Y.; et al. Disseminated infections with Talaromyces marneffei in non-AIDS patients given monoclonal antibodies against CD20 and kinase inhibitors. Emerg. Infect. Dis. 2015, 21, 1101–1106. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.W.; Lau, S.K.P.; Yuen, K.-Y.; Woo, P.C.Y. Talaromyces (Penicillium) marneffei infection in non-HIV-infected patients. Emerg. Microbes Infect. 2016, 5, e19. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lau, S.K.P.; Liu, B.; Cai, J.J.; Chong, K.T.K.; Tse, H.; Kao, R.Y.T.; Chan, C.-M.; Chow, W.-N.; Yuen, K.-Y. Draft genome sequence of Penicillium marneffei strain PM1. Eukaryot. Cell 2011, 10, 1740–1741. [Google Scholar] [CrossRef] [PubMed]
- Tam, E.W.T.; Tsang, C.-C.; Lau, S.K.P.; Woo, P.C.Y. Comparative mitogenomic and phylogenetic characterization on the complete mitogenomes of Talaromyces (Penicillium) marneffei. Mitochondrial DNA B Resour. 2016, 1, 941–942. [Google Scholar] [CrossRef]
- Xu, Z.; Hao, B. CVTree update: A newly designed phylogenetic study platform using composition vectors and whole genomes. Nucleic Acids Res. 2009, 37, W174–W178. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.C.; Hanage, W.P.; de Hoog, S.; Johnson, E.; Smith, M.D.; White, N.J.; Vanittanakom, N. Low effective dispersal of asexual genotypes in heterogeneous landscapes by the endemic pathogen Penicillium marneffei. PLoS Pathog. 2005, 1, e20. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Chong, K.T.K.; Tse, H.; Cai, J.J.; Lau, C.C.Y.; Zhou, A.C.; Lau, S.K.P.; Yuen, K.-Y. Genomic and experimental evidence for a potential sexual cycle in the pathogenic thermal dimorphic fungus Penicillium marneffei. FEBS Lett. 2006, 580, 3409–3416. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Chan, C.-M.; Lee, C.; Wong, S.S.Y.; Yuen, K.-Y. MP1 encodes an abundant and highly antigenic cell wall mannoprotein in the pathogenic fungus Penicillium marneffei. Infect. Immun. 1998, 66, 966–973. [Google Scholar] [PubMed]
- Cao, L.; Chan, K.-M.; Chen, D.; Vanittanakom, N.; Lee, C.; Chan, C.-M.; Sirisanthana, T.; Tsang, D.N.C.; Yuen, K.-Y. Detection of cell wall mannoprotein Mp1p in culture supernatants of Penicillium marneffei and in sera of penicilliosis patients. J. Clin. Microbiol. 1999, 37, 981–986. [Google Scholar] [PubMed]
- Yuen, K.-Y.; Chan, C.-M.; Chan, K.-M.; Woo, P.C.Y.; Che, X.-Y.; Leung, A.S.P.; Cao, L. Characterization of AFMP1: A novel target for serodiagnosis of Aspergillosis. J. Clin. Microbiol. 2001, 39, 3830–3837. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Chan, C.-M.; Leung, A.S.P.; Lau, S.K.P.; Che, X.-Y.; Wong, S.S.Y.; Cao, L.; Yuen, K.-Y. Detection of cell wall galactomannoprotein AFMP1P in culture supernatants of Aspergillus fumigatus and in sera of aspergillosis patients. J. Clin. Microbiol. 2002, 40, 4382–4387. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Chong, K.T.K.; Leung, A.S.P.; Wong, S.S.Y.; Lau, S.K.P.; Yuen, K.-Y. AFLMP1 encodes an antigenic cell wall protein in Aspergillus flavus. J. Clin. Microbiol. 2003, 41, 845–850. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lau, S.K.P.; Lau, C.C.Y.; Tung, E.T.K.; Chong, K.T.K.; Yang, F.; Zhang, H.; Lo, R.K.C.; Cai, J.-P.; Au-Yeung, R.K.H.; et al. Mp1p is a virulence factor in Talaromyces (Penicillium) marneffei. PLoS Negl. Trop. Dis. 2016, 10, e0004907. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.; Tung, E.T.K.; Zheng, W.; Chong, K.; Xu, Y.; Dai, P.; Guo, Y.; Bartlam, M.; Yuen, K.-Y.; Rao, Z. Crystal structure of the Mp1p ligand binding domain 2 reveals its function as a fatty acid-binding protein. J. Biol. Chem. 2010, 285, 9211–9220. [Google Scholar] [CrossRef] [PubMed]
- Sze, K.-H.; Lam, W.-H.; Zhang, H.; Ke, Y.-H.; Tse, M.-K.; Woo, P.C.Y.; Lau, S.K.P.; Lau, C.C.Y.; Cai, J.-P.; Tung, E.T.K.; et al. Talaromyces marneffei Mp1p Is a virulence factor that binds and sequesters a key proinflammatory lipid to dampen host innate immune response. Cell Chem. Biol. 2017, 24, 182–194. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Tsang, C.-C.; Xue, S.; Yang, F.; Tan, Y.-P.; Cai, J.-P.; Kok, K.-H.; Yuen, K.-Y.; Lau, S.K.P. Talaromyces marneffei Mp1p binds a variety of proteins, sphingolipids and phospholipids: Implication on virulence mechanism. 2017; unpublished, manuscript in preparation. [Google Scholar]
- Maiden, M.C.J.; Bygraves, J.A.; Feil, E.; Morelli, G.; Russell, J.E.; Urwin, R.; Zhang, Q.; Zhou, J.; Zurth, K.; Caugant, D.A.; et al. Multilocus sequence typing: A portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 1998, 95, 3140–3145. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lau, C.C.Y.; Chong, K.T.K.; Tse, H.; Tsang, D.N.C.; Lee, R.A.; Tse, C.W.S.; Que, T.-L.; Chung, L.M.W.; Ngan, A.H.Y.; et al. MP1 homologue-based multilocus sequence system for typing the pathogenic fungus Penicillium marneffei: A novel approach using lineage-specific genes. J. Clin. Microbiol. 2007, 45, 3647–3654. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lam, C.-W.; Tam, E.W.T.; Leung, C.K.F.; Wong, S.S.Y.; Lau, S.K.P.; Yuen, K.-Y. First discovery of two polyketide synthase genes for mitorubrinic acid and mitorubrinol yellow pigment biosynthesis and implications in virulence of Penicillium marneffei. PLoS Negl. Trop. Dis. 2012, 6, e1871. [Google Scholar] [CrossRef] [PubMed]
- Tam, E.W.T.; Tsang, C.-C.; Lau, S.K.P.; Woo, P.C.Y. Polyketides, toxins and pigments in Penicillium marneffei. Toxins 2015, 7, 4421–4436. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Tam, E.W.T.; Chong, K.T.K.; Cai, J.J.; Tung, E.T.K.; Ngan, A.H.Y.; Lau, S.K.P.; Yuen, K.-Y. High diversity of polyketide synthase genes and the melanin biosynthesis gene cluster in Penicillium marneffei. FEBS J. 2010, 277, 3750–3758. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lam, C.-W.; Tam, E.W.T.; Lee, K.-C.; Yung, K.K.Y.; Leung, C.K.F.; Sze, K.-H.; Lau, S.K.P.; Yuen, K.-Y. The biosynthetic pathway for a thousand-year-old natural food colorant and citrinin in Penicillium marneffei. Sci. Rep. 2014, 4, 6728. [Google Scholar] [CrossRef] [PubMed]
- Nimmanee, P.; Woo, P.C.Y.; Kummasook, A.; Vanittanakom, N. Characterization of sakA gene from pathogenic dimorphic fungus Penicillium marneffei. Int. J. Med. Microbiol. 2015, 305, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Nimmanee, P.; Tam, E.W.T.; Woo, P.C.Y.; Vanittanakom, P.; Vanittanakom, N. Role of the Talaromyces marneffei (Penicillium marneffei) sakA gene in nitrosative stress response, conidiation and red pigment production. FEMS Microbiol. Lett. 2017, 364, fnw292. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-C.; Li, L.; Gu, W.; Xue, Z.; Crosthwaite, S.K.; Pertsemlidis, A.; Lewis, Z.A.; Freitag, M.; Selker, E.U.; Mello, C.C.; et al. Diverse pathways generate microRNA-like RNAs and Dicer-independent small interfering RNAs in fungi. Mol. Cell 2010, 38, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Fu, Y.; Xie, J.; Li, B.; Jiang, D.; Li, G.; Cheng, J. Identification of microRNA-like RNAs in a plant pathogenic fungus Sclerotinia sclerotiorum by high-throughput sequencing. Mol. Genet. Genom. 2012, 287, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Wang, Z.; Zhang, J.; Meng, H.; Huang, B. Genome-wide identification and profiling of microRNA-like RNAs from Metarhizium anisopliae during development. Fungal Biol. 2012, 116, 1156–1162. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Yang, Y.; Janbon, G.; Pan, J.; Zhu, X. Identification and functional demonstration of miRNAs in the fungus Cryptococcus neoformans. PLoS ONE 2012, 7, e52734. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.P.; Chow, W.-N.; Wong, A.Y.P.; Yeung, J.M.Y.; Bao, J.; Zhang, N.; Lok, S.; Woo, P.C.Y.; Yuen, K.-Y. Identification of microRNA-like RNAs in mycelial and yeast phases of the thermal dimorphic fungus Penicillium marneffei. PLoS Negl. Trop. Dis. 2013, 7, e2398. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Ran, Y.; Gou, L.; He, F.; Zhang, R.; Wang, P.; Dai, Y. Comprehensive transcription analysis of human pathogenic fungus Penicillium marneffei in mycelial and yeast cells. Med. Mycol. 2012, 50, 835–842. [Google Scholar] [CrossRef] [PubMed]
- Pasricha, S.; Payne, M.; Canovas, D.; Pase, L.; Ngaosuwankul, N.; Beard, S.; Oshlack, A.; Smyth, G.K.; Chaiyaroj, S.C.; Boyce, K.J.; et al. Cell-type–specific transcriptional profiles of the dimorphic pathogen Penicillium marneffei reflect distinct reproductive, morphological, and environmental demands. G3 (Bethesda) 2013, 3, 1997–2014. [Google Scholar] [CrossRef] [PubMed]
- Yang, E.; Wang, G.; Woo, P.C.Y.; Lau, S.K.P.; Chow, W.-N.; Chong, K.T.K.; Tse, H.; Kao, R.Y.T.; Chan, C.-M.; Che, X.; et al. Unraveling the molecular basis of temperature-dependent genetic regulation in Penicillium marneffei. Eukaryot. Cell 2013, 12, 1214–1224. [Google Scholar] [CrossRef] [PubMed]
- Yang, E.; Chow, W.-N.; Wang, G.; Woo, P.C.Y.; Lau, S.K.P.; Yuen, K.-Y.; Lin, X.; Cai, J.J. Signature gene expression reveals novel clues to the molecular mechanisms of dimorphic transition in Penicillium marneffei. PLoS Genet. 2014, 10, e1004662. [Google Scholar] [CrossRef] [PubMed]
- Xi, L.; Xu, X.; Liu, W.; Li, X.; Liu, Y.; Li, M.; Zhang, J.; Li, M. Differentially expressed proteins of pathogenic Penicillium marneffei in yeast and mycelial phases. J. Med. Microbiol. 2007, 56, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.M.; Treece, E.R.; Trenary, H.R.; Brenneman, J.L.; Flickner, T.J.; Frommelt, J.L.; Oo, Z.M.; Patterson, M.M.; Rundle, W.T.; Valle, O.V.; et al. Protein profiling of the dimorphic, pathogenic fungus, Penicillium marneffei. Proteome Sci. 2008, 6, 17. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.P.; Tse, H.; Chan, J.S.Y.; Zhou, A.C.; Curreem, S.O.T.; Lau, C.C.Y.; Yuen, K.-Y.; Woo, P.C.Y. Proteome profiling of the dimorphic fungus Penicillium marneffei extracellular proteins and identification of glyceraldehyde-3-phosphate dehydrogenase as an important adhesion factor for conidial attachment. FEBS J. 2013, 280, 6613–6626. [Google Scholar] [CrossRef] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lau, S.K.P.; Tsang, C.-C.; Woo, P.C.Y. Talaromyces marneffei Genomic, Transcriptomic, Proteomic and Metabolomic Studies Reveal Mechanisms for Environmental Adaptations and Virulence. Toxins 2017, 9, 192. https://doi.org/10.3390/toxins9060192
Lau SKP, Tsang C-C, Woo PCY. Talaromyces marneffei Genomic, Transcriptomic, Proteomic and Metabolomic Studies Reveal Mechanisms for Environmental Adaptations and Virulence. Toxins. 2017; 9(6):192. https://doi.org/10.3390/toxins9060192
Chicago/Turabian StyleLau, Susanna K. P., Chi-Ching Tsang, and Patrick C. Y. Woo. 2017. "Talaromyces marneffei Genomic, Transcriptomic, Proteomic and Metabolomic Studies Reveal Mechanisms for Environmental Adaptations and Virulence" Toxins 9, no. 6: 192. https://doi.org/10.3390/toxins9060192







