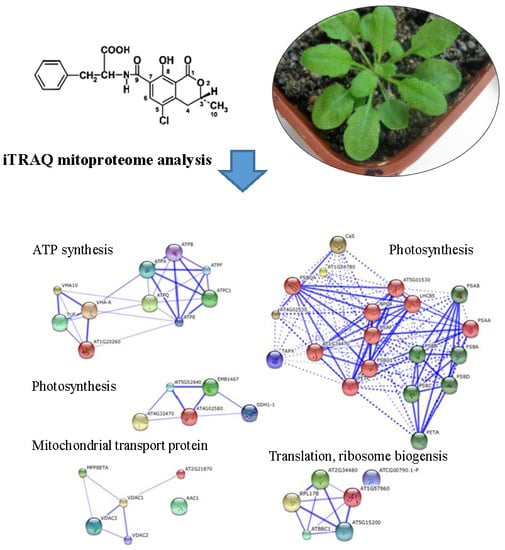
iTRAQ Mitoproteome Analysis Reveals Mechanisms of Programmed Cell Death in Arabidopsis thaliana Induced by Ochratoxin A
Abstract
:1. Introduction
2. Results
2.1. Effect of OTA on mtROS Content and Respiration Rate
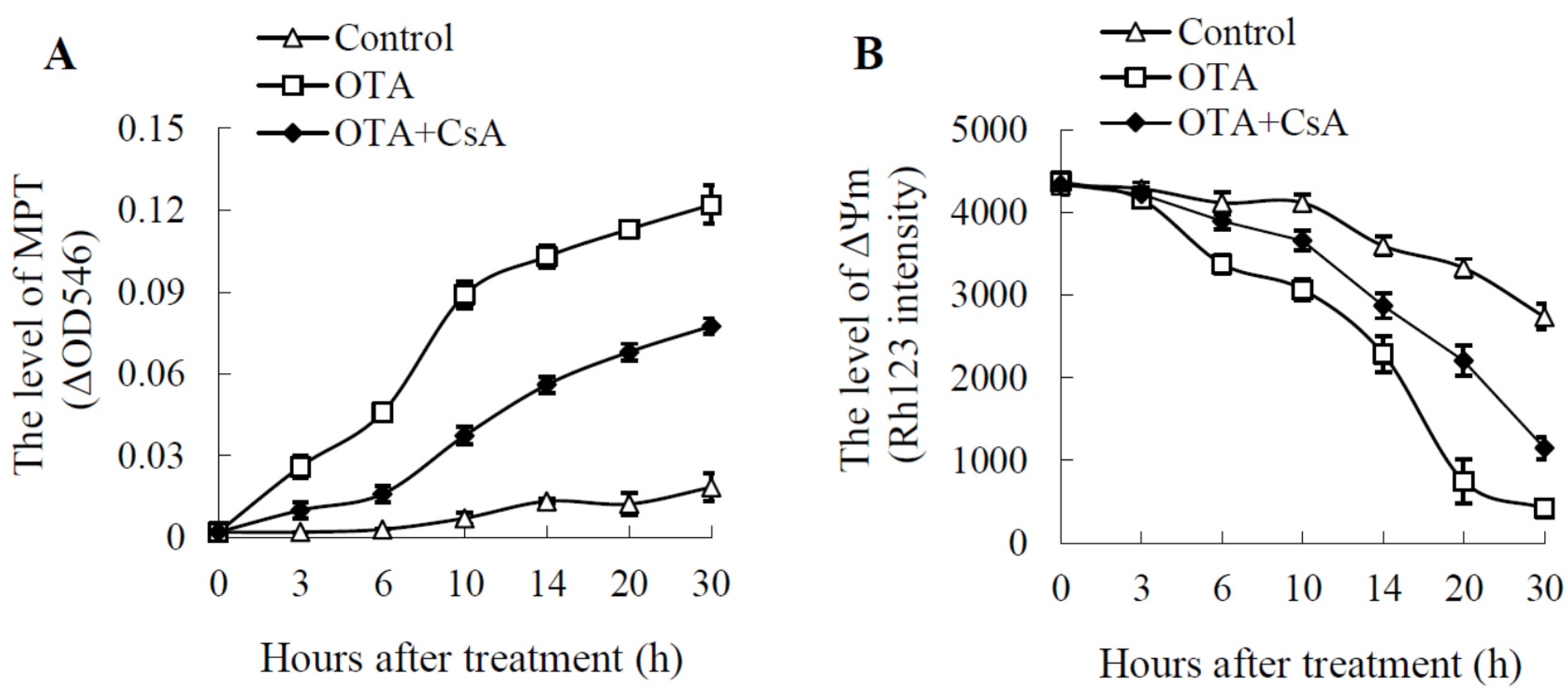
2.2. Mitochondrial Swelling and Decreased ΔΨm in OTA Treatment
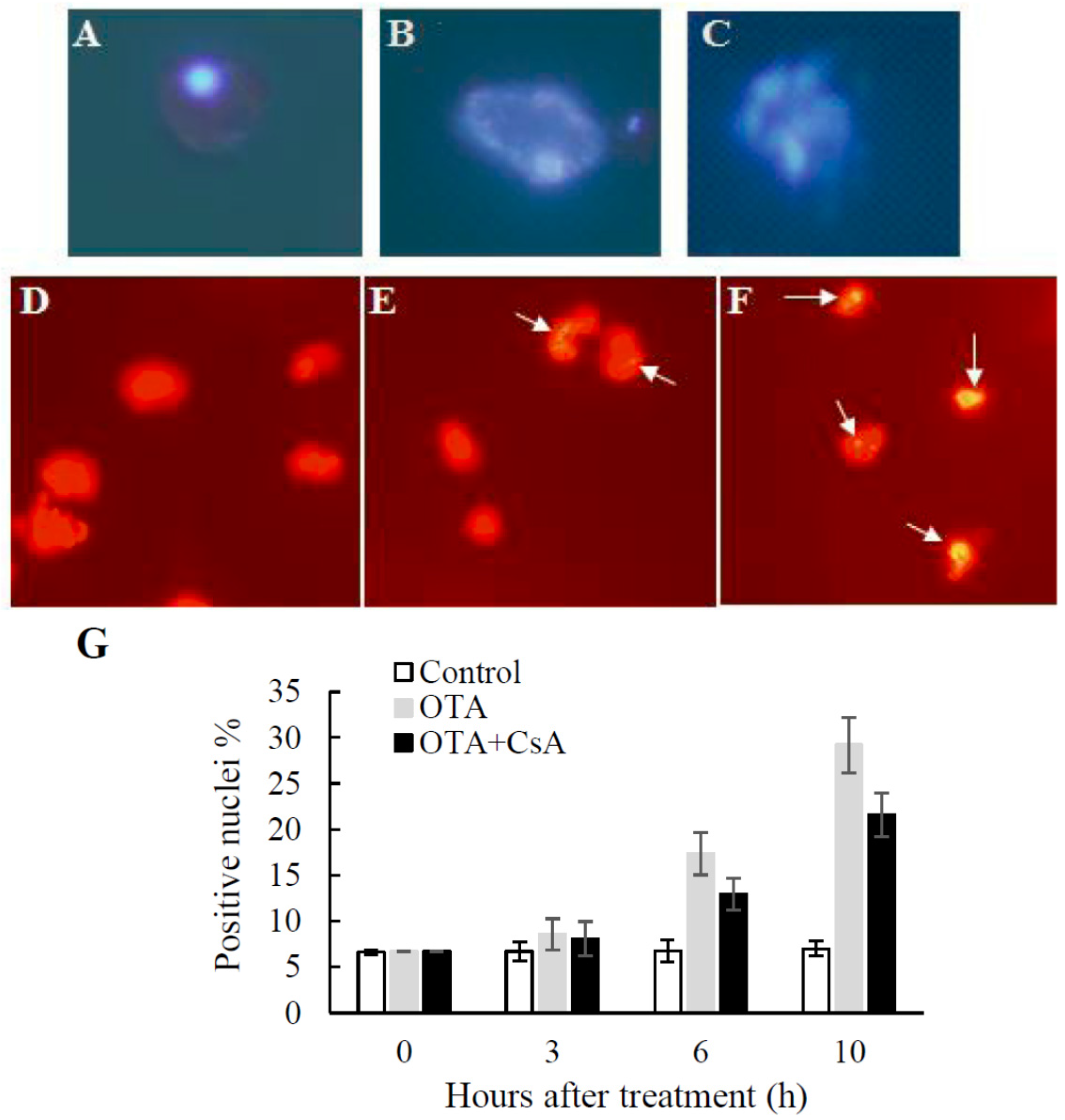
2.3. PCD-like Features in OTA-treated Arabidopsis Protoplasts
2.4. Cyt c Release Occurs in OTA-induced PCD of Arabidopsis Leaves
2.5. iTRAQ Analysis of OTA-Responsive Mitochondrial Proteins in Arabidopsis Leaves
2.6. Classification of Differentially Expressed Mitoproteome
2.7. DGE Profiling Analysis
2.8. The Expression Profile of Nucleic Acid Metabolism
2.9. The Expression Profile of Oxidative Phosphorylation and Citric Acid Cycle in Mitochondria
3. Discussion
3.1. OTA Disturbed the Mitochondrial Electron Transport Chain
3.2. OTA Interferes with ATP Synthesis
3.3. OTA Induces PCD in Arabidopsis thaliana
4. Experimental Section
4.1. Plant Material, Growth Conditions and OTA Treatments
4.2. Mitochondria Isolation
4.3. Mitochondrial ROS (mtROS) Measurement
4.4. Assessment of Mitochondrial Permeability Transition (MPT)
4.5. Measurement of Mitochondrial Membrane Potential (ΔΨm)
4.6. Respiration Assay
4.7. Nuclear Morphology and DNA Fragmentation
4.8. Measurement of Cytochrome c/a Ratio
4.9. Cyt c Western Blot
4.10. iTRAQ Labeling and Mass Spectrometry Identification
4.11. Database Identification and iTRAQ Quantification
4.12. Mitoproteome Analysis
4.13. DGE Tag Profiling
4.14. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wang, H.; Li, J.; Bostock, R.M.; Gilchrist, D.G. Apoptosis: a functional paradigm for programmed plant cell death induced by a host-selective phytotoxin and invoked during development. Plant Cell 1996, 8, 375–391. [Google Scholar] [CrossRef] [PubMed]
- Campbell, N.A.; Brad, W.; Robin, J.H. Biology: Exploring Life; Pearson Prentice Hall: Boston, MA, USA, 2006; ISBN 0-13-250882-6. [Google Scholar]
- Yoshinaga, K.; Arimura, S.I.; Niwa, Y.; Tsutsumi, N.; Uchimiya, H.; Kawai-Yamada, M. Mitochondrial behaviour in the early stages of ROS stress leading to cell death in Arabidopsis thaliana. Ann. Bot. 2005, 96, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Vacca, R.A.; De Pinto, M.C.; Valenti, D.; Passarella, S.; Marra, E.; De Gara, L. Production of reactive oxygen species, alteration of cytosolic ascorbate peroxidase, and impairment of mitochondrial metabolism are early events in heat shock-induced programmed cell death in tobacco Bright-Yellow 2 cells. Plant Physiol. 2004, 134, 1100–1112. [Google Scholar] [CrossRef] [PubMed]
- Lam, E.; Kato, N.; Lawton, M. Programmed cell death, mitochondria and the plant hypersensitive response. Nature 2001, 411, 848–853. [Google Scholar] [CrossRef] [PubMed]
- Yao, N.; Tada, Y.; Sakamoto, M.; Nakayashiki, H.; Park, P.; Tosa, Y.; Mayama, S. Mitochondrial oxidative burst involved in apoptotic response in oats. Plant J. 2002, 30, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, B.S.; Belenghi, B.; Levine, A. Oxidative stress increased respiration and generation of reactive oxygen species, resulting in ATP depletion, opening of mitochondrial permeability transition, and programmed cell death. Plant Physiol. 2002, 128, 1271–1281. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Xing, D.; Gao, X.; Chen, W.R. High fluence low-power laser irradiation induces mitochondrial permeability transition mediated by reactive oxygen species. J. Cell. Physiol. 2008, 218, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.L.; Zhao, Y.; Hong, X.; Zhai, Z.H. Cytochrome c release and caspase activation during menadione-induced apoptosis in plants. FEBS Lett. 1999, 462, 317–321. [Google Scholar] [CrossRef]
- Vacca, R.A.; Valenti, D.; Bobba, A.; Merafina, R.S.; Passarella, S.; Marra, E. Cytochrome C is released in a reactive oxygen species-dependent manner and is degraded via caspase-like proteases in tobacco Bright-Yellow 2 cells en route to heat shock-induced cell death. Plant Physiol. 2006, 141, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Balk, J.; Chew, S.K.; Leaver, C.J.; McCabe, P.F. The intermembrane space of plant mitochondria contains a DNase activity that may be involved in programmed cell death. Plant J. 2003, 34, 573–583. [Google Scholar] [CrossRef] [PubMed]
- Scorrano, L. Opening the doors to cytochrome C: Changes in mitochondrial shape and apoptosis. Int. J. Biochem. Cell B 2009, 41, 1875–1883. [Google Scholar] [CrossRef] [PubMed]
- Van der Merwe, K.; Steyn, P.; Fourie, L.; Scott, D.B.; Theron, J. Ochratoxin A, a toxic metabolite produced by Aspergillus ochraceus Wilh. Nature 1965, 205, 1112–1113. [Google Scholar] [CrossRef] [PubMed]
- Ciegler, A.; Fennell, D.; Mintzlaff, H.-J.; Leistner, L. Ochratoxin synthesis by Penicillium species. Naturwissenschaften 1972, 59, 365–366. [Google Scholar] [CrossRef] [PubMed]
- Abarca, M.; Bragulat, M.; Castella, G.; Cabanes, F. Ochratoxin A production by strains of Aspergillus niger var niger. Appl. Environ. Microbiol. 1994, 60, 2650–2652. [Google Scholar] [PubMed]
- WHO. Safety evaluation of certain mycotoxins in food, fifty-sixth meeting of the joint FAO/WHO expert committee on food additives. WHO Food Additives Series 2001, 47, 706. [Google Scholar]
- Petrik, J.; Žanić-Grubišić, T.; Barišić, K.; Pepeljnjak, S.; Radić, B.; Ferenčić, Ž.; Čepelak, I. Apoptosis and oxidative stress induced by ochratoxin A in rat kidney. Arch. Toxicol. 2003, 77, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Boesch-Saadatmandi, C.; Lou, Y.; Wolffram, S.; Huebbe, P.; Rimbach, G. Ochratoxin A induces apoptosis in neuronal cells. Genes Nutr. 2009, 4, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.L.; Zhang, Y.; Xu, W.; Liang, R.; Zheng, J.; Luo, Y.; Wang, Y.; Huang, K. An iTRAQ-based mitoproteomics approach for profiling the nephrotoxicity mechanisms of ochratoxin A in HEK 293 cells. J. Proteom. 2013, 78, 398–415. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Xu, W.; Wang, Y.; Huang, K.; Liang, Z.; Zhao, W.; Luo, Y. Mycotoxin ochratoxin A-induced cell death and changes in oxidative metabolism of Arabidopsis thaliana. Plant Cell Rep. 2010, 29, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Peng, X.; Xu, W.; Luo, Y.; Zhao, W.; Hao, J.; Liang, Z.; Zhang, Y.; Huang, K. Transcript and protein profiling analysis of OTA-induced cell death reveals the regulation of the toxicity response process in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 2171–2187. [Google Scholar] [CrossRef] [PubMed]
- Asai, T.; Stone, J.M.; Heard, J.E.; Kovtun, Y.; Yorgey, P.; Sheen, J.; Ausubel, F.M. Fumonisin B1–induced cell death in arabidopsis protoplasts requires jasmonate-, ethylene-, and salicylate-dependent signaling pathways. Plant Cell 2000, 12, 1823–1835. [Google Scholar] [CrossRef] [PubMed]
- Stone, J.M.; Heard, J.E.; Asai, T.; Ausubel, F.M. Simulation of fungal-mediated cell death by fumonisin B1 and selection of fumonisin B1-resistant (fbr) Arabidopsis mutants. Plant Cell 2000, 12, 1811–1822. [Google Scholar] [CrossRef] [PubMed]
- Gechev, T.; Gadjev, I.; Hille, J. An extensive microarray analysis of AAL-toxin-induced cell death in Arabidopsis thaliana brings new insights into the complexity of programmed cell death in plants. Cell Mol. Life Sci. 2004, 61, 1185–1197. [Google Scholar] [PubMed]
- Poppenberger, B.; Berthiller, F.; Lucyshyn, D.; Sieberer, T.; Schuhmacher, R.; Krska, R.; Kuchler, K.; Glössl, J.; Luschnig, C.; Adam, G. Detoxification of the Fusarium mycotoxin deoxynivalenol by a UDP-glucosyltransferase from Arabidopsis thaliana. J. Biol. Chem. 2003, 278, 47905–47914. [Google Scholar] [CrossRef] [PubMed]
- Masuda, D.; Ishida, M.; Yamaguchi, K.; Yamaguchi, I.; Kimura, M.; Nishiuchi, T. Phytotoxic effects of trichothecenes on the growth and morphology of Arabidopsis thaliana. J. Exp. Bot. 2007, 58, 1617–1626. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, W.; Hao, J.; Xu, W.; Luo, Y.; Wu, W.; Liang, Z.; Yang, Z.; Huang, K. Changes in biosynthesis and metabolism of glutathione upon ochratoxin A stress in Arabidopsis thaliana. Plant Physiol. Biochem. 2014, 79, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hao, J.; Zhao, W.; Yang, Z.; Wu, W.; Zhang, Y.; Xu, W.; Luo, Y.; Huang, K. Comparative proteomics and physiological characterization of Arabidopsis thaliana seedlings in responses to Ochratoxin A. Plant Mol. Biol. 2013, 82, 321–337. [Google Scholar] [CrossRef] [PubMed]
- Fukao, Y.; Ferjani, A.; Tomioka, R.; Nagasaki, N.; Kurata, R.; Nishimori, Y.; Fujiwara, M.; Maeshima, M. iTRAQ Analysis Reveals Mechanisms of Growth Defects Due to Excess Zinc in Arabidopsis. Plant Physiol. 2011, 155, 1893–1907. [Google Scholar] [CrossRef] [PubMed]
- Lan, P.; Li, W.; Wen, T.N.; Shiau, J.Y.; Wu, Y.C.; Lin, W.; Schmidt, W. iTRAQ Protein Profile Analysis of Arabidopsis Roots Reveals New Aspects Critical for Iron Homeostasis. Plant Physiol. 2011, 155, 821–834. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Kobayashi, Y.; Devi, S.R.; Rikiishi, S.; Matsumoto, H. Aluminum toxicity is associated with mitochondrial dysfunction and the production of reactive oxygen species in plant cells. Plant Physiol. 2002, 128, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Rogiers, S.Y.; Knowles, N.R. Effects of storage temperature and atmosphere on saskatoon (Amelanchier alnifolia Nutt.) fruit quality, respiration and ethylene production. Postharvest Biol. Technol. 1998, 13, 183–190. [Google Scholar] [CrossRef]
- Millar, A.; Liddell, A.; Leaver, C. Isolation and subfractionation of mitochondria from plants. Methods Cell Biol. 2001, 65, 53–74. [Google Scholar] [PubMed]
- De Pinto, M.C.; Tommasi, F.; Gara, L.D. Changes in the antioxidant systems as part of the signaling pathway responsible for the programmed cell death activated by nitric oxide and reactive oxygen species in tobacco Bright-Yellow 2 cells. Plant physiol. 2002, 130, 698–708. [Google Scholar] [CrossRef] [PubMed]
- Hoeberichts, F.A.; Woltering, E.J. Multiple mediators of plant programmed cell death: Interplay of conserved cell death mechanisms and plant-specific regulators. Bioessays 2002, 25, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Gechev, T.S.; Van Breusegem, F.; Stone, J.M.; Denev, I.; Laloi, C. Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. Bioessays 2006, 28, 1091–1101. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, D.M.; Umbach, A.L.; Subbaiah, C.C.; Siedow, J.N. Mitochondrial reactive oxygen species. Contribution to oxidative stress and interorganellar signaling. Plant Physiol. 2006, 141, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Brandt, U. Energy converting NADH: Quinone oxidoreductase (complex I). Annu. Rev. Biochem. 2006, 75, 69–92. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009, 417, 1. [Google Scholar] [CrossRef] [PubMed]
- Muller, F.; Liu, Y.; Abdul-Ghani, M.; Lustgarten, M.; Bhattacharya, A.; Jang, Y.; Van Remmen, H. High rates of superoxide production in skeletal-muscle mitochondria respiring on both complex I-and complex II-linked substrates. Biochem. J. 2008, 409, 491–499. [Google Scholar] [CrossRef] [PubMed]
- White, R.J.; Reynolds, I.J. Mitochondrial depolarization in glutamate-stimulated neurons: an early signal specific to excitotoxin exposure. J. Neurosci. 1996, 16, 5688–5697. [Google Scholar] [PubMed]
- Muller, F.L.; Liu, Y.; Van Remmen, H. Complex III releases superoxide to both sides of the inner mitochondrial membrane. J. Biol. Chem. 2004, 279, 49064–49073. [Google Scholar] [CrossRef] [PubMed]
- Kuthanova, A.; Opatrny, Z.; Fischer, L. Is internucleosomal DNA fragmentation an indicator of programmed death in plant cells? J. Exp. Bot. 2008, 59, 2233–2240. [Google Scholar] [CrossRef] [PubMed]
- Bissenbaev, A.K.; Keniev, A.M.; Bersimbaev, R.I. Endogenous deoxyribonucleases involved in nuclear DNA degradation of wheat aleurone cells. J. Cell Mol. Biol. 2004, 3, 77–81. [Google Scholar]
- Xu, Y.; Hanson, M.R. Programmed cell death during pollination-induced petal senescence in petunia. Plant Physiol. 2000, 122, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Qu, G.Q.; Liu, X.; Zhang, Y.L.; Yao, D.; Ma, Q.M.; Yang, M.Y.; Zhu, W.H.; Yu, S.; Luo, Y. Evidence for programmed cell death and activation of specific caspase-like enzymes in the tomato fruit heat stress response. Planta 2009, 229, 1269–1279. [Google Scholar] [CrossRef] [PubMed]
- Halestrap, A.P.; Kerr, P.M.; Javadov, S.; Woodfield, K.-Y. Elucidating the molecular mechanism of the permeability transition pore and its role in reperfusion injury of the heart. BBA-Bioenergetics 1998, 1366, 79–94. [Google Scholar] [CrossRef]
- Halestrap, A.P.; Clarke, S.J.; Javadov, S.A. Mitochondrial permeability transition pore opening during myocardial reperfusion-a target for cardioprotection. Cardiovasc. Res. 2004, 61, 372–385. [Google Scholar] [CrossRef]
- Tomasello, F.; Messina, A.; Lartigue, L.; Schembri, L.; Medina, C.; Reina, S.; Thoraval, D.; Crouzet, M.; Ichas, F.; De Pinto, V.; et al. Outer membrane VDAC1 controls permeability transition of the inner mitochondrial membrane in cellulo during stress-induced apoptosis. Cell Res. 2009, 19, 1363–1376. [Google Scholar] [CrossRef] [PubMed]
- Rosano, C. Molecular model of hexokinase binding to the outer mitochondrial membrane porin (VDAC1): implication for the design of new cancer therapies. Mitochondrion 2011, 11, 513–519. [Google Scholar] [CrossRef] [PubMed]
- Ni, C.Z.; Wang, H.Q.; Xu, T.; Zhe, Q.; Liu, G.Q. AtKP1, a kinesin-like protein, mainly localizes to mitochondria in Arabidopsis thaliana. Cell Res. 2005, 15, 725–733. [Google Scholar] [CrossRef] [PubMed]
- Tonshin, A.; Saprunova, V.; Solodovnikova, I.; Bakeeva, L.; Yaguzhinsky, L. Functional activity and ultrastructure of mitochondria isolated from myocardial apoptotic tissue. Biochemistry (Moscow) 2003, 68, 875–881. [Google Scholar] [CrossRef] [PubMed]
- Duncan, O.; Taylor, N.L.; Carrie, C.; Eubel, H.; Kubiszewski-Jakubiak, S.; Zhang, B.; Narsai, R.; Millar, A.H.; Whelan, J. Multiple Lines of Evidence Localize Signaling, Morphology, and Lipid Biosynthesis Machinery to the Mitochondrial Outer Membrane of Arabidopsis. Plant Physiol. 2011, 157, 1093–1113. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhao, C.; Li, X.; Zhu, Y.; Gan, C.S.; Wang, Y.; Ravasi, T.; Qian, P.-Y.; Wong, S.C.; Sze, S.K. Study of monocyte membrane proteome perturbation during lipopolysaccharide-induced tolerance using iTRAQ-based quantitative proteomic approach. Proteomics 2010, 10, 2780–2789. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2008, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Banci, L.; Bertini, I.; Ciofi-Baffoni, S.; D’Alessandro, A.; Jaiswal, D.; Marzano, V.; Neri, S.; Ronci, M.; Urbani, A. Copper exposure effects on yeast mitochondrial proteome. J. Proteom. 2011, 74, 2522–2535. [Google Scholar] [CrossRef] [PubMed]



| Protein Name | UniProtKB Accession | Gene Names | Unused Protein Score | % of Coverage | No. of Unique Peptides | Mean ± SD (8 h) | Mean ± SD (24 h) |
|---|---|---|---|---|---|---|---|
| 23.6 kDa heat shock protein | Q96331 | HSP23.6 | 13.45 | 20.00 | 5 | - | 4.68 ± 0.43 |
| 15.7 kDa heat shock protein | Q9FHQ3 | HSP15.7 | 3.92 | 15.33 | 2 | - | 3.72 ± 0.29 |
| 12-oxophytodienoate reductase 3 | Q9FUP0 | OPR3 | 4.67 | 5.37 | 2 | - | 2.56 ± 0.10 |
| Catalase-3 | Q42547 | CAT3 | 17.43 | 22.56 | 9 | - | 2.38 ± 0.22 |
| Plasma membrane-associated cation-binding protein 1 | Q96262 | PCAP1 | 7.70 | 27.11 | 5 | 2.22 ± 0.26 | - |
| V-type proton ATPase subunit G1 | O82628 | VHA-G1 | 7.10 | 43.64 | 7 | 1.82 ± 0.41 | 0.56 ± 0.05 |
| Catalase-2 | P25819 | CAT2 | 8.99 | 10.98 | 5 | - | 1.98 ± 0.45 |
| Glycerate dehydrogenase | Q9C9W5 | HRP1 | 8.10 | 11.14 | 4 | - | 1.83 ± 0.32 |
| Oxygen-evolving enhancer protein 3-1 | Q9XFT3 | PSBQ1 | 10.88 | 20.98 | 5 | - | 1.81 ± 0.23 |
| Homocysteine methyltransferase | O50008 | CIMS | 5.14 | 5.62 | 3 | - | 1.79 ± 0.02 |
| Peroxisomal (S)-2-hydroxy-acid oxidase | Q9LRR9 | GLO1 | 10.35 | 19.62 | 6 | - | 1.71 ± 0.35 |
| NADH dehydrogenase [ubiquinone] flavoprotein 2 | O22769 | At4g02580 | 8.40 | 18.04 | 4 | - | 1.67 ± 0.15 |
| Adenylate kinase 1 | O82514 | ADK1 | 15.95 | 36.99 | 7 | 1.55 ± 0.14 | - |
| Patellin-1 | Q56WK6 | PATL1 | 7.97 | 4.89 | 3 | 1.50 ± 0.10 | - |
| Glutathione S-transferase F2 | P46422 | GSTF2 | 8.99 | 10.98 | 3 | - | 1.49 ± 0.22 |
| Heat shock protein 90-3 | P51818 | HSP90-3 | 8.10 | 11.14 | 4 | - | 1.47 ± 0.10 |
| Cytochrome b-c1 complex subunit 7-1 | Q9SUU5 | QCR7-1 | 6.44 | 40.16 | 5 | 1.42 ± 0.01 | - |
| Mitochondrial outer membrane protein porin 2 | Q9FJX3 | VDAC2 | 7.10 | 11.96 | 6 | 1.40 ± 0.05 | - |
| Probable NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5 | Q9FLX7 | At5g52840 | 13.58 | 37.87 | 7 | 1.39 ± 0.04 | - |
| Malate dehydrogenase | Q9ZP05 | At5g09660 | 7.84 | 14.41 | 4 | 1.38 ± 0.17 | - |
| Mitochondrial outer membrane protein porin 1 | Q9SRH5 | VDAC1 | 7.97 | 28.62 | 6 | 1.36 ± 0.05 | - |
| NADH dehydrogenase [ubiquinone] iron-sulfur protein 1 | Q9FGI6 | EMB1467 | 10.13 | 7.89 | 6 | - | 1.36 ± 0.15 |
| Probable arginase | Q9ZPF5 | At4g08870 | 20.60 | 43.90 | 12 | 1.35 ± 0.08 | 1.35 ± 0.08 |
| ATP synthase subunit d | Q9FT52 | At3g52300 | 19.84 | 54.17 | 9 | 1.35 ± 0.05 | 1.35 ± 0.05 |
| V-type proton ATPase subunit B3 | Q8W4E2 | VHA-B3 | 21.24 | 23.82 | 10 | 1.35 ± 0.06 | 0.67 ± 0.01 |
| Glyceraldehyde-3-phosphate dehydrogenase | P25858 | GAPC | 10.50 | 21.60 | 6 | 1.34 ± 0.05 | 1.39 ± 0.01 |
| Mitochondrial outer membrane protein porin 3 | Q9SMX3 | VDAC3 | 16.93 | 35.77 | 13 | 1.34 ± 0.07 | - |
| Formate dehydrogenase | Q9S7E4 | FDH1 | 14.52 | 23.18 | 9 | - | 1.33 ± 0.04 |
| Mitochondrial-processing peptidase subunit beta | Q42290 | At3g02090 | 34.46 | 41.81 | 22 | 1.32 ± 0.00 | - |
| Dihydrolipoyl dehydrogenase 1 | Q9M5K3 | LPD1 | 4.61 | 5.72 | 2 | - | 1.31 ± 0.08 |
| Probable ATP synthase 24 kDa subunit | Q9SJ12 | At2g21870 | 19.94 | 27.08 | 8 | - | 1.31 ± 0.15 |
| Succinate dehydrogenase [ubiquinone] flavoprotein subunit 1 | O82663 | SDH1-1 | 8.86 | 8.52 | 4 | - | 1.31 ± 0.07 |
| 60S ribosomal protein L13-1 | P41127 | RPL13B | 5.68 | 19.90 | 3 | 1.31 ± 0.07 | - |
| CBS domain-containing protein | Q9LEV3 | CBSX3 | 5.47 | 18.45 | 3 | 1.30 ± 0.02 | 1.45 ± 0.06 |
| Ribulose bisphosphate carboxylase large chain | O03042 | rbcL | 32.95 | 31.94 | 19 | 0.76 ± 0.02 | - |
| 60S ribosomal protein L17-2 | P51413 | RPL17B | 5.32 | 16.57 | 2 | 0.75 ± 0.06 | - |
| UPF0603 protein | Q9ZVL6 | At1g54780 | 18.42 | 40.70 | 8 | 0.75 ± 0.07 | - |
| Chlorophyll a-b binding protein CP26 | Q9XF89 | LHCB5 | 16.85 | 27.14 | 13 | 0.73 ± 0.04 | - |
| Apocytochrome f | P56771 | petA | 40.06 | 53.44 | 23 | 0.73 ± 0.02 | - |
| Calcium sensing receptor | Q9FN48 | CAS | 6.61 | 12.14 | 4 | - | 0.73 ± 0.03 |
| Photosystem I reaction center subunit III | Q9SHE8 | PSAF | 13.04 | 23.08 | 7 | - | 0.73 ± 0.02 |
| 50S ribosomal protein L16 | P56793 | rpl16 | 4.15 | 20.74 | 2 | - | 0.70 ± 0.00 |
| V-type proton ATPase catalytic subunit A | O23654 | VHA-A | 22.44 | 17.50 | 11 | - | 0.70 ± 0.02 |
| Glycine dehydrogenase [decarboxylating] 2 | Q94B78 | At4g33010 | 14.38 | 9.35 | 7 | 0.70 ± 0.06 | - |
| Geranylgeranyl diphosphate reductase1 | Q9CA67 | CHLP | 12.10 | 12.42 | 6 | 0.70 ± 0.04 | - |
| Chlorophyll a-b binding protein CP29.1 | Q07473 | LHCB4.1 | 23.47 | 28.62 | 16 | 0.70 ± 0.01 | - |
| Cytochrome b6-f complex iron-sulfur subunit | Q9ZR03 | petC | 10.58 | 23.58 | 7 | 0.69 ± 0.01 | - |
| Photosystem II CP47 chlorophyll apoprotein | P56777 | psbB | 30.58 | 22.64 | 25 | 0.69 ± 0.03 | 0.54 ± 0.01 |
| Clathrin heavy chain 1 | Q0WNJ6 | CHC1 | 2.96 | 0.82 | 1 | 0.69 ± 0.03 | - |
| Protein TIC110 | Q8LPR9 | TIC110 | 4.49 | 1.77 | 2 | - | 0.66 ± 0.06 |
| 40S ribosomal protein S9-1 | Q9LXG1 | RPS9B | 18.15 | 34.85 | 8 | - | 0.66 ± 0.02 |
| Lipoxygenase 2 | P38418 | LOX2 | 11.99 | 16.29 | 12 | 0.66 ± 0.03 | - |
| ATP synthase subunit b | P56759 | atpF | 19.29 | 44.57 | 14 | 0.65 ± 0.04 | - |
| ADP,ATP carrier protein 1 | P31167 | AAC1 | 14.85 | 19.95 | 7 | 0.64 ± 0.05 | - |
| Photosystem II 22 kDa protein | Q9XF91 | PSBS | 3.60 | 6.04 | 2 | 0.62 ± 0.04 | - |
| L-ascorbate peroxidase T | Q42593 | APXT | 2.60 | 9.16 | 2 | 0.62 ± 0.02 | - |
| Photosystem Q(B) protein | P83755 | psbA | 8.81 | 17.28 | 13 | 0.62 ± 0.04 | - |
| 60S ribosomal protein L18a-2 | P51418 | RPL18AB | 3.25 | 5.06 | 2 | - | 0.62 ± 0.04 |
| 60S ribosomal protein L21-2 | Q9FDZ9 | RPL21E | 6.81 | 11.59 | 3 | - | 0.62 ± 0.07 |
| V-type proton ATPase subunit E1 | Q39258 | VHA-E1 | 4.40 | 11.74 | 3 | - | 0.61 ± 0.01 |
| Photosystem II CP43 chlorophyll apoprotein | P56778 | psbC | 23.73 | 28.33 | 14 | 0.61 ± 0.03 | 0.60 ± 0.03 |
| Thylakoid lumenal 16.5 kDa protein | O22773 | At4g02530 | 6.90 | 18.06 | 4 | 0.60 ± 0.09 | - |
| Photosystem I P700 chlorophyll a apoprotein A2 | P56767 | psaB | 13.90 | 10.49 | 7 | - | 0.59 ± 0.01 |
| Photosystem II D2 protein | P56761 | psbD | 16.40 | 22.38 | 15 | 0.56 ± 0.02 | 0.54 ± 0.04 |
| Photosystem I P700 chlorophyll a apoprotein A1 | P56766 | psaA | 10.60 | 6.00 | 5 | 0.55 ± 0.05 | 0.58 ± 0.05 |
| ATP synthase subunit alpha | P56757 | atpA | 43.10 | 37.87 | 25 | - | 0.52 ± 0.05 |
| Oxygen-evolving enhancer protein 1-1 | P23321 | PSBO1 | 40.9 | 58.73 | 25 | 0.49 ± 0.04 | 0.72 ± 0.01 |
| ATP synthase gamma chain 1 | Q01908 | ATPC1 | 22.92 | 26.54 | 10 | - | 0.46 ± 0.03 |
| ATP synthase epsilon chain | P09468 | atpE | 10.29 | 25.00 | 5 | - | 0.41 ± 0.02 |
| ATP synthase subunit beta | P19366 | atpB | 59.44 | 64.26 | 39 | - | 0.41 ± 0.02 |
| Aquaporin PIP2-1 | P43286 | PIP2-1 | 8.23 | 10.45 | 4 | - | 0.38 ± 0.07 |
| Protein Name | UniProtKB Accession | Mean ± SD (8 h) | Mean ± SD (24 h) | Protein Name | UniProtKB Accession | Mean ± SD (8 h) | Mean ± SD (24 h) |
|---|---|---|---|---|---|---|---|
| Group 1 Photosynthesis | Group 3 ATP synthase | ||||||
| Calcium sensing receptor | Q9FN48 | 0.73 ± 0.03 | V-type proton ATPase catalytic subunit A | O23654 | 0.67 ± 0.02 | ||
| Photosystem II CP47 chlorophyll apoprotein | P56777 | 0.69 ± 0.03 | 0.54 ± 0.01 | ATP synthase subunit alpha | P56757 | 0.52 ± 0.05 | |
| Apocytochrome f | P56771 | 0.73 ± 0.02 | ATP synthase epsilon chain | P09468 | 0.41 ± 0.02 | ||
| Cytochrome b6-f complex iron-sulfur subunit | Q9ZR03 | 0.69 ± 0.01 | ATP synthase gamma chain 1 | Q01908 | 0.46 ± 0.03 | ||
| L-ascorbate peroxidase T | Q42593 | 0.62 ± 0.02 | ATP synthase subunit b | P56759 | 0.65 ± 0.04 | ||
| Photosystem I P700 chlorophyll apoprotein A1 | P56766 | 0.55 ± 0.05 | ATP synthase subunit beta | P19366 | 0.41 ± 0.02 | ||
| Photosystem I P700 chlorophyll apoprotein A2 | P56767 | 0.59 ± 0.01 | V-type proton ATPase subunit B3 | Q8W4E2 | 1.35 ± 0.06 | ||
| Photosystem II CP43 chlorophyll apoprotein | P56778 | 0.60 ± 0.03 | V-type proton ATPase subunit E1 | Q39258 | 0.61 ± 0.01 | ||
| Chlorophyll a-b binding protein CP29.1 | Q07473 | 0.69 ± 0.01 | ATP synthase subunit d | Q9FT52 | 1.35 ± 0.05 | ||
| Chlorophyll a-b binding protein CP26 | Q9XF89 | 0.73 ± 0.04 | V-type proton ATPase subunit G1 | O82628 | 1.82 ± 0.41 | 0.56 ± 0.05 | |
| Geranylgeranyl diphosphate reductase1 | Q9CA67 | 0.70 ± 0.04 | Group 4 Translation, ribosome biogenesis | ||||
| Photosystem Q(B) protein | P83755 | 0.62 ± 0.04 | 60S ribosomal protein L13-1 | P41127 | 1.31 ± 0.07 | ||
| UPF0603 protein | Q9ZVL6 | 0.75 ± 0.07 | 60S ribosomal protein L17-2 | P51413 | 0.75 ± 0.06 | ||
| Thylakoid lumenal 16.5 kDa protein | O22773 | 0.60 ± 0.09 | 50S ribosomal protein L16 | P56793 | 0.70 ± 0.00 | ||
| Photosystem II 22 kDa protein | Q9XF91 | 0.62 ± 0.04 | 40S ribosomal protein S9-1 | Q9LXG1 | 0.66 ± 0.02 | ||
| Photosystem II D2 protein | P56761 | 0.56 ± 0.02 | 0.54 ± 0.04 | 60S ribosomal protein L21-2 | Q9FDZ9 | 0.62 ± 0.07 | |
| Oxygen-evolving enhancer protein 3-1 | Q9XFT3 | 1.81 ± 0.23 | 60S ribosomal protein L18a-2 | P51418 | 0.62 ± 0.04 | ||
| Oxygen-evolving enhancer protein 1-1 | P23321 | 0.49 ± 0.04 | 0.72 ± 0.01 | Group 5 Mitochondrial transport protein | |||
| Photosystem I reaction center subunit III | Q9SHE8 | 0.73 ± 0.02 | Probable ATP synthase 24 kDa subunit | Q9SJ12 | 1.31 ± 0.15 | ||
| Lipoxygenase 2 | P38418 | 0.66 ± 0.03 | Mitochondrial-processing peptidase subunit beta | Q42290 | 1.32 ± 0.00 | ||
| Group 2 Mitochondrial electron transport | Mitochondrial outer membrane protein porin 3 | Q9SMX3 | 1.34 ± 0.07 | ||||
| NADH dehydrogenase | Q9FGI6 | 1.36 ± 0.15 | Mitochondrial outer membrane protein porin 1 | Q9SRH5 | 1.36 ± 0.05 | ||
| Cytochrome b-c1 complex subunit 7-1 | Q9SUU5 | 1.42 ± 0.01 | Mitochondrial outer membrane protein porin 2 | Q9FJX3 | 1.40 ± 0.05 | ||
| Probable NADH dehydrogenase | Q9FLX7 | 1.39 ± 0.04 | ADP,ATP carrier protein 1 | P31167 | 0.64 ± 0.05 | ||
| Succinate dehydrogenase flavoprotein subunit 1 | O82663 | 1.31 ± 0.07 | |||||
| NADH dehydrogenase flavoprotein 2 | O22769 | 1.67 ± 0.15 | |||||
| Gene Category | ID | Gene Description | Fold Change (log2) |
|---|---|---|---|
| DNA | AT3G18500 | Endonuclease/exonuclease/phosphatase family protein | 4.0 (2.0) |
| Endonuclease | AT3G10010 | Demeter-like 2 (DML2) | 3.7 (1.9) |
| AT5G17540 | Bifunctional nuclease I (BFN1) | 90.5 (6.5) | |
| AT1G59720 | Chlororespiratory reduction 28 (CRR28) | 59.7 (5.9) | |
| AT1G53250 | Endonuclease | 59.7 (5.9) | |
| AT2G21800 | Essential meiotic endonuclease 1A (EME1A) | 59.7 (5.9) | |
| AT1G73875 | Endonuclease/exonuclease/phosphatase family protein | 2.5 (1.4) | |
| AT3G21530 | Endonuclease/exonuclease/phosphatase family protein | 2.4 (1.3) | |
| AT1G68290 | Endonuclease 2 (ENDO2) | 2.3 (1.2) | |
| DNA replicate | AT2G07690 | Minichromosome maintenance family protein (MCM5) | −2.8 (−1.5) |
| AT1G67630 | DNA polymerase alpha 2 (POLA2) | −3.0 (−1.6) | |
| AT1G67320 | DNA primase | −4.0 (−2.0) | |
| AT4G02060 | Minichromosome maintenance family protein (MCM7) | −4.6 (−2.2) | |
| AT1G10590 | DNA-binding protein-related | 2.2 (1.1) | |
| DNA repair | AT1G10590 | DNA-binding protein-related | 2.2 (1.1) |
| AT4G17020 | DNA repair related | 2.1 (1.1) | |
| RNA | AT5G60040 | DNA-directed RNA polymerase (NPRC1) | 10.3 (3.4) |
| polymerase | AT1G54250 | DNA-directed RNA polymerase II, core complex (NPR8A) | 6.1 (2.6) |
| AT3G52090 | DNA-directed RNA polymerase II, core complex (NP11) | 3.4 (1.8) | |
| AT5G60040 | DNA-directed RNA polymerase (NPRC1) | 10.3 (3.4) | |
| RNA | AT2G36530 | Copper ion binding/ phosphopyruvate hydratase (LOS2) | 2.9 (1.6) |
| degradation | AT1G17980 | Nucleotidyltransferase family protein | 2.5 (1.3) |
| Gene Category | ID | Gene Description | Fold Change (log2) |
|---|---|---|---|
| TCA | AT1G24180 | Pyruvate dehydrogenase E1a-like subunit (IAR4) | 5.9 (2.6) |
| AT1G01090 | Pyruvate dehydrogenase E1 alpha | 2.4 (1.2) | |
| AT2G44350 | ATP citrate synthase (ATCS) | 3.8 (1.9) | |
| AT3G58750 | Citrate synthase 2 (CSY2) | 2.0 (1.0) | |
| AT2G17130 | Isocitrate dehydrogenase subunit 2 (IDH2) | 9.1 (3.2) | |
| AT3G09810 | Isocitrate dehydrogenase, putative | 5.2 (2.4) | |
| AT5G03290 | Isocitrate dehydrogenase, putative | 4.7 (2.4) | |
| AT4G26910 | 2-oxoacid dehydrogenase family protein | 3.8 (1.9) | |
| AT2G05710 | Aconitate hydratase (Aconitase) | 4.0 (2.0) | |
| AT5G23250 | Succinyl-CoA ligase, putative | 2.2 (1.3) | |
| AT5G40650, | Succinate dehydrogenase (SDH2-2) | 2.5 (1.3) | |
| AT2G47510 | Fumarase 1 (FUM1) | 3.4 (1.8) | |
| AT5G09660 | NAD-malate dehydrogenase 2 (PMDH2) | −2.3 (−1.2) | |
| Oxidative phosphorylation | |||
| Complex I | AT3G12260 | Complex 1 family protein | 7.6 (2.9) |
| AT5 G47890 | NADH-ubiquinone oxidoreductase B8 subunit, putative | 9.7 (3.3) | |
| AT1G79010 | NADH-ubiquinone oxidoreductase 23 kDa subunit | 2.4 (1.3) | |
| AT4G05020 | NAD(P)H dehydrogenase B2 (NDB2) | 3.6 (1.9) | |
| AT3G06310 | NADH-ubiquinone oxidoreductase 19 kDa subunit (NDUFA8) | −2.2 (−1.2) | |
| Complex II | AT5G40650 | Succinate dehydrogenase (SDH2-2) | 2.5 (1.3) |
| Complex III | AT5G25450 | Ubiquinol-cytochrome C reductase complex 14 kDa protein | 4.7 (2.2) |
| Complex IV | AT3G15352 | Cytochrome c oxidase, Copper chaperone (COX17) | 3.7 (1.9) |
| AT2G44520 | Cytochrome c oxidase 10 (COX10) | 3.0 (1.6) | |
| AT1G22450 | Cytochrome c oxidase 6B (COX6B) | 2.1 (1.1) | |
| ATPase | AT2G33040 | ATP synthase gamma chain | 2.6 (1.1) |
| (Complex V) | AT4G30190 | Hydrogen-exporting ATPase | 2.2 (1.2) |
| AT4G02620 | ATPase subunit F family protein | 5.9 (2.6) | |
| AT4G11150 | ATP synthase subunit E1 (TUF) | 4.8 (2.3) | |
| AT3G28715 | H+-transporting two-sector ATPase, putative | 3.7 (1.9) | |
| AT2G18960 | H+-ATPase 1 (AHA1) | 2.8 (1.5) | |
| AT2G21410 | Vacuolar proton ATPase A2 (VHA-A2) | 2.8 (1.5) | |
| AT2G28520 | Vacuolar proton ATPase A1 (VHA-A1) | 2.7 (1.4) | |
| AT5G62670 | H+-ATPase 11 (AHA11) | −3.0 (−1.6) | |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Peng, X.; Yang, Z.; Zhao, W.; Xu, W.; Hao, J.; Wu, W.; Shen, X.L.; Luo, Y.; Huang, K. iTRAQ Mitoproteome Analysis Reveals Mechanisms of Programmed Cell Death in Arabidopsis thaliana Induced by Ochratoxin A. Toxins 2017, 9, 167. https://doi.org/10.3390/toxins9050167
Wang Y, Peng X, Yang Z, Zhao W, Xu W, Hao J, Wu W, Shen XL, Luo Y, Huang K. iTRAQ Mitoproteome Analysis Reveals Mechanisms of Programmed Cell Death in Arabidopsis thaliana Induced by Ochratoxin A. Toxins. 2017; 9(5):167. https://doi.org/10.3390/toxins9050167
Chicago/Turabian StyleWang, Yan, Xiaoli Peng, Zhuojun Yang, Weiwei Zhao, Wentao Xu, Junran Hao, Weihong Wu, Xiao Li Shen, Yunbo Luo, and Kunlun Huang. 2017. "iTRAQ Mitoproteome Analysis Reveals Mechanisms of Programmed Cell Death in Arabidopsis thaliana Induced by Ochratoxin A" Toxins 9, no. 5: 167. https://doi.org/10.3390/toxins9050167
APA StyleWang, Y., Peng, X., Yang, Z., Zhao, W., Xu, W., Hao, J., Wu, W., Shen, X. L., Luo, Y., & Huang, K. (2017). iTRAQ Mitoproteome Analysis Reveals Mechanisms of Programmed Cell Death in Arabidopsis thaliana Induced by Ochratoxin A. Toxins, 9(5), 167. https://doi.org/10.3390/toxins9050167