Biodegradation Mechanisms of Patulin in Candida guilliermondii: An iTRAQ-Based Proteomic Analysis
Abstract
:1. Introduction
2. Results
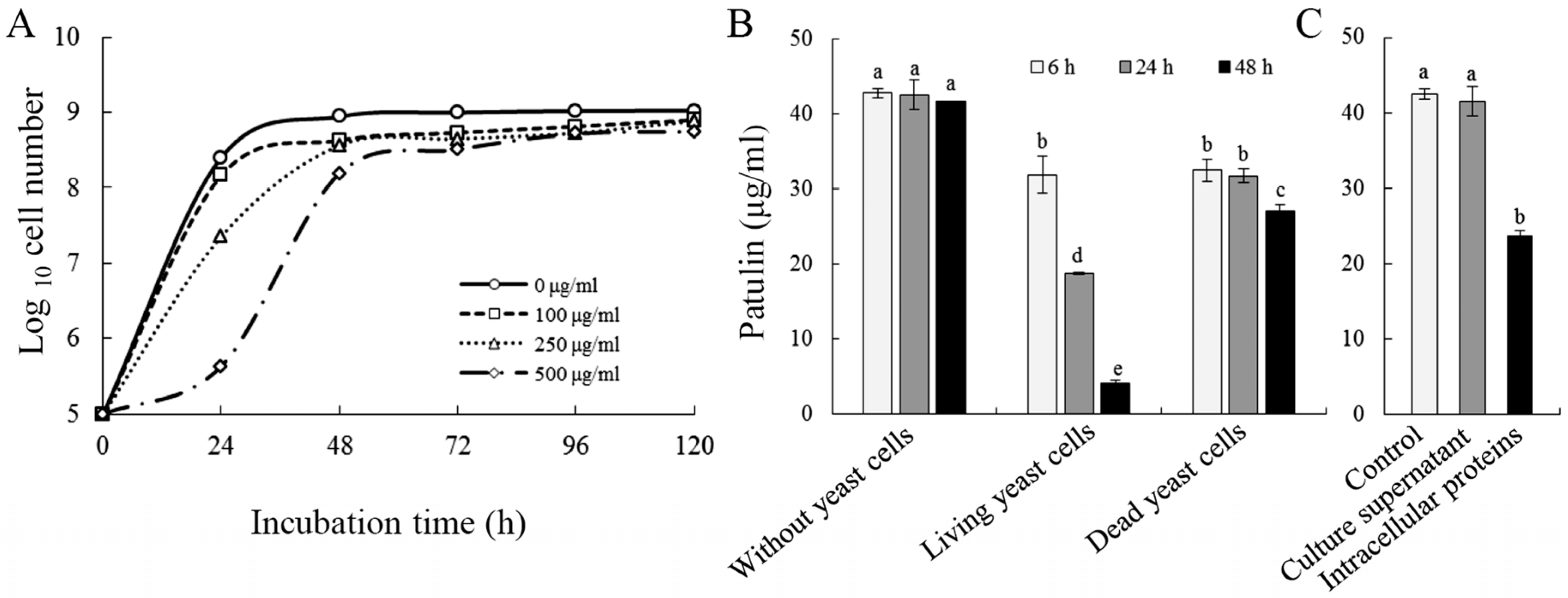
2.1. Biodegradation of Patulin by C. guilliermondii
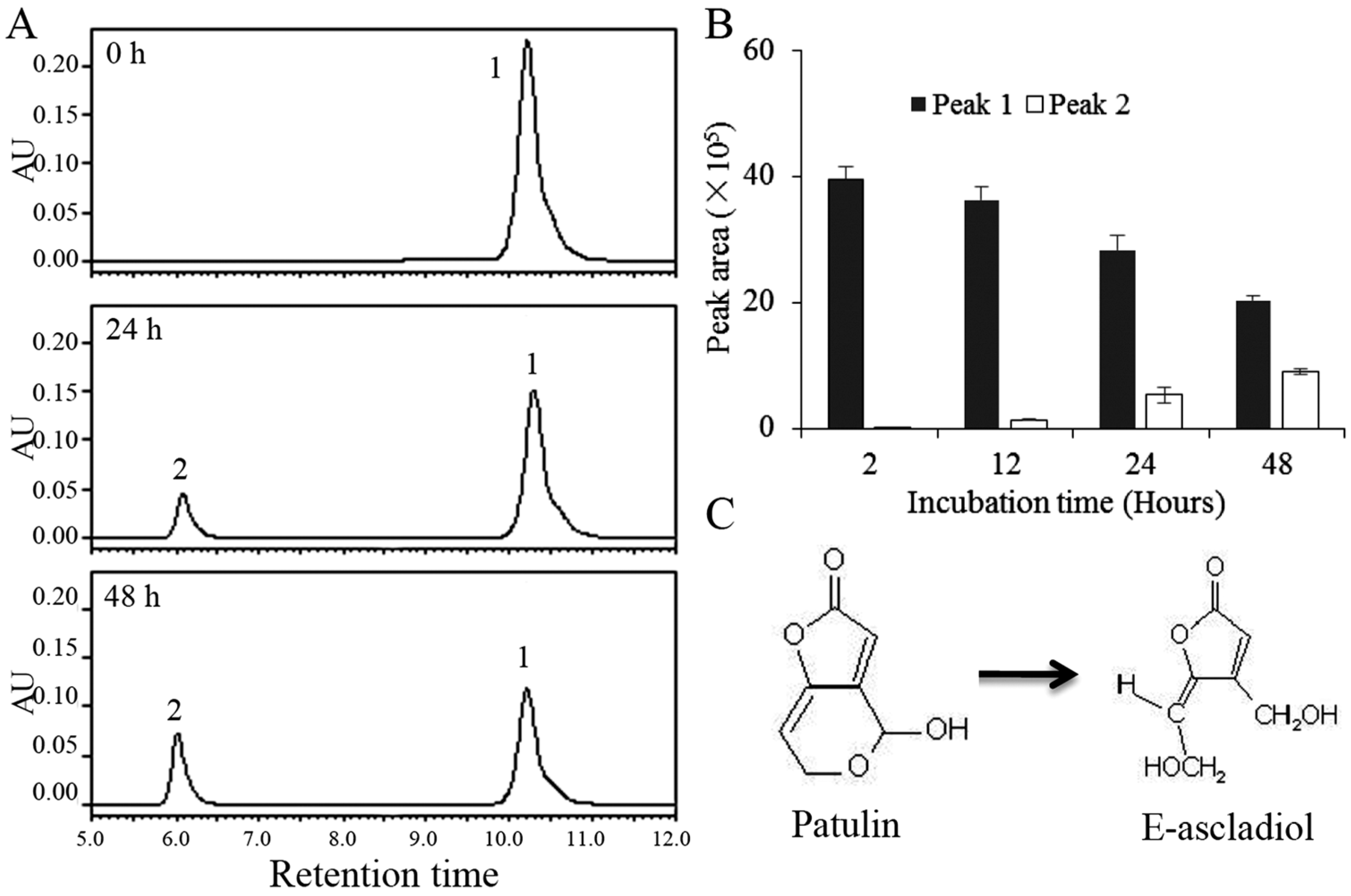
2.2. Biodegradation Products of Patulin by C. guilliermondii
2.3. Proteomic Responses of C. guilliermondii to Patulin
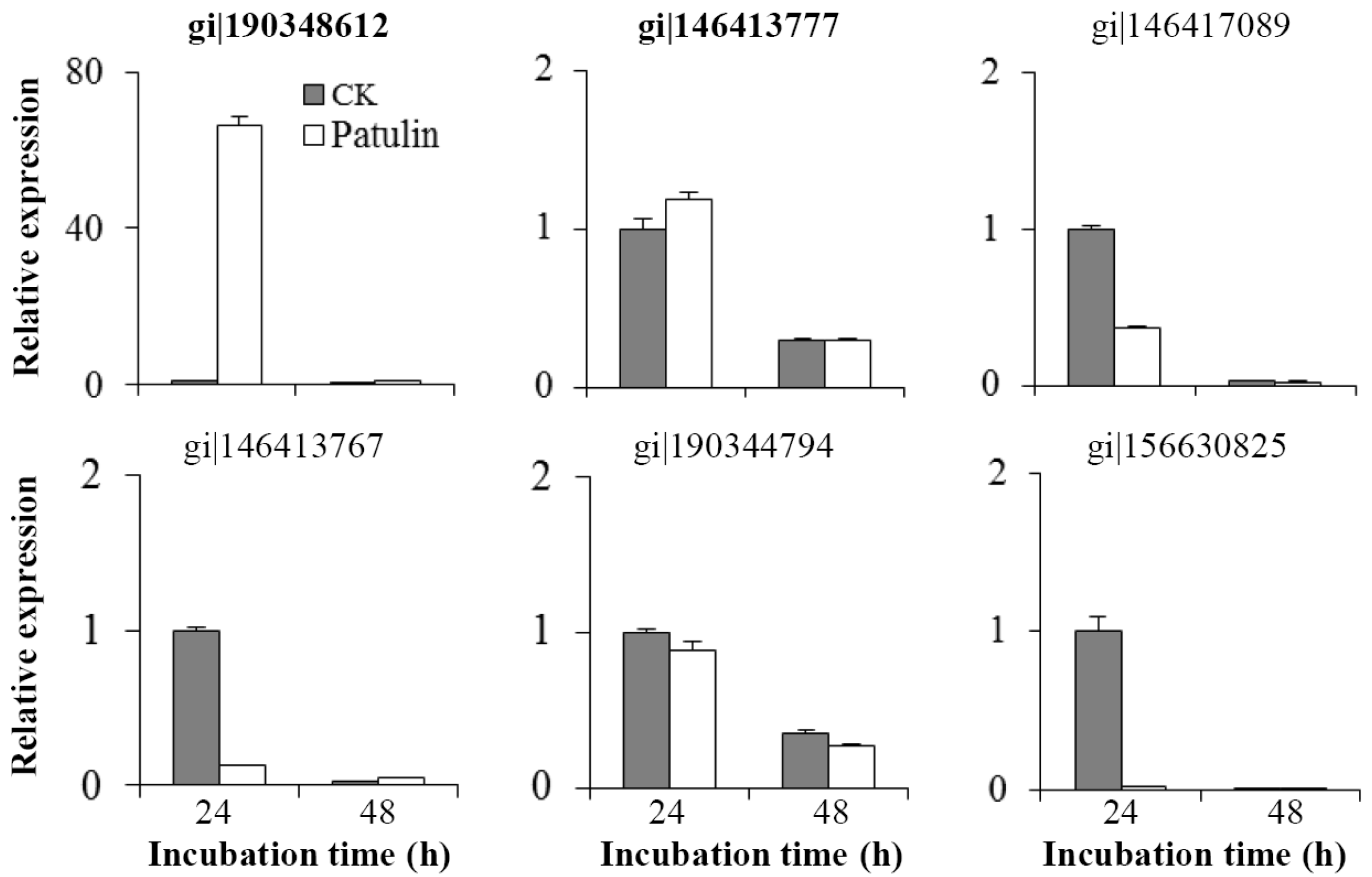
2.4. Changes in mRNA Abundances of Related Genes
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Yeast Strain
5.2. Effect of Patulin on the Growth of C. guilliermondii
5.3. Biodegradation Assay of Patulin
5.4. HPLC Analysis for Patulin and Biodegradation Products
5.5. LC-TOF/MS and LC-TOF-MS/MS Analysis for Biodegradation Products
5.6. Protein Extraction
5.7. Protein Digestion and iTRAQ Labeling
5.8. NanoLC-MS/MS and Bioinformatic Analysis
5.9. RNA Isolation and Reverse Transcription-Quantitative PCR (RT-qPCR)
5.10. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ianiri, G.; Idnurm, A.; Wright, S.A.; Durán-Patrón, R.; Mannina, L.; Ferracane, R.; Ritieni, A.; Castoria, R. Searching for genes responsible for patulin degradation in a biocontrol yeast provides insight into the basis for resistance to this mycotoxin. Appl. Environ. Microbiol. 2013, 79, 3101–3115. [Google Scholar] [CrossRef] [PubMed]
- Bosso, L.; Scelza, R.; Varlese, R.; Meca, G.; Testa, A.; Rao, M.R.; Cristinzio, G. Assessing the effectiveness of Byssochlamys. nivea and Scopulariopsis. brumptii in pentachlorophenol removal and biological control of two Phytophthora. species. Fungal Biol. 2016, 120, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Puel, O.; Galtier, P.; Oswald, I.P. Biosynthesis and toxicological effects of patulin. Toxins 2010, 2, 613–631. [Google Scholar] [CrossRef] [PubMed]
- Andersen, B.; Smedsgaard, J.; Frisvad, J.C. Penicillium. expansum: Consistent production of patulin, chaetoglobosins, and other secondary metabolites in culture and their natural occurrence in fruit products. J. Agric. Food Chem. 2004, 52, 2421–2428. [Google Scholar] [CrossRef] [PubMed]
- Tannous, J.; El Khoury, R.; Snini, S.P.; Lippi, Y.; El Khoury, A.; Atoui, A.; Lteifc, R.; Oswald, I.P.; Puel, O. Sequencing, physical organization and kinetic expression of the patulin biosynthetic gene cluster from Penicillium. expansum. Int. J. Food Microbiol. 2014, 189, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Ballester, A.R.; Marcet-Houben, M.; Levin, E.; Sela, N.; Selma-Lazaro, C.; Carmona, L.; Wisniewski, M.; Droby, S.; González-Candelas, L.; Gabaldón, T. Genome, transcriptome, and functional analyses of Penicillium. expansum provide new insights into secondary metabolism and pathogenicity. Mol. Plant-Microbe Interact. 2015, 28, 232–248. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zong, Y.; Du, Z.; Chen, Y.; Zhang, Z.; Qin, G.; Zhao, W.; Tian, S. Genomic characterization reveals insights into patulin biosynthesis and pathogenicity in Penicillium. species. Mol. Plant-Microbe Interact. 2015, 28, 635–647. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.W.; Klich, M. Mycotoxins. Clin. Microbiol. Rev. 2003, 16, 497–516. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, E.; Gross, K.; Metzler, M. Aneuploidogenic and clastogenic potential of the mycotoxins citrinin and patulin. Carcinogenesis 1998, 19, 1313–1318. [Google Scholar] [CrossRef] [PubMed]
- Commission Regulation. European Commission Regulation. EC 1881/2006, setting maximum levels for certain contaminants in foodstuffs. Available online: http://ec.europa.eu/dgs/health_food-safety/index_en.htm (accessed on 24 January 2017).
- Kabak, B.; Dobson, A.D.W.; Var, I. Strategies to prevent mycotoxin contamination of food and animal feed: A review. Crit. Rev. Food Sci. Nutr. 2006, 46, 593–619. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Jiang, W.; Li, C.; Ma, N.; Xu, Y.; Meng, X. Patulin biodegradation by marine yeast Kodameae. ohmeri. Food Addit. Contam. Part A 2015, 32, 352–360. [Google Scholar]
- Bata, A.; Lásztity, R. Detoxification of mycotoxin-contaminated food and feed by microorganisms. Trends Food Sci. Technol. 1999, 10, 223–228. [Google Scholar] [CrossRef]
- Shetty, P.H.; Jespersen, L. Saccharomyces cerevisiae and lactic acid bacteria as potential mycotoxin decontaminating agents. Trends Food Sci. Technol. 2006, 17, 48–55. [Google Scholar] [CrossRef]
- Moake, M.M.; Padilla-Zakour, O.I.; Worobo, R.W. Comprehensive review of patulin control methods in foods. Compr. Rev. Food Sci. Food Saf. 2005, 4, 8–21. [Google Scholar] [CrossRef]
- Ricelli, A.; Baruzzi, F.; Solfrizzo, M.; Morea, M.; Fanizzi, F.P. Biotransformation of patulin by Gluconobacter oxydans. Appl. Environ. Microbiol. 2007, 73, 785–792. [Google Scholar] [CrossRef] [PubMed]
- Castoria, R.; Morena, V.; Caputo, L.; Panfili, G.; De Curtis, F.; De Cicco, V. Effect of the biocontrol yeast Rhodotorula. glutinis strain LS11 on patulin accumulation in stored apples. Phytopathology 2005, 95, 1271–1278. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Zhang, H.; Yang, Q.; Ren, R. Efficacy of Pichia. caribbica in controlling blue mold rot and patulin degradation in apples. Int. J. Food Microbiol. 2013, 162, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Castoria, R.; Mannina, L.; Durán-Patrón, R.; Maffei, F.; Sobolev, A.P.; De Felice, D.V.; Pinedo-Rivilla, C.; Ritieni, A.; Ferracane, R.; Wright, S.A.I. Conversion of the mycotoxin patulin to the less toxic desoxypatulinic acid by the biocontrol yeast Rhodosporidium. kratochvilovae strain LS11. J. Agric. Food Chem. 2011, 59, 11571–11578. [Google Scholar] [CrossRef] [PubMed]
- Papon, N.; Savini, V.; Lanoue, A.; Simkin, A.J.; Crèche, J.; Giglioli-Guivarc’h, N.; Clastre, M.; Courdavault, V.; Sibirny, A.A. Candida guilliermondii: Biotechnological applications, perspectives for biological control, emerging clinical importance and recent advances in genetics. Curr. Genet. 2013, 59, 73–90. [Google Scholar] [CrossRef] [PubMed]
- Mcguire, R.G. Application of Candida guilliermondii in commercial citrus coatings for biocontrol of Penicillium. digitatum on grapefruits. Biol. Control. 1994, 4, 1–7. [Google Scholar] [CrossRef]
- Fan, Q.; Tian, S.P.; Xu, Y.; Wang, Y.; Jiang, A.L. Biological control of Rhizopus. rot of peach fruits by Candida guilliermondii. Acta Bot. Sin. 2000, 42, 1033–1038. [Google Scholar]
- Saligkariasa, I.D.; Gravanis, F.T.; Epton, H.A.S. Biological control of Botrytis cinerea on tomato plants by the use of epiphytic yeasts Candida guilliermondii strains 101 and US 7 and Candida oleophila strain I-182: II. A study on mode of action. Biol. Control. 2002, 25, 151–161. [Google Scholar] [CrossRef]
- Xu, X.; Qin, G.; Tian, S. Effect of microbial biocontrol agents on alleviating oxidative damage of peach fruit subjected to fungal pathogen. Int. J. Food Microbiol. 2008, 126, 153–158. [Google Scholar] [CrossRef] [PubMed]
- McCormick, S. Microbial detoxification of mycotoxins. J. Chem. Ecol. 2013, 39, 907–918. [Google Scholar] [CrossRef] [PubMed]
- Mahunu, G.K.; Zhang, H.; Yang, Q.; Li, C.; Zheng, X. Biological control of patulin by antagonistic yeast: A case study and possible model. Crit. Rev. Microbiol. 2015, 42, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Zhu, R.; Feussner, K.; Wu, T.; Yan, F.; Karlovsky, P.; Zheng, X. Detoxification of mycotoxin patulin by the yeast Rhodosporidium. paludigenum. Food Chem. 2015, 179, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Moss, M.O.; Long, M.T. Fate of patulin in the presence of the yeast Saccharomyces cerevisiae. Food Addit. Contam. 2002, 19, 387–399. [Google Scholar] [CrossRef] [PubMed]
- Ianiri, G.; Idnurm, A.; Castoria, R. Transcriptomic responses of the basidiomycete yeast Sporobolomyces. sp. to the mycotoxin patulin. BMC Genom. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Sumbu, Z.L.; Thonart, P.; Bechet, J. Action of patulin on a yeast. Appl. Environ. Microbiol. 1983, 45, 110–115. [Google Scholar] [PubMed]
- Papp, G.; Horváth, E.; Mike, N.; Gazdag, Z.; Belágyi, J.; Gyöngyi, Z.; Bánfalvi, G.; Hornok, L.; Pesti, M. Regulation of patulin-induced oxidative stress processes in the fission yeast Schizosaccharomyces. pombe. Food Chem. Toxicol. 2012, 50, 3792–3798. [Google Scholar] [CrossRef] [PubMed]
- Iwahashi, Y.; Hosoda, H.; Park, J.H.; Lee, J.H.; Suzuki, Y.; Kitagawa, E.; Murata, S.M.; Jwa, N.S.; Gu, M.B.; Iwahashi, H. Mechanisms of patulin toxicity under conditions that inhibit yeast growth. J. Agric. Food Chem. 2006, 54, 1936–1942. [Google Scholar] [CrossRef] [PubMed]
- Kavanagh, K.L.; Jörnvall, H.; Persson, B.; Oppermann, U. Medium- and short-chain dehydrogenase/reductase gene and protein families: The SDR superfamily: Functional and structural diversity within a family of metabolic and regulatory enzymes. Cell. Mol. Life Sci. 2008, 65, 3895–3906. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Yang, Q.; Zhang, H.; Cao, J.; Zhang, X.; Apaliya, M.T. The Possible Mechanisms Involved in Degradation of Patulin by Pichia. caribbica. Toxins 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, K.P.; Borges, J.C. The molecular chaperone Hsp70 family members function by a bidirectional heterotrophic allosteric mechanism. Protein Pept. Lett. 2011, 18, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Zong, Y.; Li, B.; Tian, S. Effects of carbon, nitrogen and ambient pH on patulin production and related gene expression in Penicillium. expansum. Int. J. Food Microbiol. 2015, 206, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Wang, Y.; Qin, G.; Tian, S. iTRAQ-based quantitative proteomic analysis reveals the role of the tonoplast in fruit senescence. J. Proteom. 2016, 146, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Blast2GO. Available online: https://www.blast2go.com/ (accessed on 20 April 2016).
- GO annotation. Available online: http://www.geneontology.org/ (accessed on 22 April 2016).
- Li, Q.Q.; Skinner, J.; Bennett, J.E. Evaluation of reference genes for real-time quantitative PCR studies in Candida glabrata following azole treatment. BMC Mol. Biol. 2012, 13. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

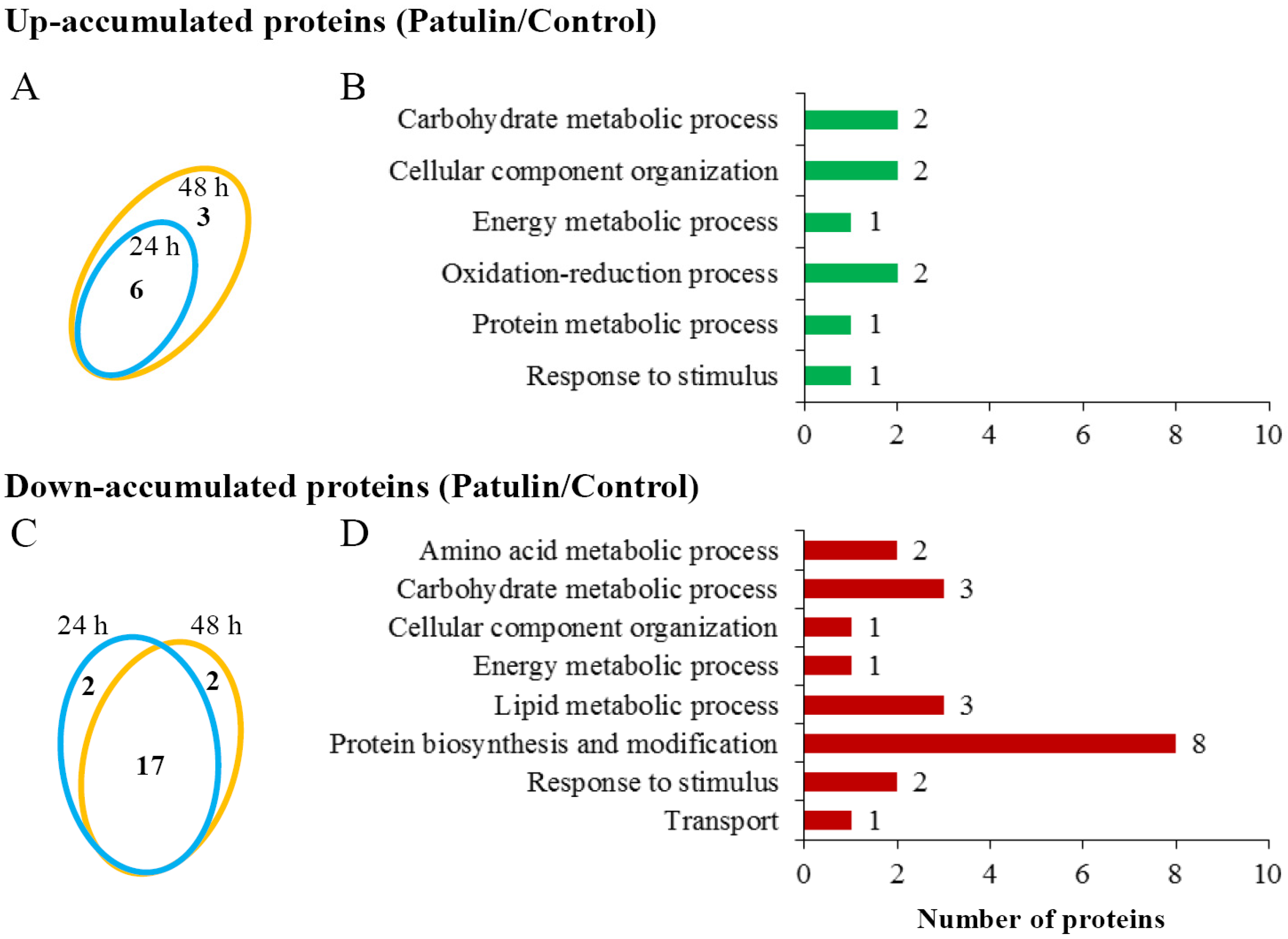
| Protein ID | GI Number | Protein Name | Fold Change (Patulin/Control) | |
|---|---|---|---|---|
| 24 h | 48 h | |||
| Carbohydrate metabolic process | ||||
| U1 | gi|190345021 | Glucose-6-phosphate isomerase | 1.39 | 1.56 |
| U2 | gi|190348104 | Glyceraldehyde 3-phosphate dehydrogenase | 2.97 | 4.09 |
| Cellular component organization | ||||
| U3 | gi|146411959 | NADPH-dependent methylglyoxal reductase | 6.70 | 4.33 |
| U4 | gi|146420860 | NADPH-dependent methylglyoxal reductase | 1.89 | 1.64 |
| Energy metabolic process | ||||
| U5 | gi|146416915 | Phosphoglycerate kinase | 1.10 | 1.83 |
| Oxidation-reduction process | ||||
| U6 | gi|146414467 | Potential oxidoreductase | 6.83 | 9.54 |
| U7 | gi|190348612 | Short-chain dehydrogenase | 51.1 | 24.0 |
| Protein metabolic process | ||||
| U8 | gi|190347960 | Cell division control protein 48 | 1.51 | 1.61 |
| Response to stimulus | ||||
| U9 | gi|146413777 | Heat shock protein 70 | 1.42 | 1.63 |
| Protein ID | GI Number | Protein Name | Fold Change (Patulin/Control) | |
|---|---|---|---|---|
| 24 h | 48 h | |||
| Amino acid metabolic process | ||||
| D1 | gi|146417089 | NADP-specific glutamate dehydrogenase | 0.52 | 0.45 |
| D2 | gi|146417813 | Serine hydroxymethyltransferase | 0.24 | 0.65 |
| Carbohydrate metabolic process | ||||
| D3 | gi|146413757 | Isocitrate lyase | 0.60 | 0.57 |
| D4 | gi|146414542 | Mannose-1-phosphate guanyltransferase | 0.63 | 0.49 |
| D5 | gi|146415901 | S-adenosylmethionine synthase | 0.54 | 0.53 |
| Cellular component organization | ||||
| D6 | gi|146413767 | Histone H4 | 0.50 | 0.38 |
| Energy metabolic process | ||||
| D7 | gi|190345530 | Ketol-acid mitochondrial | 0.13 | 0.63 |
| Lipid metabolic process | ||||
| D8 | gi|190344794 | Acyl-Coenzyme A oxidase 4 | 0.24 | 0.46 |
| D9 | gi|146416129 | Cytochrome P450 (Lanosterol 14-alpha demethylase) | 0.39 | 0.60 |
| D10 | gi|190346072 | Fatty acid synthase subunit beta | 0.39 | 0.74 |
| Protein biosynthesis and modification | ||||
| D11 | gi|146423689 | 3-isopropylmalate dehydratase | 0.66 | 0.58 |
| D12 | gi|269969601 | 40S ribosomal protein s1 | 0.38 | 0.66 |
| D13 | gi|146415016 | 40S ribosomal protein S19-B | 0.08 | 0.91 |
| D14 | gi|146419667 | 60S ribosomal protein L17-B | 0.60 | 0.95 |
| D15 | gi|146418206 | 60S ribosomal protein L30 | 0.28 | 0.45 |
| D16 | gi|146417697 | 60S ribosomal protein L4-b | 0.10 | 0.57 |
| D17 | gi|152032427 | Elongation factor 1-alpha | 0.48 | 0.31 |
| D18 | gi|190346643 | Nucleolar protein 56 | 0.90 | 0.58 |
| Response to stimulus | ||||
| D19 | gi|146420955 | 5-methyltetrahydropteroyltriglutamate homocysteine methyltransferase | 0.37 | 0.60 |
| D20 | gi|156630825 | Histone H3.1/H3.2 | 0.46 | 0.48 |
| Transport | ||||
| D21 | gi|146412540 | ABC transporter ATP-binding protein ARB1 | 0.98 | 0.48 |
| Gene | Primer Sequences (5′–3′) |
|---|---|
| 5.8S rRNA | F: GCTGTCGACCTCTCAATGTATTAGG |
| R: TAAGGCCGGGCCAACAAT | |
| gi|190348612 | F: TCTTTCGGCCCACTTTTGAA |
| R: AAGCGAATGGGTCCCAAGAC | |
| gi|146413777 | F: CCACCAACCAAAGAGCATTGA |
| R: GGTTTGAGCAGAAGAAGACAAGGT | |
| gi|146417089 | F: AAAGGCCACTGGTGGAAAGG |
| R: CAGCGTATTGGGCAACGTTA | |
| gi|146413767 | F: ATTAGGAAAGGGAGGCGCTAA |
| R: TTCTAATAGCTGGCTTGGTGATACC | |
| gi|190344794 | F: CGAAGCCACCGAGGAGTTT |
| R: AAGAATGAGCAGCACCACCAA | |
| gi|156630825 | F: CAAGAAAGTCCACTGGTGGTAAGG |
| R: TCCAGTAGAAGGAGCGGATTTT |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Peng, H.-M.; Wang, X.; Li, B.-Q.; Long, M.-Y.; Tian, S.-P. Biodegradation Mechanisms of Patulin in Candida guilliermondii: An iTRAQ-Based Proteomic Analysis. Toxins 2017, 9, 48. https://doi.org/10.3390/toxins9020048
Chen Y, Peng H-M, Wang X, Li B-Q, Long M-Y, Tian S-P. Biodegradation Mechanisms of Patulin in Candida guilliermondii: An iTRAQ-Based Proteomic Analysis. Toxins. 2017; 9(2):48. https://doi.org/10.3390/toxins9020048
Chicago/Turabian StyleChen, Yong, Huai-Min Peng, Xiao Wang, Bo-Qiang Li, Man-Yuan Long, and Shi-Ping Tian. 2017. "Biodegradation Mechanisms of Patulin in Candida guilliermondii: An iTRAQ-Based Proteomic Analysis" Toxins 9, no. 2: 48. https://doi.org/10.3390/toxins9020048
APA StyleChen, Y., Peng, H.-M., Wang, X., Li, B.-Q., Long, M.-Y., & Tian, S.-P. (2017). Biodegradation Mechanisms of Patulin in Candida guilliermondii: An iTRAQ-Based Proteomic Analysis. Toxins, 9(2), 48. https://doi.org/10.3390/toxins9020048








