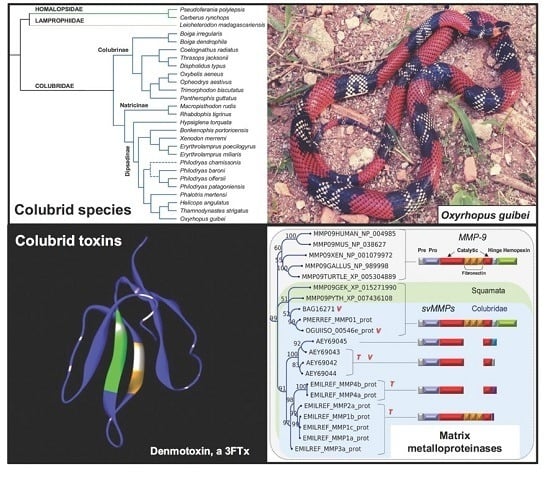
Colubrid Venom Composition: An -Omics Perspective
Abstract
:1. Introduction
2. Results and Discussion
2.1. Compiling the Venom Components of Colubrid Snakes
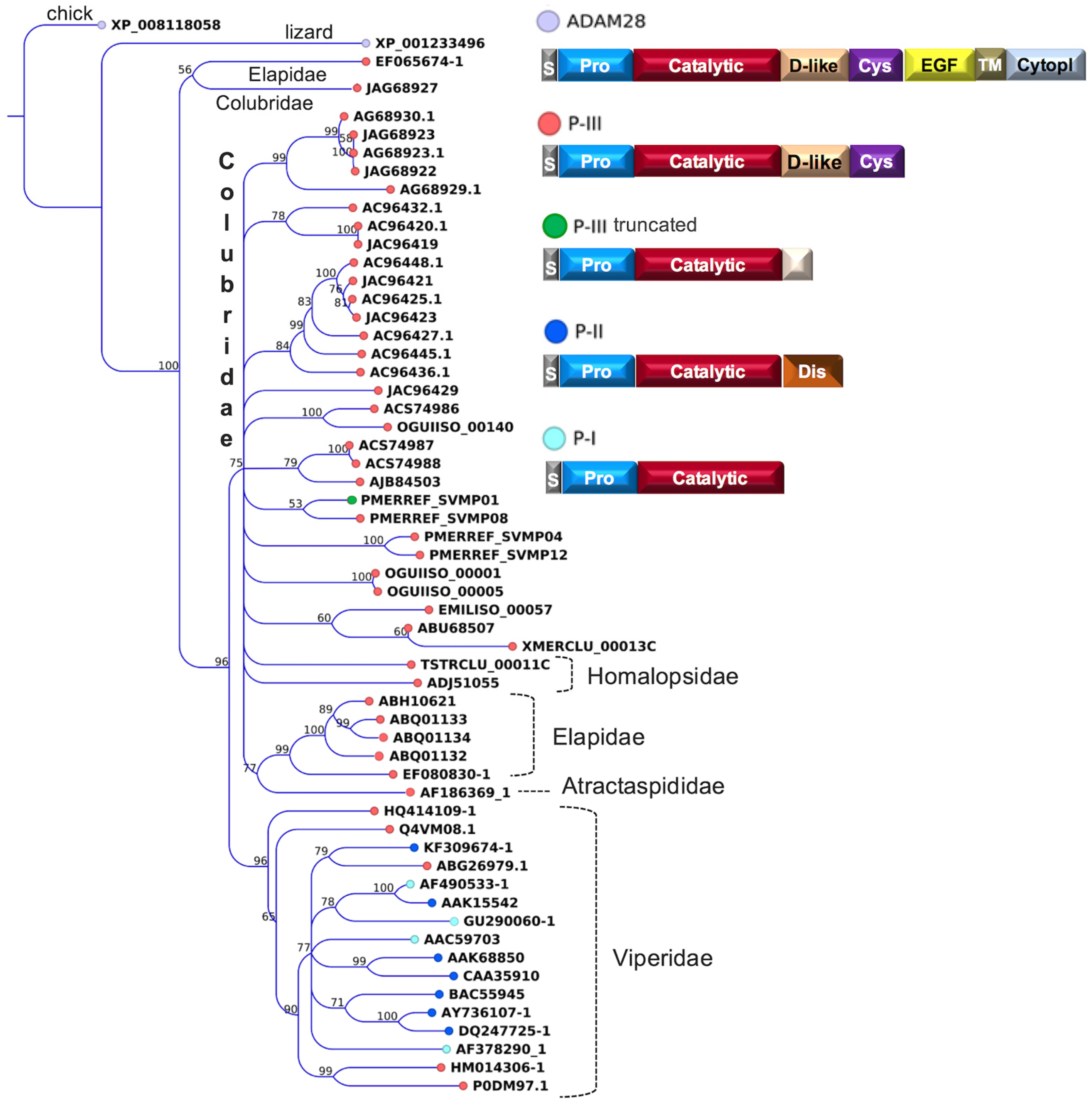
2.2. Major Snake Venom Enzymatic Components
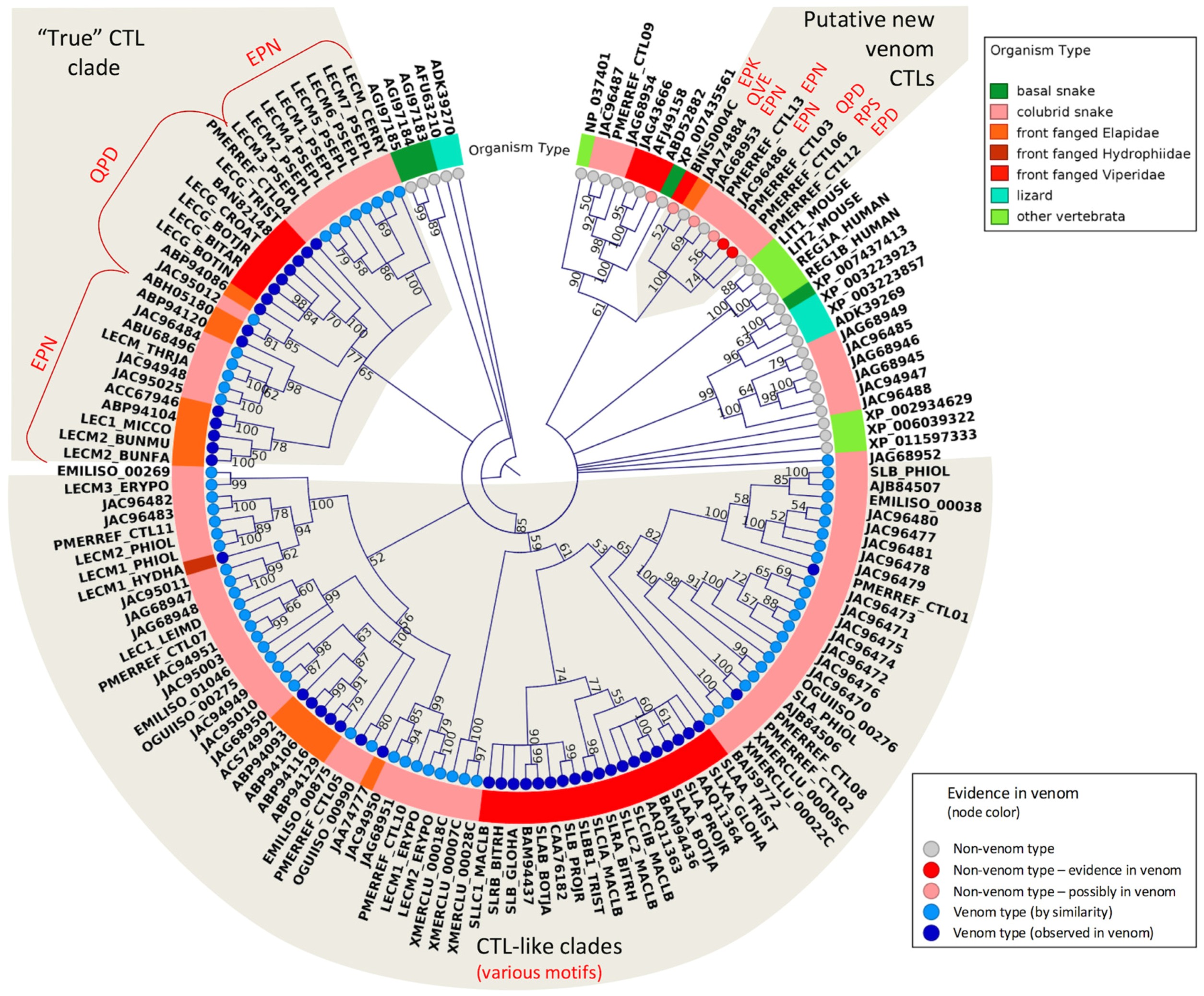
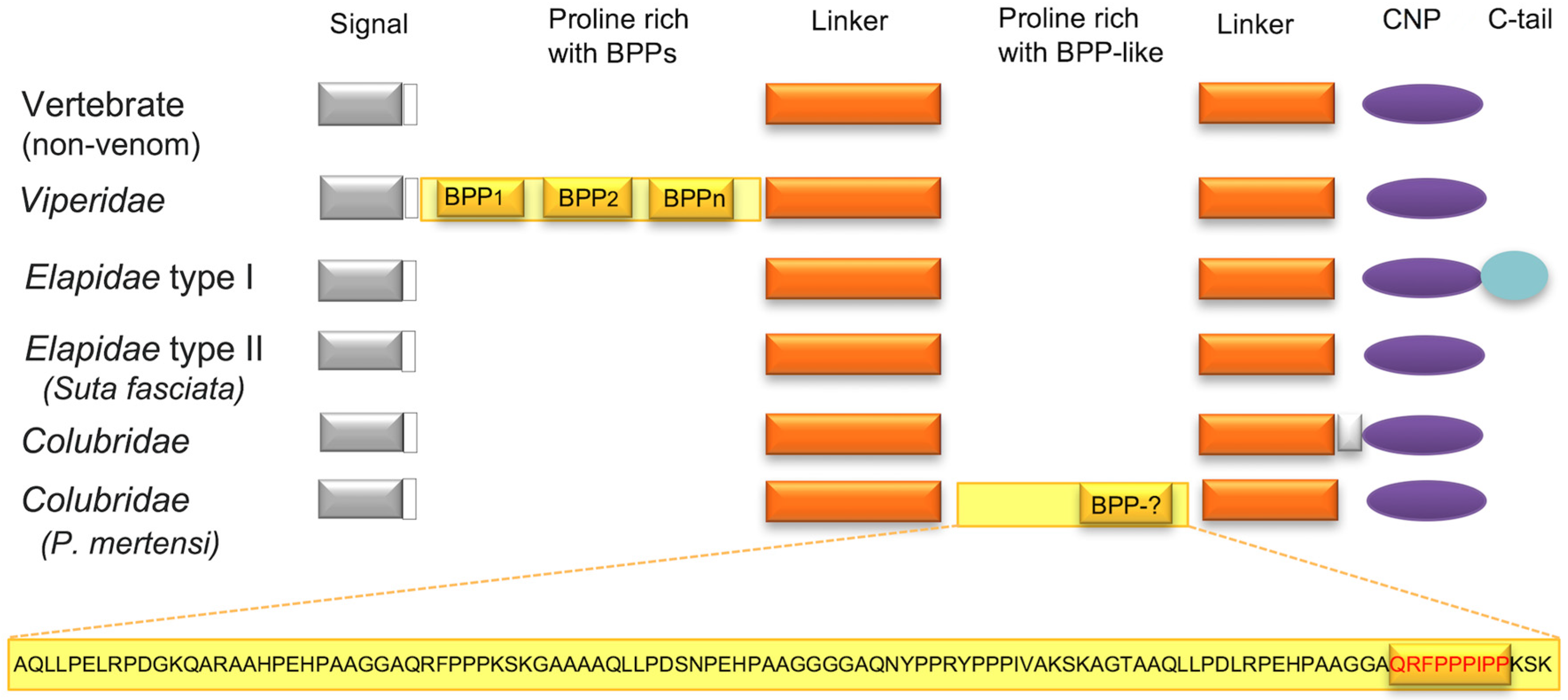
2.3. Major Snake Venom Non-Enzymatic Components
2.4. Minor or Arguably Actual Venom Components
2.5. Putative New Snake Toxins Suggested from Colubrid Venoms
3. Conclusions
4. Materials and Methods
4.1. Original Transcriptomic Data
4.1.1. Animals
4.1.2. RNA-Seq
4.1.3. Expressed Sequence Tags (ESTs) Generation
4.2. Public Database Sequence Retrieval
4.3. Sequence Comparisons and Evolutionary Analyses
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
References
- Calvete, J.J. Snake venomics: From the inventory of toxins to biology. Toxicon 2013, 75, 44–62. [Google Scholar] [CrossRef] [PubMed]
- Ducancel, F.; Durban, J.; Verdenaud, M. Transcriptomics and venomics: Implications for medicinal chemistry. Future Med. Chem. 2014, 15, 1629–1643. [Google Scholar] [CrossRef] [PubMed]
- Brahma, R.K.; McCleary, R.J.; Kini, R.M.; Doley, R. Venom gland transcriptomics for identifying, cataloging, and characterizing venom proteins in snakes. Toxicon 2015, 93, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Gilles, N.; Servent, D. The European FP7 Venomics Project. Future Med. Chem. 2014, 15, 1611–1612. [Google Scholar] [CrossRef] [PubMed]
- Aird, S.D.; Watanabe, Y.; Villar-Briones, A.; Roy, M.C.; Terada, K.; Mikheyev, A.S. Quantitative high-throughput profiling of snake venom gland transcriptomes and proteomes (Ovophis okinavensis and Protobothrops flavoviridis). BMC Genom. 2013, 14. [Google Scholar] [CrossRef] [PubMed]
- Aird, S.D.; Aggarwal, S.; Villar-Briones, A.; Tin, M.M.; Terada, K.; Mikheyev, A.S. Snake venoms are integrated systems, but abundant venom proteins evolve more rapidly. BMC Genom. 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Chapeaurouge, A.; Reza, M.A.; Mackessy, S.P.; Carvalho, P.C.; Valente, R.H.; Teixeira-Ferreira, A.; Perales, J.; Lin, Q.; Kini, R.M. Interrogating the venom of the viperid snake Sistrurus catenatus edwardsii by a combined approach of electrospray and MALDI mass spectrometry. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Durban, J.; Juárez, P.; Angulo, Y.; Lomonte, B.; Flores-Diaz, M.; Alape-Girón, A.; Sasa, M.; Sanz, L.; Gutiérrez, J.M.; Dopazo, J.; et al. Profiling the venom gland transcriptomes of Costa Rican snakes by 454 pyrosequencing. BMC Genom. 2011, 12. [Google Scholar] [CrossRef] [PubMed]
- Hargreaves, A.D.; Swain, M.T.; Logan, D.W.; Mulley, J.F. Testing the Toxicofera: Comparative transcriptomics casts doubt on the single, early evolution of the reptile venom system. Toxicon 2014, 92, 140–156. [Google Scholar] [CrossRef] [PubMed]
- Junqueira-de-Azevedo, I.L.; Bastos, C.M.; Ho, P.L.; Luna, M.S.; Yamanouye, N.; Casewell, N.R. Venom-related transcripts from Bothrops jararaca tissues provide novel molecular insights into the production and evolution of snake venom. Mol. Biol. Evol. 2015, 32, 754–766. [Google Scholar] [CrossRef] [PubMed]
- Margres, M.J.; McGivern, J.J.; Wray, K.P.; Seavy, M.; Calvin, K.; Rokyta, D.R. Linking the transcriptome and proteome to characterize the venom of the eastern diamondback rattlesnake (Crotalus adamanteus). J. Proteom. 2014, 96, 145–158. [Google Scholar] [CrossRef] [PubMed]
- McGivern, J.J.; Wray, K.P.; Margres, M.J.; Couch, M.E.; Mackessy, S.P.; Rokyta, D.R. RNA-seq and high-definition mass spectrometry reveal the complex and divergent venoms of two rear-fanged colubrid snakes. BMC Genom. 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- Petras, D.; Heiss, P.; Süssmuth, R.D.; Calvete, J.J. Venom Proteomics of Indonesian King cobra, Ophiophagus. hannah: Integrating top-down and bottom-up approaches. J. Proteome Res. 2015, 14, 2539–2556. [Google Scholar] [CrossRef] [PubMed]
- Reeks, T.; Lavergne, V.; Sunagar, K.; Jones, A.; Undheim, E.; Dunstan, N.; Fry, B.; Alewood, P.F. Deep venomics of the Pseudonaja genus reveals inter- and intra-specific variation. J. Proteom. 2016, 133, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Rokyta, D.R.; Wray, K.P.; Margres, M.J. The genesis of an exceptionally lethal venom in the timber rattlesnake (Crotalus horridus) revealed through comparative venom-gland transcriptomics. BMC Genom. 2013, 14. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.H.; Fung, S.Y.; Yap, M.K.; Leong, P.K.; Liew, J.L.; Tan, N.H. Unveiling the elusive and exotic: Venomics of the Malayan blue coral snake (Calliophis bivirgata flaviceps). J. Proteom. 2016, 132, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Viala, V.L.; Hildebrand, D.; Trusch, M.; Fucase, T.M.; Sciani, J.M.; Pimenta, D.C.; Arni, R.K.; Schlüter, H.; Betzel, C.; Mirtschin, P.; et al. Venomics of the Australian eastern brown snake (Pseudonaja textilis): Detection of new venom proteins and splicing variants. Toxicon 2015, 107, 252–265. [Google Scholar] [CrossRef] [PubMed]
- Terrat, Y.; Sunagar, K.; Fry, B.G.; Jackson, T.N.; Scheib, H.; Fourmy, R.; Verdenaud, M.; Blanchet, G.; Antunes, A.; Ducancel, F. Atractaspis aterrima toxins: The first insight into the molecular evolution of venom in side-stabbers. Toxins (Basel) 2013, 5, 1948–1964. [Google Scholar] [CrossRef] [PubMed]
- Casewell, N.R.; Wagstaff, S.C.; Wüster, W.; Cook, D.A.; Bolton, F.M.; King, S.I.; Pla, D.; Sanz, L.; Calvete, J.J.; Harrison, R.A. Medically important differences in snake venom composition are dictated by distinct postgenomic mechanisms. Proc. Natl. Acad. Sci. USA 2014, 111, 9205–9210. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, H.L.; Mackessy, S.P. Functional basis of a molecular adaptation: Prey-specific toxic effects of venom from Sistrurus rattlesnakes. Toxicon 2009, 53, 672–679. [Google Scholar] [CrossRef] [PubMed]
- Rokyta, D.R.; Wray, K.P.; McGivern, J.J.; Margres, M.J. The transcriptomic and proteomic basis for the evolution of a novel venom phenotype within the Timber Rattlesnake (Crotalus horridus). Toxicon 2015, 98, 34–48. [Google Scholar] [CrossRef] [PubMed]
- Margres, M.J.; McGivern, J.J.; Seavy, M.; Wray, K.P.; Facente, J.; Rokyta, D.R. Contrasting modes and tempos of venom expression evolution in two snake species. Genetics 2015, 199, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Modahl, C.; Saviola, A.J.; Mackessy, S.P. Proteomic and genomic approaches to the study of rear-fanged (“colubrid”) snake venoms. In Handbooks of Toxinology. Venom Genomics and Proteomics; Gopalakrishnakone, P., Ed.; Springer Science: Dordrecht, The Netherlands, 2015; p. 23. [Google Scholar]
- Kardong, K.V. Evolutionary patterns in advanced snakes. Am. Zool. 1980, 20, 269–282. [Google Scholar] [CrossRef]
- Savitzky, A.H. The role of venom delivery strategies in snake evolution. Evolution 1980, 34, 1194–1204. [Google Scholar] [CrossRef]
- Vidal, N. Colubroid systematics: Evidence for an early appearance of the venom apparatus followed by extensive evolutionary tinkering. J. Toxicol.-Toxin Rev. 2002, 21, 27–47. [Google Scholar] [CrossRef]
- Mackessy, S.P.; Saviola, A.J. Venoms from “non-venomous” snakes: Rear-fanged snake venoms as sources of novel compounds. In Snake Venoms and Envenomation: Modern Trends and Future Prospects; Utkin, Y., Krivoshein, A.V., Eds.; Nova Science Publishers, Inc.: New York, NY, USA, 2016; p. 29. [Google Scholar]
- Dowling, H.G.; Hass, C.A.; Hedges, S.B.; Highton, R. Snake relationships revealed by slow-evolving proteins: A preliminary survey. J. Zool. Lond. 1996, 240, 1–28. [Google Scholar] [CrossRef]
- Heise, P.J.; Maxson, L.R.; Dowling, H.G.; Hedges, S.B. Higherlevel snake phylogeny inferred from mitochondrial DNA sequences of 12S rRNA and 16S rRNA genes. Mol. Biol. Evol. 1995, 12, 259–265. [Google Scholar] [PubMed]
- Kelly, C.M.R.; Barker, N.P.; Villet, M.H. Phylogenetics of advanced snakes (Caenophidia) based on four mitochondrial genes. Syst. Biol. 2003, 52, 439–459. [Google Scholar] [CrossRef] [PubMed]
- Kraus, F.; Brown, W.M. Phylogenetic relationships of colubroid snakes based on mitochondrial DNA sequences. Zool. J. Linn. Soc. 1998, 122, 455–487. [Google Scholar] [CrossRef]
- Lee, M.S.Y.; Hugall, A.F.; Lawson, R.; Scanlon, J.D. Snake phylogeny based on multiple morphological and molecular data sets. Syst. Biodivers. 2007, 5, 371–389. [Google Scholar] [CrossRef]
- Pyron, R.A.; Burbrink, F.T.; Colli, G.R.; de Oca, A.N.; Vitt, L.J.; Kuczynski, C.A.; Wiens, J.J. The phylogeny of advanced snakes (Colubroidea), with discovery of a new subfamily and comparison of support methods for likelihood trees. Mol. Phylogenet. Evol. 2011, 58, 329–342. [Google Scholar] [CrossRef] [PubMed]
- Pyron, R.A.; Burbrink, F.T.; Wiens, J.J. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol. Biol. 2013, 13. [Google Scholar] [CrossRef] [PubMed]
- Vidal, N.; Delmas, A.S.; David, P.; Cruaud, C.; Couloux, A.; Hedges, S.B. The phylogeny and classification of caenophidian snakes inferred from seven nuclear protein-coding genes. C. R. Biol. 2007, 330, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Uetz, P. The original descriptions of reptiles. Zootaxa 2010, 2334, 59–68. [Google Scholar]
- Uetz, P. The Reptile Database. Available online: http://www.reptile-database.org (accessed on 3 April 2016).
- Weinstein, S.A.; Smith, T.L.; Kardong, K.V. Reptile venom glands. Form, function, and future. In Handbook of Venoms and Toxins of Reptiles; Mackessy, S.P., Ed.; CRC Press/Taylor & Francis Group: Boca Raton, FL, USA, 2009; pp. 65–91. [Google Scholar]
- Saviola, A.J.; Peichoto, M.E.; Mackessy, S.P. Rear-fanged snake venoms: An untapped source of novel compounds and potential drug leads. Toxin Rev. 2014, 33, 185–201. [Google Scholar] [CrossRef]
- Zaher, H.; de Oliveira, L.; Grazziotin, F.G.; Campagner, M.; Jared, C.; Antoniazzi, M.M.; Prudente, A.L. Consuming viscous prey: A novel protein-secreting delivery system in neotropical snail-eating snakes. BMC Evol. Biol. 2014, 14. [Google Scholar] [CrossRef] [PubMed]
- Mackessy, S.P. Biochemistry and pharmacology of colubrid snake venoms. J. Toxicol.-Toxin Rev. 2002, 21, 43–83. [Google Scholar] [CrossRef]
- Pawlak, J.; Mackessy, S.P.; Fry, B.G.; Bhatia, M.; Mourier, G.; Fruchart-Gaillard, C.; Servent, D.; Ménez, R.; Stura, E.; Ménez, A.; et al. Denmotoxin, a three-finger toxin from the colubrid snake Boiga dendrophila (Mangrove Catsnake) with bird-specific activity. J. Biol. Chem. 2006, 281, 29030–29041. [Google Scholar] [CrossRef] [PubMed]
- Lumsden, N.G.; Fry, B.G.; Ventura, S.; Kini, R.M.; Hodgson, W.C. Pharmacological characterisation of a neurotoxin from the venom of Boiga dendrophila (mangrove catsnake). Toxicon 2005, 45, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Mackessy, S.P.; Sixberry, N.M.; Heyborne, W.H.; Fritts, T. Venom of the Brown Treesnake, Boiga irregularis: Ontogenetic shifts and taxa-specific toxicity. Toxicon 2006, 47, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Pawlak, J.; Mackessy, S.P.; Sixberry, N.M.; Stura, E.A.; Le Du, M.H.; Ménez, R.; Foo, C.S.; Ménez, A.; Nirthanan, S.; Kini, R.M. Irditoxin, a novel covalently linked heterodimeric three-finger toxin with high taxon-specific neurotoxicity. FASEB J. 2009, 23, 534–545. [Google Scholar] [CrossRef] [PubMed]
- Weldon, C.L.; Mackessy, S.P. Biological and proteomic analysis of venom from the Puerto Rican Racer (Alsophis portoricensis: Dipsadidae). Toxicon 2010, 55, 558–569. [Google Scholar] [CrossRef] [PubMed]
- Weldon, C.L.; Mackessy, S.P. Alsophinase, a new P-III metalloproteinase with alpha-fibrinogenolytic and hemorrhagic activity from the venom of the Puerto Rican Racer Alsophis portoricensis (Serpentes: Dipsadidae). Biochimie 2012, 94, 1189–1198. [Google Scholar] [CrossRef] [PubMed]
- OmPraba, G.; Chapeaurouge, A.; Doley, R.; Devi, K.R.; Padmanaban, P.; Venkatraman, C.; Velmurugan, D.; Lin, Q.; Kini, R.M. Identification of a novel family of snake venom proteins Veficolins from Cerberus rynchops using a venom gland transcriptomics and proteomics approach. J. Proteome Res. 2010, 9, 1882–1893. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Lumsden, N.G.; Wüster, W.; Wickramaratna, J.C.; Hodgson, W.C.; Kini, R.M. Isolation of a neurotoxin (alpha-colubritoxin) from a nonvenomous colubrid: Evidence for early origin of venom in snakes. J. Mol. Evol. 2003, 57, 446–452. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Scheib, H.; van der Weerd, L.; Young, B.; McNaughtan, J.; Ramjan, S.F.; Vidal, N.; Poelmann, R.E.; Norman, J.A. Evolution of an arsenal: Structural and functional diversification of the venom system in the advanced snakes (Caenophidia). Mol. Cell. Proteom. 2008, 7, 215–246. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Scheib, H.; Junqueira-de-Azevedo, I.L.M.; Silva, D.A.; Casewell, N.R. Novel transcripts in the maxillary venom glands of advanced snakes. Toxicon 2012, 59, 696–708. [Google Scholar] [CrossRef] [PubMed]
- Kamiguti, A.S.; Theakston, R.D.; Sherman, N.; Fox, J.W. Mass spectrophotometric evidence for P-III/P-IV metalloproteinases in the venom of the Boomslang (Dispholidus typus). Toxicon 2000, 38, 1613–1620. [Google Scholar] [CrossRef]
- Estrella, A.; Sánchez, E.E.; Galán, J.A.; Tao, W.A.; Guerrero, B.; Navarrete, L.F.; Rodríguez-Acosta, A. Characterization of toxins from the broad-banded water snake Helicops angulatus (Linnaeus, 1758): Isolation of a cysteine-rich secretory protein, Helicopsin. Arch. Toxicol. 2011, 85, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Peichoto, M.E.; Tavares, F.L.; Santoro, M.L.; Mackessy, S.P. Venom proteomes of South and North American opisthoglyphous (Colubridae and Dipsadidae) snake species: A preliminary approach to understanding their biological roles. Comp. Biochem. Physiol. Part D 2012, 7, 361–369. [Google Scholar]
- Zhang, Z.; Zhang, X.; Hu, T.; Zhou, W.; Cui, Q.; Tian, J.; Zheng, Y.; Fan, Q. Discovery of toxin-encoding genes from the false viper Macropisthodon rudis, a rear-fanged snake, by transcriptome analysis of venom gland. Toxicon 2015, 106, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Heyborne, W.H.; Mackessy, S.P. Identification and characterization of a taxon-specific three-finger toxin from the venom of the Green Vinesnake (Oxybelis fulgidus; family Colubridae). Biochimie 2013, 95, 1923–1932. [Google Scholar] [CrossRef] [PubMed]
- Campos, P.F.; Silva, D.A.; Zelanis, A.; Paes Leme, A.F.; Rocha, M.M.T.; Menezes, M.C.; Serrano, S.M.; Junqueira-de-Azevedo, I.L. Trends in the evolution of snake toxins underscored by an integrative omics approach to profile the venom of the colubrid Phalotris mertensi. Genome Biol. Evol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Urra, F.A.; Pulgar, R.; Gutiérrez, R.; Hodar, C.; Cambiazo, V.; Labra, A. Identification and molecular characterization of five putative toxins from the venom gland of the snake Philodryas chamissonis (Serpentes: Dipsadidae). Toxicon 2015, 108, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Ching, A.T.; Rocha, M.M.; Paes Leme, A.F.; Pimenta, D.C.; de Furtado, M.F.; Serrano, S.M.; Ho, P.L.; Junqueira-de-Azevedo, I.L. Some aspects of the venom proteome of the Colubridae snake Philodryas olfersii revealed from a Duvernoy’s (venom) gland transcriptome. FEBS Lett. 2006, 580, 4417–4422. [Google Scholar] [CrossRef] [PubMed]
- Ching, A.T.; Paes Leme, A.F.; Zelanis, A.; Rocha, M.M.; de Furtado, M.F.; Silva, D.A.; Trugilho, M.R.; da Rocha, S.L.; Perales, J.; Ho, P.L.; et al. Venomics profiling of Thamnodynastes strigatus unveils matrix metalloproteinases and other novel proteins recruited to the toxin arsenal of rear-fanged snakes. J. Proteome Res. 2012, 11, 1152–1162. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Mackessy, S.P. Biochemical characterization of phospholipase A2 (trimorphin) from the venom of the Sonoran Lyre Snake Trimorphodon biscutatus lambda (family Colubridae). Toxicon 2004, 44, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Wiens, J.J.; Kuczynski, C.A.; Smith, S.A.; Mulcahy, D.G.; Sites, J.W., Jr.; Townsend, T.M.; Reeder, T.W. Branch lengths, support, and congruence: Testing the phylogenomic approach with 20 nuclear loci in snakes. Syst. Biol. 2008, 57, 420–431. [Google Scholar] [CrossRef] [PubMed]
- Taub, A.M. Ophidian cephalic glands. J. Morphol. 1966, 118, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Salomão, M.G.; Albolea, A.B.P.; Santos, S.M.A. Colubrid snakebite: A public health problem in Brazil. Herpetol. Rev. 2003, 34, 307–312. [Google Scholar]
- Weinstein, S.A.; Griffin, R.; Ismail, A.K. Non-front-fanged colubroid (“colubrid”) snakebites: Three cases of local envenoming by the mangrove or ringed cat-eyed snake (Boiga dendrophila; Colubridae, Colubrinae), the Western beaked snake (Rhamphiophis oxyrhynchus; Lamprophiidae, Psammophinae) and the rain forest cat-eyed snake (Leptodeira frenata; Dipsadidae). Clin. Toxicol. (Phila.) 2014, 52, 277–282. [Google Scholar] [PubMed]
- Komori, K.; Konishi, M.; Maruta, Y.; Toriba, M.; Sakai, A.; Matsuda, A.; Hori, T.; Nakatani, M.; Minamino, N.; Akizawa, T. Characterization of a novel metalloproteinase in Duvernoy’s gland of Rhabdophis tigrinus tigrinus. J. Toxicol. Sci. 2006, 31, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Modahl, C.M.; Mackessy, S.P. Full-length venom protein cDNA sequences from venom-derived mRNA: Exploring compositional variation and adaptive multigene evolution. PLoS Negl. Trop. Dis. 2016, 10, e0004587. [Google Scholar] [CrossRef] [PubMed]
- Moura-da-Silva, A.M.; Theakston, R.D.; Crampton, J.M. Evolution of disintegrin cysteine-rich and mammalian matrix-degrading metalloproteinases: Gene duplication and divergence of a common ancestor rather than convergent evolution. J. Mol. Evol. 1996, 43, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Casewell, N.R.; Wagstaff, S.C.; Harrison, R.A.; Renjifo, C.; Wüster, W. Domain loss facilitates accelerated evolution and neofunctionalization of duplicate snake venom metalloproteinase toxin genes. Mol. Biol. Evol. 2011, 28, 2637–2649. [Google Scholar]
- Vaiyapuri, S.; Sunagar, K.; Gibbins, J.M.; Jackson, T.N.W.; Reeks, T.; Fry, B.G. Kallikrein enzymes. In Venomous Reptiles and Their Toxins: Evolution, Pathophysiology and Biodiscovery, 1st ed.; Fry, B.G., Ed.; Oxford University Press: Oxford, UK, 2015; p. 576. [Google Scholar]
- Weinstein, S.A.; Kardong, K.V. Properties of Duvernoy’s secretions from opisthoglyphous and aglyphous colubrid snakes. Toxicon 1994, 32, 1161–1185. [Google Scholar] [CrossRef]
- Vest, D.K.; Mackessy, S.P.; Kardong, K.V. The unique Duvernoy’s secretion of the brown tree snake (Boiga irregularis). Toxicon 1991, 29, 532–535. [Google Scholar] [CrossRef]
- Hill, R.E.; Mackessy, S.P. Characterization of venom (Duvernoy’s secretion) from twelve species of colubrid snakes and partial sequence of four venom proteins. Toxicon 2000, 38, 1663–1687. [Google Scholar] [CrossRef]
- Reyes-Velasco, J.; Card, D.C.; Andrew, A.L.; Shaney, K.J.; Adams, R.H.; Schield, D.R.; Casewell, N.R.; Mackessy, S.P.; Castoe, T.A. Expression of venom gene homologs in diverse python tissues suggests a new model for the evolution of snake venom. Mol. Biol. Evol. 2015, 32, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Drickamer, K. C-type lectin-like domains. Curr. Opin. Struct. Biol. 1999, 9, 585–590. [Google Scholar] [CrossRef]
- Arlinghaus, F.T.; Fry, B.G.; Sunagar, K.; Jackson, T.N.W.; Eble, J.A.; Reeks, T.; Clemetson, K.J. Lectin proteins. In Venomous Reptiles and Their Toxins: Evolution, Pathophysiology and Biodiscovery, 1st ed.; Fry, B.G., Ed.; Oxford University Press: Oxford, UK, 2015; p. 576. [Google Scholar]
- Junqueira-de-Azevedo, I.L.; Ho, P.L. A survey of gene expression and diversity in the venom glands of the pitviper snake Bothrops insularis through the generation of expressed sequence tags (ESTs). Gene 2002, 299, 279–291. [Google Scholar] [CrossRef]
- Jackson, T.N.; Sunagar, K.; Undheim, E.A.; Koludarov, I.; Chan, A.H.; Sanders, K.; Ali, S.A.; Hendrikx, I.; Dunstan, N.; Fry, B.G. Venom down under: Dynamic evolution of Australian elapid snake toxins. Toxins 2013, 5, 2621–2655. [Google Scholar] [CrossRef] [PubMed]
- Peichoto, M.E.; Mackessy, S.P.; Teibler, P.; Tavares, F.L.; Burckhardt, P.L.; Breno, M.C.; Acosta, O.; Santoro, M.L. Purification and characterization of a cysteine-rich secretory protein from Philodryas patagoniensis (Dipsadidae) snake venom. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2009, 150, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Sunagar, K.; Johnson, W.E.; O’Brien, S.J.; Vasconcelos, V.; Antunes, A. Evolution of CRISPs associated with toxicoferan-reptilian venom and mammalian reproduction. Mol. Biol. Evol. 2012, 29, 1807–1822. [Google Scholar] [CrossRef] [PubMed]
- Ondetti, M.A.; Rubin, B.; Cushman, D.W. Design of specific inhibitors of angiotensin-converting enzyme: New class of orally active anti-hypertensive agents. Science 1977, 196, 441–444. [Google Scholar] [CrossRef] [PubMed]
- Munekiyo, S.M.; Mackessy, S.P. Presence of peptide inhibitors in rattlesnake venoms and their effects on endogenous metalloproteases. Toxicon 2005, 45, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, S.; Murayama, N.; Saguchi, K.; Ohi, H.; Fujita, Y.; da Silva, N.J., Jr.; de Siqueira, R.J.; Lahlou, S.; Aird, S.D. A novel peptide from the ACEI/BPP-CNP precursor in the venom of Crotalus durissus collilineatus. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2006, 144, 107–121. [Google Scholar] [CrossRef] [PubMed]
- Wagstaff, S.C.; Favreau, P.; Cheneval, O.; Laing, G.D.; Wilkinson, M.C.; Miller, R.L.; Stöcklin, R.; Harrison, R.A. Molecular characterisation of endogenous snake venom metalloproteinase inhibitors. Biochem. Biophys. Res. Commun. 2008, 365, 650–656. [Google Scholar] [CrossRef] [PubMed]
- Graham, R.L.; Graham, C.; McClean, S.; Chen, T.; O’Rourke, M.; Hirst, D.; Theakston, D.; Shaw, C. Identification and functional analysis of a novel bradykinin inhibitory peptide in the venoms of New World Crotalinae pit vipers. Biochem. Biophys. Res. Commun. 2005, 338, 1587–1592. [Google Scholar] [CrossRef] [PubMed]
- Correa, P.G.; Oguiura, N. Phylogenetic analysis of β-defensin-like genes of Bothrops, Crotalus and Lachesis snakes. Toxicon 2013, 69, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Doley, R.; Pahari, S.; Reza, M.A.; Mackessy, S.P.; Kini, K.M. The gene structure and evolution of ku-wap-fusin (Kunitz Waprin Fusion Protein), a novel evolutionary intermediate of the Kunitz serine protease inhibitors and waprins from Sistrurus catenatus (Massasauga Rattlesnake) venom glands. Open Evol. J. 2010, 4, 31–41. [Google Scholar]
- Laskowski, M., Jr.; Kato, I. Protein inhibitors of proteinases. Annu. Rev. Biochem. 1980, 49, 593–626. [Google Scholar] [CrossRef] [PubMed]
- Millers, E.K.; Trabi, M.; Masci, P.P.; Lavin, M.F.; de Jersey, J.; Guddat, L.W. Crystal structure of textilinin-1, a Kunitz-type serine protease inhibitor from the venom of the Australian common brown snake (Pseudonaja textilis). FEBS J. 2009, 276, 3163–3175. [Google Scholar] [CrossRef] [PubMed]
- Rao, V.S.; Joseph, J.S.; Kini, R.M. Group D prothrombin activators from snake venom are structural homologues of mammalian blood coagulation factor Xa. Biochem. J. 2003, 369, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Junqueira-de-Azevedo, I.L.; Farsky, S.H.; Oliveira, M.L.; Ho, P.L. Molecular cloning and expression of a functional snake venom vascular endothelium growth factor (VEGF) from the Bothrops insularis pit viper. A new member of the VEGF family of proteins. J. Biol. Chem. 2001, 276, 39836–39842. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Hattori, S.; Iwamatsu, A.; Takizawa, H.; Shibuya, M. A novel snake venom vascular endothelial growth factor (VEGF) predominantly induces vascular permeability through preferential signaling via VEGF receptor-1. J. Biol. Chem. 2004, 279, 46304–46314. [Google Scholar] [CrossRef] [PubMed]
- Vogel, C.W.; Bredehorst, R.; Fritzinger, D.C.; Grunwald, T.; Ziegelmüller, P.; Kock, M.A. Structure and function of cobra venom factor, the complement-activating protein in cobra venom. Adv. Exp. Med. Biol. 1996, 391, 97–114. [Google Scholar] [PubMed]
- Okumura, K.; Masui, K.; Inoue, S.; Ikeda, K.; Hayashi, K. Purification, characterization and cDNA cloning of a phospholipase A2 inhibitor from the serum of the non-venomous snake Elaphe quadrivirgata. Biochem. J. 1999, 341, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Brillard-Bourdet, M.; Nguyên, V.; Ferrer-di Martino, M.; Gauthier, F.; Moreau, T. Purification and characterization of a new cystatin inhibitor from Taiwan cobra (Naja naja atra) venom. Biochem. J. 1998, 331, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Ducancel, F.; Matre, V.; Dupont, C.; Lajeunesse, E.; Wollberg, Z.; Bdolah, A.; Kochva, E.; Boulain, J.C.; Ménez, A. Cloning and sequence analysis of cDNAs encoding precursors of sarafotoxins. Evidence for an unusual “rosary-type” organization. J. Biol. Chem. 1993, 268, 3052–3055. [Google Scholar] [PubMed]
- Casewell, N.R.; Harrison, R.A.; Wüster, W.; Wagstaff, S.C. Comparative venom gland transcriptome surveys of the saw-scaled vipers (Viperidae: Echis) reveal substantial intra-family gene diversity and novel venom transcripts. BMC Genom. 2009, 10. [Google Scholar] [CrossRef] [PubMed]
- Corrêa-Netto, C.; Junqueira-de-Azevedo, I.L.; Silva, D.A.; Ho, P.L.; Leitão-de-Araújo, M.; Alves, M.L.; Sanz, L.; Foguel, D.; Zingali, R.B.; Calvete, J.J. Snake venomics and venom gland transcriptomic analysis of Brazilian coral snakes, Micrurus altirostris and M. corallinus. J. Proteom. 2011, 74, 1795–1809. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Roelants, K.; Norman, J.; King, G.; Tyndal, J.; Lewis, R.; Norton, R.; Renjifo, C.; Rodriguez de la Vega, R.C. Toxicogenomic multiverse: Convergent recruitment of proteins into animal venoms. Annu. Rev. Genom. Hum. Genet. 2009, 10, 483–511. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Wells, R.G.; Reed, R.R. Isolation of an olfactory cDNA: Similarity to retinol-binding protein suggests a role in olfaction. Science 1987, 235, 1053–1056. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by rna-seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Madan, A. CAP3: A DNA sequence assembly program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef] [PubMed]
- Junqueira-de-Azevedo, I.L.; Ching, A.T.; Carvalho, E.; Faria, F.; Nishiyama, M.Y., Jr.; Ho, P.L.; Diniz, M.R. Lachesis muta (Viperidae) cDNAs reveal diverging pit viper molecules and scaffolds typical of cobra (Elapidae) venoms: implications for snake toxin repertoire evolution. Genetics 2006, 173, 877–889. [Google Scholar] [CrossRef] [PubMed]



| Species | Enzymatic | Non-Enzymatic | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LAAO | PLA2 (IA) | SVMP | SVSP | 3FTx | CNP | CRISP | CTL | DEFEN | KUN-1 | KUN-2 | ||
| Boiga dendrophila | B | [42,43] | ||||||||||
| Boiga irregularis | TPB | TPB | T | TP | T | t | [12,44,45] | |||||
| Borikenophis portoricensis | B | BP | [46,47] | |||||||||
| Cerberus rynchops | TP | TP | TP | [48] | ||||||||
| Coelognathus radiatus | B | [49] | ||||||||||
| Dispholidus typus | xP | x | x | [50,51,52] | ||||||||
| Erythrolamprus miliaris | T | t | T | T | This work; [50] | |||||||
| Erythrolamprus poecilogyrus | x | x | x | x | [50,51] | |||||||
| Helicops angulatus | BP | [53] | ||||||||||
| Hypsiglena sp. | TP | T | T | TP | TP | t | [13] | |||||
| Hypsiglena torquata | P | [54] | ||||||||||
| Leoiheterodon madagascarensis | x | x | x | [50] | ||||||||
| Macropisthodon rudis | t | [55] | ||||||||||
| Opheodrys aestivus | x | t | t | t | [9] | |||||||
| Oxybelis fulgidus | B | [56] | ||||||||||
| Oxyrhopus guibei | T | t | T | T | t | This work | ||||||
| Phalotris mertensi | TP | T | tP | t | t | t | T | TP | TP | [57] | ||
| Pantherophis guttatus | t | x | t | t | t | [9] | ||||||
| Philodryas baroni | P | [54] | ||||||||||
| Philodryas chamissonis | x | x | x | x | x | [58] | ||||||
| Philodryas olfersii | xTP | xTP | T | xTP | TP | x | [51,59] | |||||
| Philodryas patagoniensis | P | [54] | ||||||||||
| Pseudoferania polylepis | x | x | x | x | [50] | |||||||
| Rhabdophis tigrinus | x | x | t | [50] | ||||||||
| Telescopus dhara | x | x | x | x | [50,51] | |||||||
| Thamnodynastes strigatus | TP | t * | t | TP | TP | T | [60] | |||||
| Thrasops jacksonii | x | x | x | [51] | ||||||||
| Trimorphodon biscutatus | B | B | B | [54,61] | ||||||||
| Xenodon merremi | T | T | T * | T | T | This work | ||||||
| Species | Enzymatic | Non-Enzymatic | Reference | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5NUCLEO | AChE | DPP | FactV | FactX | HYAL | PDE | AVIT | bPLA2i | CVF | CYST | gPLA2i | KU-WA-FU | NGF * | OHA | VEGF-A ** | WAP | ||
| Boiga dendrophila | [42,43] | |||||||||||||||||
| Boiga irregularis | T | t | t | t | t | t | t | t | t | [12,44,45] | ||||||||
| Borikenophis portoricensis | B | B | [46,47] | |||||||||||||||
| Cerberus rynchops | [48] | |||||||||||||||||
| Coelognathus radiatus | [49] | |||||||||||||||||
| Dispholidus typus | [50,51,52] | |||||||||||||||||
| Erythrolamprus miliaris | t | t | t | t | T | t | xt | This work; [50] | ||||||||||
| Erythrolamprus poecilogyrus | x | [50,51] | ||||||||||||||||
| Helicops angulatus | [53] | |||||||||||||||||
| Hypsiglena sp. | t | tP | t | t | t | [13] | ||||||||||||
| Hypsiglena torquata | [54] | |||||||||||||||||
| Leoiheterodon madagascarensis | [50] | |||||||||||||||||
| Macropisthodon rudis | t | [55] | ||||||||||||||||
| Opheodrys aestivus | t | t | t | x | t | x | t | t | t | [9] | ||||||||
| Oxybelis fulgidus | [56] | |||||||||||||||||
| Oxyrhopus guibei | t | t | t | t | t | t | This work | |||||||||||
| Phalotris mertensi | tP | t | t | t | tP | t | tP | t | T | [57] | ||||||||
| Pantherophis guttatus | t | t | *** | t | t | *** | t | x | t | t | t | [9] | ||||||
| Philodryas baroni | [54] | |||||||||||||||||
| Philodryas chamissonis | [58] | |||||||||||||||||
| Philodryas olfersii | t | T | x | [51,59] | ||||||||||||||
| Philodryas patagoniensis | [54] | |||||||||||||||||
| Pseudoferania polylepis | [50] | |||||||||||||||||
| Rhabdophis tigrinus | x | [50] | ||||||||||||||||
| Telescopus dhara | [50,51] | |||||||||||||||||
| Thamnodynastes strigatus | t | t | [60] | |||||||||||||||
| Thrasops jacksonii | x | [51] | ||||||||||||||||
| Trimorphodon biscutatus | x | [54,61] | ||||||||||||||||
| Xenodon merremi | t * | This work | ||||||||||||||||
| Species | Enzymatic | Non-Enzymatic | Reference | ||||||
|---|---|---|---|---|---|---|---|---|---|
| svLIPA | PLA2 (IIE) | PLB | svMMP | EGFr | Lacta | LIPO | Vefico | ||
| Boiga dendrophila | [42,43] | ||||||||
| Boiga irregularis | t | t | t | t | [12,44,45] | ||||
| Borikenophis portoricensis | [46,47] | ||||||||
| Cerberus rynchops | TP | [48] | |||||||
| Coelognathus radiatus | [49] | ||||||||
| Dispholidus typus | x | x | [50,51,52] | ||||||
| Erythrolamprus miliaris | t | T | T | This work; [50] | |||||
| Erythrolamprus poecilogyrus | x | [50,51] | |||||||
| Helicops angulatus | [53] | ||||||||
| Hypsiglena sp. | t | [13] | |||||||
| Hypsiglena torquata | [54] | ||||||||
| Leoiheterodon madagascarensis | x | [50] | |||||||
| Macropisthodon rudis | [55] | ||||||||
| Opheodrys aestivus | t | t | t | [9] | |||||
| Oxybelis fulgidus | [56] | ||||||||
| Oxyrhopus guibei | t | T | t | t | T | T | t | This work | |
| Phalotris mertensi | TP | t | tP | t | [57] | ||||
| Pantherophis guttatus | t | t | t | [9] | |||||
| Philodryas baroni | [54] | ||||||||
| Philodryas chamissonis | [58] | ||||||||
| Philodryas olfersii | t | T | t | t | [51,59] | ||||
| Philodryas patagoniensis | [54] | ||||||||
| Pseudoferania polylepis | x | [50] | |||||||
| Rhabdophis tigrinus | TB | x | [50] | ||||||
| Telescopus dhara | [50,51] | ||||||||
| Thamnodynastes strigatus | TP | T | TP | [60] | |||||
| Thrasops jacksonii | [51] | ||||||||
| Trimorphodon biscutatus | x | [54,61] | |||||||
| Xenodon merremi | This work | ||||||||
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Junqueira-de-Azevedo, I.L.M.; Campos, P.F.; Ching, A.T.C.; Mackessy, S.P. Colubrid Venom Composition: An -Omics Perspective. Toxins 2016, 8, 230. https://doi.org/10.3390/toxins8080230
Junqueira-de-Azevedo ILM, Campos PF, Ching ATC, Mackessy SP. Colubrid Venom Composition: An -Omics Perspective. Toxins. 2016; 8(8):230. https://doi.org/10.3390/toxins8080230
Chicago/Turabian StyleJunqueira-de-Azevedo, Inácio L. M., Pollyanna F. Campos, Ana T. C. Ching, and Stephen P. Mackessy. 2016. "Colubrid Venom Composition: An -Omics Perspective" Toxins 8, no. 8: 230. https://doi.org/10.3390/toxins8080230
APA StyleJunqueira-de-Azevedo, I. L. M., Campos, P. F., Ching, A. T. C., & Mackessy, S. P. (2016). Colubrid Venom Composition: An -Omics Perspective. Toxins, 8(8), 230. https://doi.org/10.3390/toxins8080230