Selenium, Selenoprotein Genes and Crohn’s Disease in a Case-Control Population from Auckland, New Zealand
Abstract
:1. Introduction
2. Experimental Section
3. Results
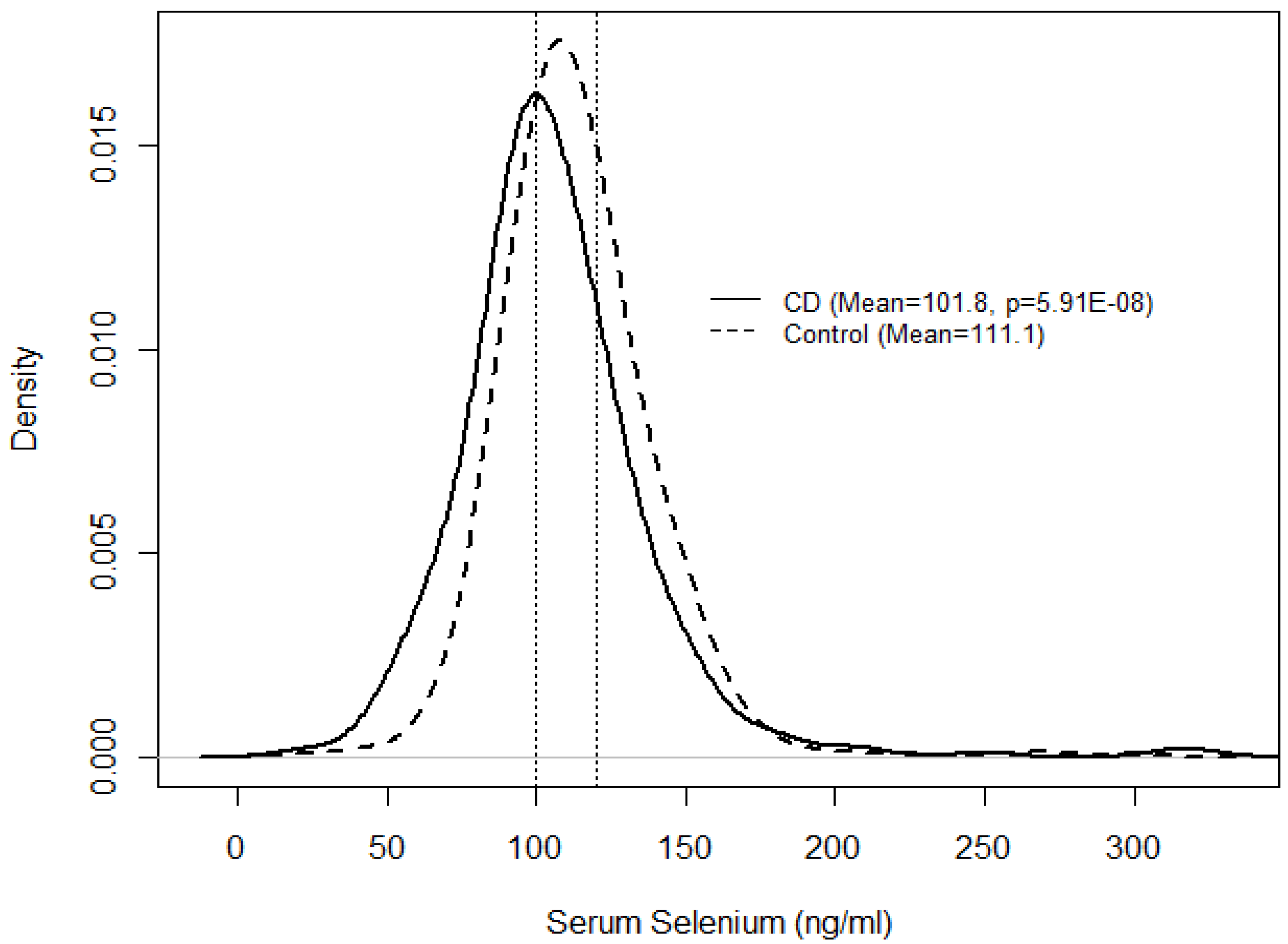
| N (%) | Serum selenium (ng/mL) | |||
|---|---|---|---|---|
| Mean (SE) | OR (95% CI) | p | ||
| CD | 351 (29.2) | 101.8 (1.02) | 0.25 (0.15–0.42) | 5.91 × 10−8 |
| Control | 853 (70.8) | 111.1 (1.01) | 1.00 | |
| Grouped serum selenium (ng/mL) | CD N (%) | Control N (%) | OR (95% CI) | p | OR (95% CI) | p |
|---|---|---|---|---|---|---|
| Low (<100.5) | 170 (48.4) | 234 (27.5) | 2.35(1.73–3.20) | 5.10 × 10−12 | 2.61(1.92–3.55) | 8.67 × 10−10 |
| Medium (100.5–118.5) | 89 (25.4) | 320 (37.6) | 0.9(0.65–1.25) | 0.996 | 1.00 | |
| High (>118.5) | 92 (26.2) | 298 (35.0) | 1.00 |
| N (%) * | Mean serum selenium (ng/mL) (SE) | Estimate (95% CI) | p | |
|---|---|---|---|---|
| Age at first diagnosis | ||||
| 0–16 years | 43 (12.9) | 91.2 (1.04) | 0.821 (0.765–0.881) | 6.57 × 10−8 |
| 17–40 years | 239 (71.5) | 103.4 (1.02) | 0.931 (0.899–0.965) | 7.56 × 10−5 |
| Over 40 years | 52 (15.6) | 104.5 (1.05) | 0.941 (0.881–1.006) | 0.073 |
| CD location | ||||
| Ileal | 122 (36.5) | 101.2 (1.03) | 0.911 (0.870–0.954) | 7.80 × 10−5 |
| Colonic | 108 (32.3) | 107.9 (1.03) | 0.971 (0.927–1.018) | 0.226 |
| Ileocolonic | 104 (31.2) | 97.0 (1.03) | 0.873 (0.832–0.916) | 4.10 × 10−8 |
| CD Behaviour | ||||
| Inflammatory | 182 (54.7) | 103.5 (1.02) | 0.932 (0.897–0.969) | 3.22 × 10−4 |
| Stricturing | 110 (33.0) | 97.5 (1.04) | 0.878 (0.836–0.923) | 2.85 × 10−7 |
| Penetrating | 41 (12.3) | 106.8 (1.03) | 0.962 (0.895–1.033) | 0.285 |
| CD location/Behaviour | ||||
| Colonic/Inflammatory | 78 (60.5) | 109.3 (1.03) | 0.984 (0.932–1.039) | 0.559 |
| Ileal/Stricturing | 51 (39.5) | 103.5 (1.06) | 0.932 (0.870–0.998) | 0.043 |
| Any relative with IBD (Yes) | 101 (30.2) | 105.1 (1.03) | 0.947 (0.902–0.993) | 0.026 |
| Bowel resection (Yes) | 119 (35.5) | 99.4 (1.03) | 0.895 (0.854–0.938) | 3.49 × 10−6 |
| Any EIMs (Yes) | 63 (18.8) | 106.6 (1.04) | 0.960 (0.905–1.019) | 0.182 |
| Perianal disease (Yes) | 48 (14.4) | 100.3 (1.04) | 0.903 (0.844–0.966) | 0.003 |
| Smoker at diagnosis (CD) or recruitment (Controls) | ||||
| CD | 106 (45.1) | 100.0 (1.03) | 0.905 (0.850–0.958) | 6.54 × 10−4 |
| Control | 224 (29.3) | 110.5 (1.02) | 1.00 |
| Gene | Name | Chr | SNP | Tested allele |
|---|---|---|---|---|
| DIO1 | deiodinase, iodothyronine, type I | 1 | rs12095080 | A |
| rs2294511 | A | |||
| rs2294512 | A | |||
| rs731828 | A | |||
| DIO2 | deiodinase, iodothyronine, type II | 14 | rs10136454 | C |
| rs12885300 | C | |||
| rs225011 | C | |||
| rs225012 | G | |||
| rs225014 | C | |||
| rs1800668 | C | |||
| GPX1 | glutathione peroxidase 1 | 3 | rs3763015 | A |
| GPX3 | glutathione perosidase 3 (plasma) | 5 | rs3792796 | C |
| rs3792797 | A | |||
| rs3828599 | A | |||
| rs8177412 | C | |||
| rs8177425 | C | |||
| rs870407 | A | |||
| rs10752294 | C | |||
| SEPHS1 | selenophosphate synthetase 1 | 10 | rs11258337 | A |
| rs17529609 | G | |||
| rs7901303 | T | |||
| rs11937742 | A | |||
| SEPSECS | Sep ( O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase | 4 | rs13123725 | A |
| rs1553153 | A | |||
| rs17480524 | C | |||
| rs2302565 | C | |||
| rs7666342 | A | |||
| rs1548357 | T | |||
| TXNRD2 | thioredoxin reductase 2 | 22 | rs5748469 | A |
| Interaction serum selenium with Gene | Tested allele | CD | ||
|---|---|---|---|---|
| Gene | SNP | Estimate (SE) | p | |
| GPX1 | rs1800668 | C | −1.685 (0.762) | 0.0270 |
| GPX3 | rs3792797 | A | −1.526 (0.755) | 0.0431 |
| DIO1 | rs12095080 | A | −1.902 (0.852) | 0.0255 |
| DIO1 | rs2294511 | A | −0.942 (0.480) | 0.0497 |
| DIO1 | rs2294512 | A | −1.005 (0.486) | 0.0388 |
| DIO2 | rs225011 | C | −1.242 (0.555) | 0.0253 |
| DIO2 | rs225012 | G | −1.179 (0.552) | 0.0327 |
| SEPHS1 | rs17529609 | G | −1.740 (0.617) | 0.0048 * |
| SEPHS1 | rs7901303 | T | −1.117 (0.420) | 0.0078 * |
| SEPSECS | rs11937742 | A | −1.000 (0.435) | 0.0214 |
| SEPSECS | rs1553153 | A | −1.754 (0.623) | 0.0048 * |
| SEPSECS | rs2302565 | C | −0.891 (0.428) | 0.0374 |
| TXNRD2 | rs1548357 | T | −0.792 (0.403) | 0.0490 |
4. Discussion
5. Conclusions
Acknowledgments
Conflict of Interest
References
- Cho, J.H.; Brant, S.R. Recent insights into the genetics of inflammatory bowel disease. Gastroenterology 2011, 140, 1704–1712. [Google Scholar] [CrossRef]
- Han, D.Y.; Fraser, A.G.; Dryland, P.; Ferguson, L.R. Environmental factors in the development of chronic inflammation: A case-control study on risk factors for Crohn’s disease within New Zealand. Mutat. Res. 2010, 690, 116–122. [Google Scholar] [CrossRef]
- Petermann, I.; Huebner, C.; Browning, B.L.; Gearry, R.B.; Barclay, M.L.; Kennedy, M.; Roberts, R.; Shelling, A.N.; Philpott, M.; Han, D.Y.; et al. Interactions among genes influencing bacterial recognition increase IBD risk in a population-based New Zealand cohort. Hum. Immunol. 2009, 70, 440–446. [Google Scholar] [CrossRef]
- Ferguson, L.R.; Han, D.Y.; Fraser, A.G.; Huebner, C.; Lam, W.J.; Morgan, A.R.; Duan, H.; Karunasinghe, N. Genetic factors in chronic inflammation: Single nucleotide polymorphisms in the STAT-JAK pathway, susceptibility to DNA damage and Crohn’s disease in a New Zealand population. Mutat. Res. 2010, 690, 108–115. [Google Scholar] [CrossRef]
- Foster, C.B.; Aswath, K.; Chanock, S.J.; McKay, H.F.; Peters, U. Polymorphism analysis of six selenoprotein genes: Support for a selective sweep at the glutathione peroxidase 1 locus (3p21) in Asian populations. BMC Genet. 2006, 7, 56. [Google Scholar]
- Ringstad, J.; Kildebo, S.; Thomassen, Y. Serum selenium, copper, and zinc concentrations in Crohn’s disease and ulcerative colitis. Scand. J. Gastroenterol. 1993, 28, 605–608. [Google Scholar] [CrossRef]
- Rayman, M.P. The importance of selenium to human health. Lancet 2000, 356, 233–241. [Google Scholar] [CrossRef] [Green Version]
- Ashrafi, M.R.; Shams, S.; Nouri, M.; Mohseni, M.; Shabanian, R.; Yekaninejad, M.S.; Chegini, N.; Khodadad, A.; Safaralizadeh, R. A probable causative factor for an old problem: Selenium and glutathione peroxidase appear to play important roles in epilepsy pathogenesis. Epilepsia 2007, 48, 1750–1755. [Google Scholar] [CrossRef]
- Zhang, S.; Rocourt, C.; Cheng, W.H. Selenoproteins and the aging brain. Mech. Ageing Dev. 2010, 131, 253–260. [Google Scholar] [CrossRef]
- De Jong, N.; Gibson, R.S.; Thomson, C.D.; Ferguson, E.L.; McKenzie, J.E.; Green, T.J.; Horwath, C.C. Selenium and zinc status are suboptimal in a sample of older New Zealand women in a community-based study. J. Nutr. 2001, 131, 2677–2684. [Google Scholar]
- Thomson, C.D.; McLachlan, S.K.; Parnell, W.R.; Wilson, N.; Wohlers, M.; Scragg, R.; Schaaf, D.; Fitzgerald, E.D. Serum selenium concentrations and dietary selenium intake of New Zealand children aged 5-14 years. Br. J. Nutr. 2007, 97, 357–364. [Google Scholar] [CrossRef]
- Gentschew, L.; Ferguson, L.R. Role of nutrition and microbiota in susceptibility to inflammatory bowel diseases. Mol. Nutr. Food Res. 2012, 56, 524–535. [Google Scholar] [CrossRef]
- Rayman, M.P. Food-chain selenium and human health: Emphasis on intake. Br. J. Nutr. 2008, 100, 254–268. [Google Scholar]
- Thomson, C.D. Selenium and iodine intakes and status in New Zealand and Australia. Br. J. Nutr. 2004, 91, 661–672. [Google Scholar] [CrossRef]
- Thomson, C.D.; Robinson, M.F. Selenium content of foods consumed in Otago, New Zealand. N. Z. Med. J. 1990, 103, 130–135. [Google Scholar]
- Thomson, B.M.; Vannoort, R.W.; Haslemore, R.M. Dietary exposure and trends of exposure to nutrient elements iodine, iron, selenium and sodium from the 2003-4 New Zealand Total Diet Survey. Br. J. Nutr. 2008, 99, 614–625. [Google Scholar]
- Fairweather-Tait, S.J.; Bao, Y.; Broadley, M.R.; Collings, R.; Ford, D.; Hesketh, J.E.; Hurst, R. Selenium in human health and disease. Antioxid. Redox Signal. 2011, 14, 1337–1383. [Google Scholar] [CrossRef]
- Wastney, M.E.; Combs, G.F., Jr.; Canfield, W.K.; Taylor, P.R.; Patterson, K.Y.; Hill, A.D.; Moler, J.E.; Patterson, B.H. A human model of selenium that integrates metabolism from selenite and selenomethionine. J. Nutr. 2011, 141, 708–717. [Google Scholar] [CrossRef]
- Satsangi, J.; Silverberg, M.S.; Vermeire, S.; Colombel, J.F. The Montreal classification of inflammatory bowel disease: Controversies, consensus, and implications. Gut 2006, 55, 749–753. [Google Scholar] [CrossRef]
- Silverberg, M.S.; Satsangi, J.; Ahmad, T.; Arnott, I.D.; Bernstein, C.N.; Brant, S.R.; Caprilli, R.; Colombel, J.F.; Gasche, C.; Geboes, K.; et al. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: Report of a Working Party of the 2005 Montreal World Congress of Gastroenterology. Can. J. Gastroenterol. 2005, 19, 5–36. [Google Scholar]
- Morgan, A.R.; Lam, W.J.; Han, D.Y.; Fraser, A.G.; Ferguson, L.R. Association analysis of ULK1 with Crohn’s Disease in a New Zealand population. Gastroenterol. Res. Pract. 2012, 2012. [Google Scholar] [CrossRef]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef]
- Morgan, A.R.; Han, D.Y.; Lam, W.J.; Fraser, A.G.; Ferguson, L.R. Association analysis of 3p21 with Crohn’s disease in a New Zealand population. Hum. Immunol. 2010, 71, 602–609. [Google Scholar] [CrossRef]
- The R Project for Statistical Computing Website. Available online: http://www.R-project.org/ (accessed on 30 September 2011).
- SAS Institute, SAS Software, version 9.2, SAS Institute: Cary, NC, USA, 2008.
- Ferguson, L.R.; Browning, B.L.; Huebner, C.; Petermann, I.; Shelling, A.N.; Demmers, P.; McCulloch, A.; Gearry, R.B.; Barclay, M.L.; Philpott, M. Single nucleotide polymorphisms in human Paneth cell defensin A5 may confer susceptibility to inflammatory bowel disease in a New Zealand Caucasian population. Dig. Liver Dis. 2008, 40, 723–730. [Google Scholar]
- Ferguson, L.R.; Han, D.Y.; Fraser, A.G.; Huebner, C.; Lam, W.J.; Morgan, A.R. IL23R and IL12B SNPs and haplotypes strongly associate with Crohn’s Disease risk in a New Zealand population. Gastroenterol. Res. Pract. 2010, 2010. [Google Scholar] [CrossRef]
- Morgan, A.R.; Lam, W.J.; Han, D.Y.; Fraser, A.G.; Ferguson, L.R. Genetic variation within TLR10 is associated with Crohn’s Disease in a New Zealand population. Hum. Immunol. 2012, 73, 416–420. [Google Scholar] [CrossRef]
- Ojuawo, A.; Keith, L. The serum concentrations of zinc, copper and selenium in children with inflammatory bowel disease. Cent. Afr. J. Med. 2002, 48, 116–119. [Google Scholar]
- Thomas, A.G.; Miller, V.; Shenkin, A.; Fell, G.S.; Taylor, F. Selenium and glutathione peroxidase status in paediatric health and gastrointestinal disease. J. Pediatr. Gastroenterol. Nutr. 1994, 19, 213–219. [Google Scholar] [CrossRef]
- Karunasinghe, N.; Ryan, J.; Tuckey, J.; Masters, J.; Jamieson, M.; Clarke, L.C.; Marshall, J.R.; Ferguson, L.R. DNA stability and serum selenium levels in a high-risk group for prostate cancer. Cancer Epidemiol. Biomarkers Prev. 2004, 13, 391–397. [Google Scholar]
- Kuroki, F.; Matsumoto, T.; Iida, M. Selenium is depleted in Crohn’s disease on enteral nutrition. Dig. Dis. 2003, 21, 266–270. [Google Scholar] [CrossRef]
- Rayman, M.P. Selenium and human health. Lancet 2012, 379, 1256–1268. [Google Scholar] [CrossRef]
- Lippman, S.M.; Klein, E.A.; Goodman, P.J.; Lucia, M.S.; Thompson, I.M.; Ford, L.G.; Parnes, H.L.; Minasian, L.M.; Gaziano, J.M.; Hartline, J.A.; et al. Effect of selenium and vitamin E on risk of prostate cancer and other cancers: The Selenium and Vitamin E Cancer Prevention Trial (SELECT). JAMA 2009, 301, 39–51. [Google Scholar]
- Rayman, M.P. Selenoproteins and human health: Insights from epidemiological data. Biochim. Biophys. Acta 1790, 1533–1540. [Google Scholar]
- Abraham, C.; Cho, J.H. Inflammatory bowel disease. N. Engl. J. Med. 2009, 361, 2066–2078. [Google Scholar] [CrossRef]
- Sopori, M. Effects of cigarette smoke on the immune system. Nat. Rev. Immunol. 2002, 2, 372–377. [Google Scholar] [CrossRef]
- Vander Heide, F.; Dijkstra, A.; Weersma, R.K.; Albersnagel, F.A.; van der Logt, E.M.; Faber, K.N.; Sluiter, W.J.; Kleibeuker, J.H.; Dijkstra, G. Effects of active and passive smoking on disease course of Crohn’s disease and ulcerative colitis. Inflamm. Bowel Dis. 2009, 15, 1199–1207. [Google Scholar] [CrossRef]
- Brigelius-Flohé, R.; Kipp, A.P. Physiological functions of GPx2 and its role in inflammation-triggered carcinogenesis. Ann. N. Y. Acad. Sci. 1259, 19–25. [Google Scholar]
- Hawkes, W.C.; Alkan, Z. Regulation of redox signaling by selenoproteins. Biol. Trace Elem. Res. 2010, 134, 235–251. [Google Scholar] [CrossRef]
- Papp, L.V.; Holmgren, A.; Khanna, K.K. Selenium and selenoproteins in health and disease. Antioxid. Redox Signal. 2010, 12, 793–795. [Google Scholar] [CrossRef]
- Hansen, R.D.; Krath, B.N.; Frederiksen, K.; Tjonneland, A.; Overvad, K.; Roswall, N.; Loft, S.; Dragsted, L.O.; Vogel, U.; Raaschou-Nielsen, O. GPX1 Pro198Leu polymorphism, erythrocyte GPX activity, interaction with alcohol consumption and smoking, and risk of colorectal cancer. Mutat. Res. 2009, 664, 13–19. [Google Scholar] [CrossRef]
- Boelen, A.; Kwakkel, J.; Alkemade, A.; Renckens, R.; Kaptein, E.; Kuiper, G.; Wiersinga, W.M.; Visser, T.J. Induction of type 3 deiodinase activity in inflammatory cells of mice with chronic local inflammation. Endocrinology 2005, 146, 5128–5134. [Google Scholar] [CrossRef]
- Debski, M.G.; Pachucki, J.; Ambroziak, M.; Olszewski, W.; Bar-Andziak, E. Human breast cancer tissue expresses high level of type 1 5′-deiodinase. Thyroid 2007, 17, 3–10. [Google Scholar] [CrossRef]
- Olivares, E.L.; Carvalho, D.P. Thyroid hormone metabolism in heart failure: Iodothyronine deiodinases in focus. Curr. Opin. Endocrinol. Diabetes Obes. 2010, 17, 414–417. [Google Scholar] [CrossRef]
- Zou, L.; Burmeister, L.A.; Styren, S.D.; Kochanek, P.M.; DeKosky, S.T. Up-regulation of type 2 iodothyronine deiodinase mRNA in reactive astrocytes following traumatic brain injury in the rat. J. Neurochem. 1998, 71, 887–890. [Google Scholar]
- Berger, M.M.; Reymond, M.J.; Shenkin, A.; Rey, F.; Wardle, C.; Cayeux, C.; Schindler, C.; Chioléro, R.L. Influence of selenium supplements on the post-traumatic alterations of the thyroid axis: A placebo-controlled trial. Intensive Care Med. 2001, 27, 91–100. [Google Scholar] [CrossRef]
- Gaertner, R. Selenium and thyroid hormone axis in critical ill states: An overview of conflicting view points. J. Trace Elem. Med.Biol. 2009, 23, 71–74. [Google Scholar] [CrossRef]
- Ravaglia, G.; Forti, P.; Maioli, F.; Nesi, B.; Pratelli, L.; Savarino, L.; Cucinotta, D.; Cavalli, G. Blood micronutrient and thyroid hormone concentrations in the oldest-old. J. Clin. Endocrinol. Metab. 2000, 85, 2260–2265. [Google Scholar] [CrossRef]
- Anastasilakis, A.D.; Toulis, K.A.; Nisianakis, P.; Goulis, D.G.; Kampas, L.; Valeri, R.M.; Oikonomou, D.; Tzellos, T.G.; Delaroudis, S. Selenomethionine treatment in patients with autoimmune thyroiditis: A prospective, quasi-randomised trial. Int. J. Clin. Pract. 2012, 66, 378–383. [Google Scholar] [CrossRef]
- Koenig, R.J. Regulation of type 1 iodothyronine deiodinase in health and disease. Thyroid 2005, 15, 835–840. [Google Scholar] [CrossRef]
- Guyot, H.; de Oliveira, L.A.; Ramery, E.; Beckers, J.F.; Rollin, F. Effect of a combined iodine and selenium supplementation on I and Se status of cows and their calves. J. Trace Elem. Med. Biol. 2011, 25, 118–124. [Google Scholar] [CrossRef]
- Xu, J.; Liu, X.L.; Yang, X.F.; Guo, H.L.; Zhao, L.N.; Sun, X.F. Supplemental selenium alleviates the toxic effects of excessive iodine on thyroid. Biol. Trace Elem. Res. 2011, 141, 110–118. [Google Scholar] [CrossRef]
- Qin, F.; Zhu, X.; Zhang, W.; Zhou, J.F.; Zhang, S.; Jia, Z. Effects of dietary iodine and selenium on nutrient digestibility, serum thyroid hormones, and antioxidant status of Liaoning Cashmere Goats. Biol. Trace Elem. Res. 2011, 143, 1480–1488. [Google Scholar] [CrossRef]
- Xu, X.-M.; Carlson, B.; Zhang, Y.; Mix, H.; Kryukov, G.; Glass, R.; Berry, M.; Gladyshev, V.; Hatfield, D. New developments in selenium biochemistry: Selenocysteine biosynthesis in eukaryotes and archaea. Biol. Trace Elem. Res. 2007, 119, 234–241. [Google Scholar] [CrossRef]
- Kim, J.Y.; Lee, K.H.; Shim, M.S.; Shin, H.; Xu, X.-M.; Carlson, B.A.; Hatfield, D.L.; Lee, B.J. Human selenophosphate synthetase 1 has five splice variants with unique interactions, subcellular localizations and expression patterns. Biochem. Biophys. Res. Commun. 2010, 397, 53–58. [Google Scholar] [CrossRef]
- Chung, H.J.; Yoon, S.I.; Shin, S.H.; Koh, Y.A.; Lee, S.J.; Lee, Y.S.; Bae, S. p53-mediatedenhancement of radiosensitivity by selenophosphate synthetase 1 overexpression. J. Cell. Physiol. 2006, 209, 131–141. [Google Scholar] [CrossRef]
- Zimmer, V.; Widmann, T.; Muller, M.; Ong, M.F.; Stein, J.M.; Pfreundschuh, M.; Lammert, F.; Roemer, K.; Assmann, G. Genotypic interaction and gender specificity of common genetic variants in the p53/mdm2 network in Crohn’s disease. Digestion 2010, 81, 246–251. [Google Scholar] [CrossRef]
- Pozza, A.; Scarpa, M.; Ruffolo, C.; Polese, L.; Erroi, F.; Bridda, A.; Norberto, L.; Frego, M. Colonic carcinogenesis in IBD: Molecular events. Ann. Ital. Chir. 2011, 82, 19–28. [Google Scholar]
- Ullman, T.A.; Itzkowitz, S.H. Intestinal inflammation and cancer. Gastroenterology 2011, 140, 1807–1816. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Gentschew, L.; Bishop, K.S.; Han, D.Y.; Morgan, A.R.; Fraser, A.G.; Lam, W.J.; Karunasinghe, N.; Campbell, B.; Ferguson, L.R. Selenium, Selenoprotein Genes and Crohn’s Disease in a Case-Control Population from Auckland, New Zealand. Nutrients 2012, 4, 1247-1259. https://doi.org/10.3390/nu4091247
Gentschew L, Bishop KS, Han DY, Morgan AR, Fraser AG, Lam WJ, Karunasinghe N, Campbell B, Ferguson LR. Selenium, Selenoprotein Genes and Crohn’s Disease in a Case-Control Population from Auckland, New Zealand. Nutrients. 2012; 4(9):1247-1259. https://doi.org/10.3390/nu4091247
Chicago/Turabian StyleGentschew, Liljana, Karen S. Bishop, Dug Yeo Han, Angharad R. Morgan, Alan G. Fraser, Wen Jiun Lam, Nishi Karunasinghe, Bobbi Campbell, and Lynnette R. Ferguson. 2012. "Selenium, Selenoprotein Genes and Crohn’s Disease in a Case-Control Population from Auckland, New Zealand" Nutrients 4, no. 9: 1247-1259. https://doi.org/10.3390/nu4091247







