Drosophila: Retrotransposons Making up Telomeres
Abstract
:1. Introduction
2. Telomere Transposition
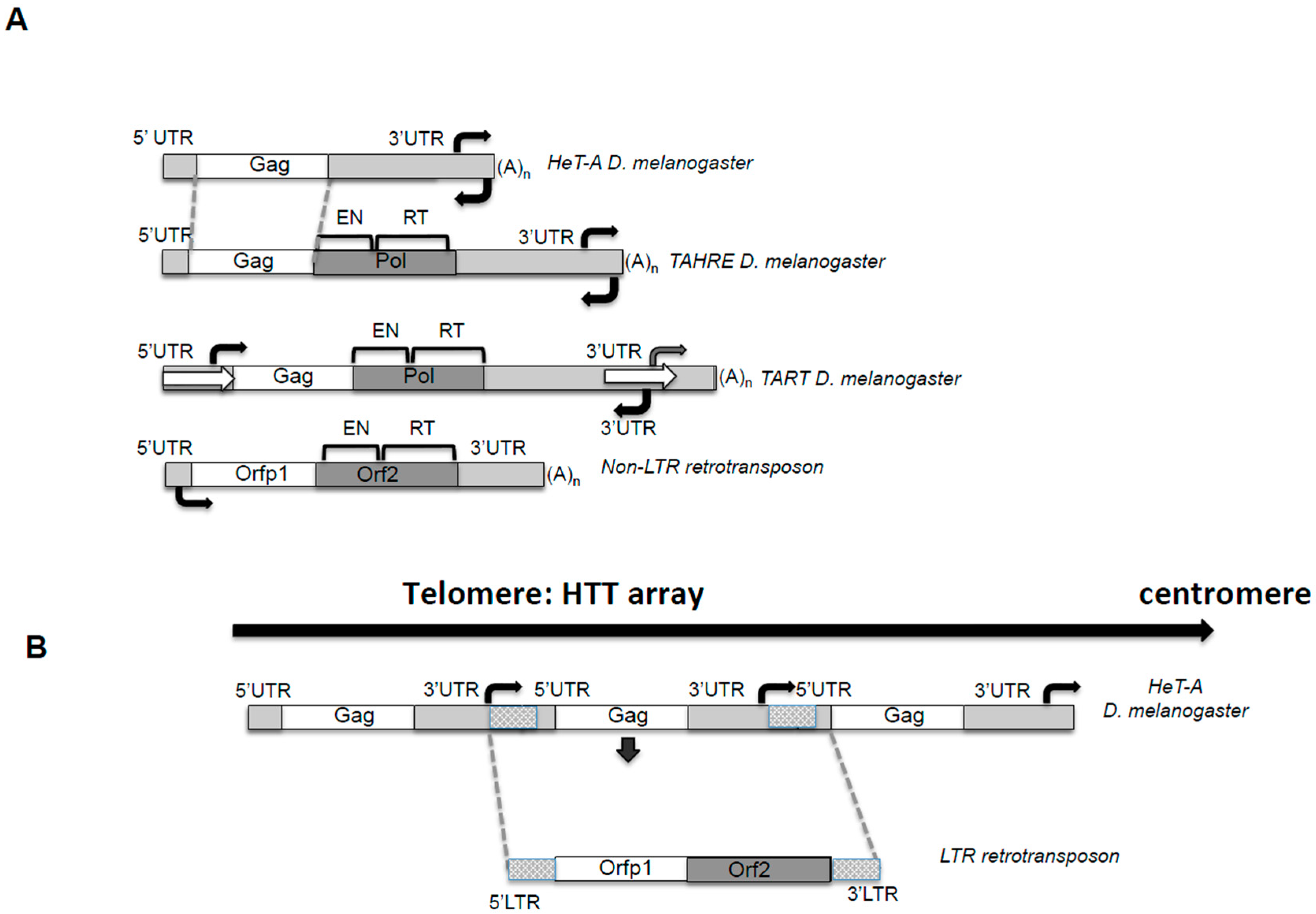
2.1. The Players
2.1.1. The UTRs and the Bidirectional Transcription
2.1.2. The Unusual Length of the 3′UTRs and Its Bias Composition
2.1.3. Coding Capacities of the HTT Elements
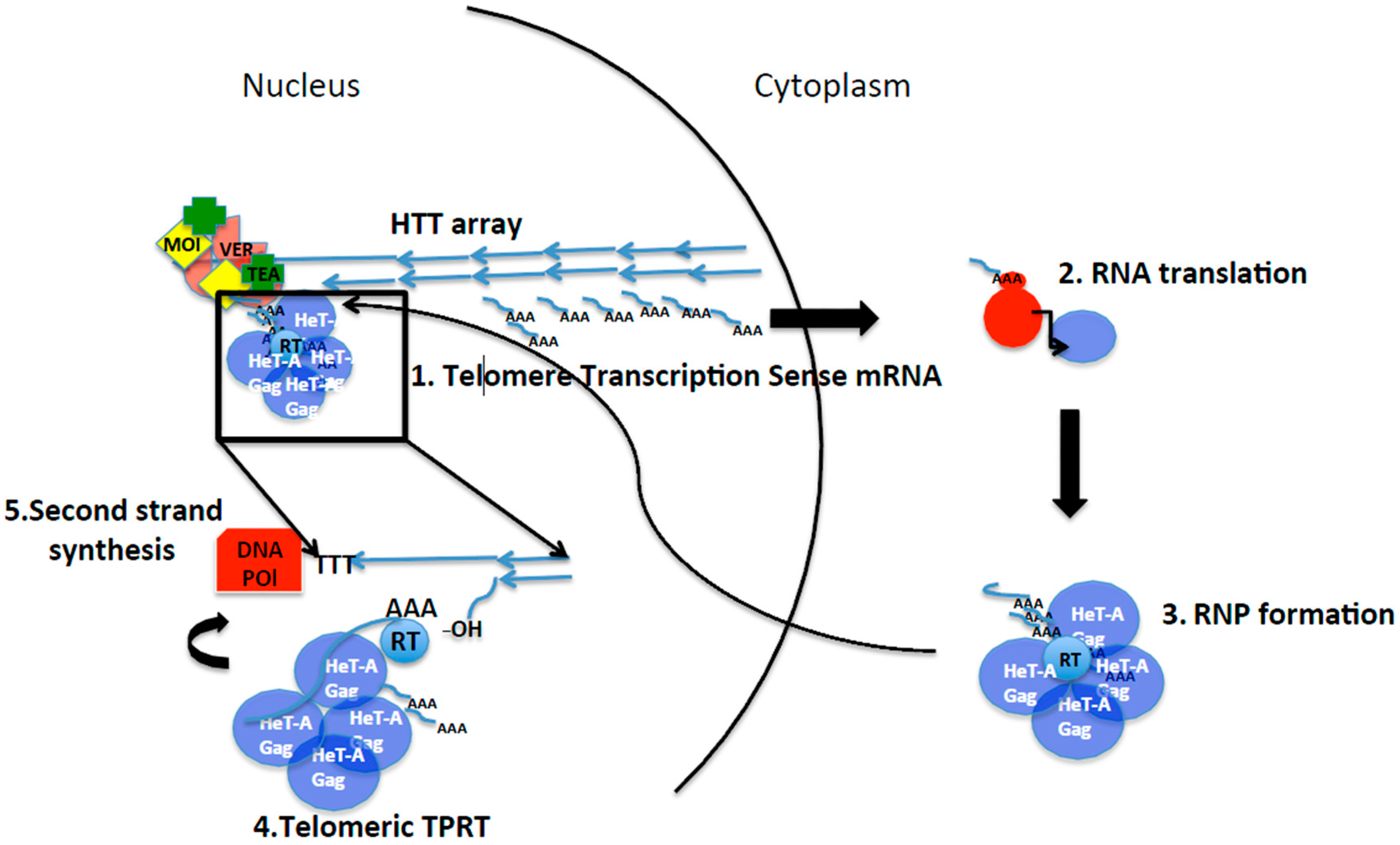
2.2. Telomere Transposition and Life Cycle
Terminal Target Primed Reverse Transcription (TPRT)
2.3. Telomere Targeting
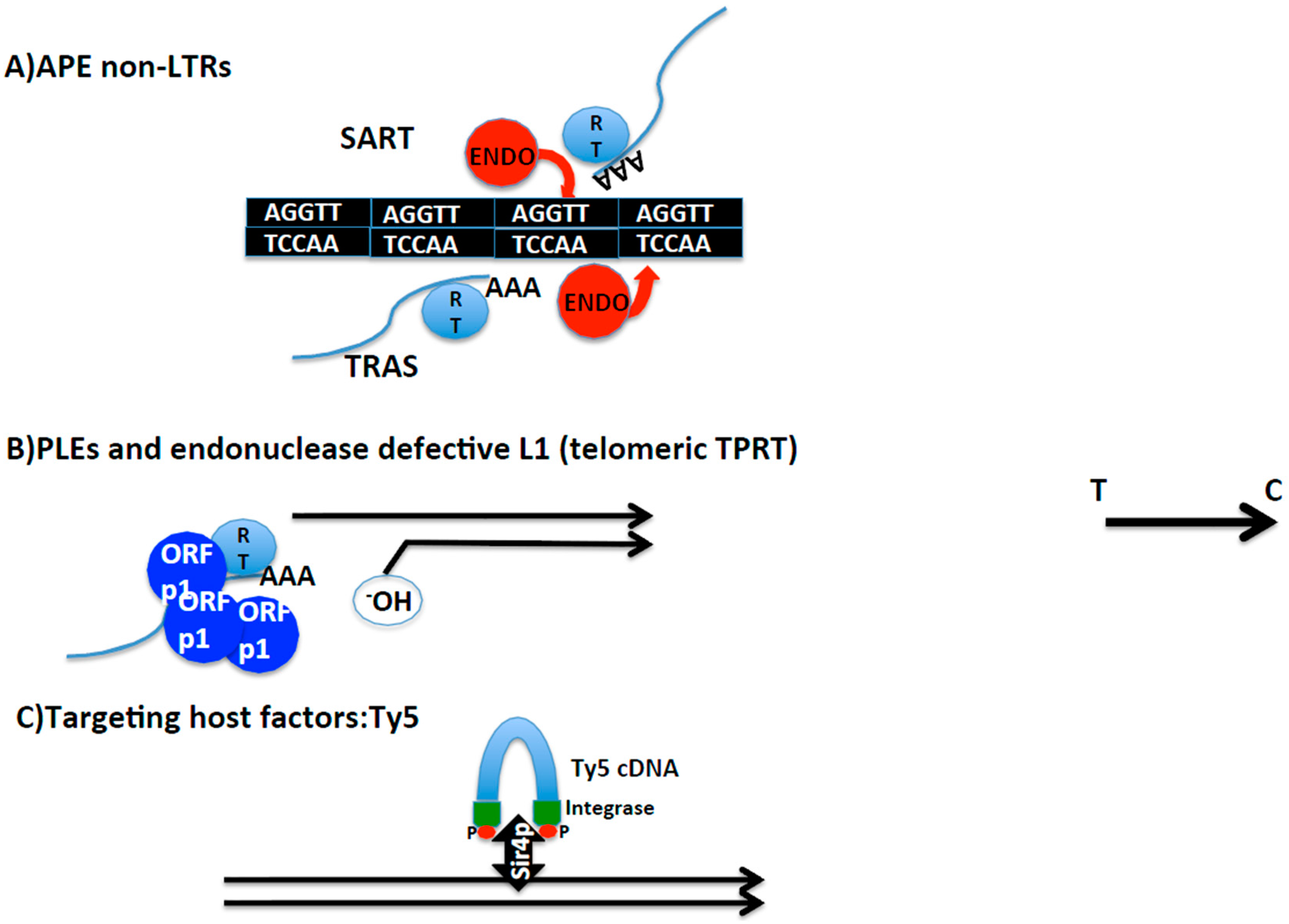
2.4. Other Cases of Telomere Transposition and Integration
2.4.1. Transposition Inside Telomere Repeats
2.4.2. Targeting of Telomere Chromatin
2.4.3. Occasional End-Transposition
3. Similarities between the Telomeric Retrotransposons in Drosophila and Telomerase
4. Evolutionary Considerations; Telomeres and Mobile Elements
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Muller, H.J. The remaking of chromosomes. Collect. Net 1938, 8, 182–198. [Google Scholar]
- McClintock, B. The Behavior in Successive Nuclear Divisions of a Chromosome Broken at Meiosis. Proc. Natl. Acad. Sci. USA 1939, 25, 405–416. [Google Scholar] [CrossRef] [PubMed]
- De Lange, T.; Blackburn, E.H.; Lundblad, V. Telomeres; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2006. [Google Scholar]
- Pardue, M.L.; DeBaryshe, P.G. Retrotransposons provide an evolutionarily robust non-telomerase mechanism to maintain telomeres. Annu. Rev. Genet. 2003, 37, 485–511. [Google Scholar] [CrossRef] [PubMed]
- George, J.A.; Pardue, M.L. The promoter of the heterochromatic Drosophila telomeric retrotransposon, HeT-A, is active when moved into euchromatic locations. Genetics 2003, 163, 625–635. [Google Scholar] [PubMed]
- Silva-Sousa, R.L.; López-Panadès, E.; Casacuberta, E. Drosophila telomeres: An example of co-evolution with transposable elements. Genome Dyn. 2012, 7, 46–67. [Google Scholar] [CrossRef] [PubMed]
- Traverse, K.L.; George, J.A.; Debaryshe, P.G.; Pardue, M.L. Evolution of species-specific promoter-associated mechanisms for protecting chromosome ends by Drosophila Het-A telomeric transposons. Proc. Natl. Acad. Sci. USA 2010, 107, 5064–5069. [Google Scholar] [CrossRef] [PubMed]
- Abad, J.P.; De Pablos, B.; Osoegawa, K.; De Jong, P.J.; Martin-Gallardo, A.; Villasante, A. TAHRE, a novel telomeric retrotransposon from Drosophila melanogaster, reveals the origin of Drosophila telomeres. Mol. Biol. Evol. 2004, 21, 1620–1624. [Google Scholar] [CrossRef] [PubMed]
- Pardue, M.L.; DeBaryshe, P.G. Drosophila telomeres: A variation on the telomerase theme. Fly 2008, 2, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Rong, Y.S. Retrotransposons at Drosophila telomeres: Host domestication of a selfish element for the maintenance of genome integrity. Biochim. Biophys. Acta 2012, 1819, 771–775. [Google Scholar] [CrossRef] [PubMed]
- Servant, G.; Deininger, P.L. Insertion of Retrotransposons at Chromosome Ends: Adaptive Response to Chromosome Maintenance. Front Genet. 2015, 6, 358. [Google Scholar] [CrossRef] [PubMed]
- Raffa, G.D.; Cenci, G.; Ciapponi, L.; Gatti, M. Organization and Evolution of Drosophila Terminin: Similarities and Differences between Drosophila and Human Telomeres. Front. Oncol. 2013, 3, 112. [Google Scholar] [CrossRef] [PubMed]
- Rong, Y.S. Telomere capping in Drosophila: Dealing with chromosome ends that most resemble DNA breaks. Chromosoma 2008, 117, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Cui, M.; Rong, Y.S. MTV sings jubilation for telomere biology in Drosophila. Fly 2017, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Silva-Sousa, R.; López-Panadès, E.; Pineyro, D.; Casacuberta, E. The chromosomal proteins JIL-1 and Z4/Putzig regulate the telomeric chromatin in Drosophila melanogaster. PLoS Genet. 2012, 8, e1003153. [Google Scholar] [CrossRef] [PubMed]
- Rashkova, S.; Karam, S.E.; Kellum, R.; Pardue, M.L. Gag proteins of the two Drosophila telomeric retrotransposons are targeted to chromosome ends. J. Cell Biol. 2002, 159, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Villasante, A.; de Pablos, B.; Méndez-Lago, M.; Abad, J.P. Telomere maintenance in Drosophila: Rapid transposon evolution at chromosome ends. Cell Cycle 2008, 7, 2134–2138. [Google Scholar] [CrossRef] [PubMed]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Mason, J.M.; Biessmann, H. The unusual telomeres of Drosophila. Trends Genet. 1995, 11, 58–62. [Google Scholar] [CrossRef]
- Casacuberta, E.; Pardue, M.L. HeT-A and TART, two Drosophila retrotransposons with a bona fide role in chromosome structure for more than 60 million years. Cytogenet. Genome Res. 2005, 110, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Danilevskaya, O.N.; Arkhipova, I.R.; Traverse, K.L.; Pardue, M.L. Promoting in tandem: The promoter for telomere transposon HeT-A and implications for the evolution of retroviral LTRs. Cell 1997, 88, 647–655. [Google Scholar] [CrossRef]
- Casacuberta, E.; Pardue, M.L. Transposon telomeres are widely distributed in the Drosophila genus: TART elements in the virilis group. Proc. Natl. Acad. Sci. USA 2003, 100, 3363–3368. [Google Scholar] [CrossRef] [PubMed]
- George, J.A.; Traverse, K.L.; DeBaryshe, P.G.; Kelley, K.J.; Pardue, M.L. Evolution of diverse mechanisms for protecting chromosome ends by Drosophila TART telomere retrotransposons. Proc. Natl. Acad. Sci. USA 2010, 107, 21052–21057. [Google Scholar] [CrossRef] [PubMed]
- Shpiz, S.; Kwon, D.; Rozovsky, Y.; Kalmykova, A. rasiRNA pathway controls antisense expression of Drosophila telomeric retrotransposons in the nucleus. Nucleic Acids Res. 2009, 37, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Pineyro, D.; López-Panadès, E.; Lucena-Pérez, M.; Casacuberta, E. Transcriptional analysis of the HeT-A retrotransposon in mutant and wild type stocks reveals high sequence variability at Drosophila telomeres and other unusual features. BMC Genom. 2011, 12, 573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azzalin, C.M.; Reichenbach, P.; Khoriauli, L.; Giulotto, E.; Lingner, J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 2007, 318, 798–801. [Google Scholar] [CrossRef] [PubMed]
- Rashkova, S.; Athanasiadis, A.; Pardue, M.L. Intracellular targeting of Gag proteins of the Drosophila telomeric retrotransposons. J. Virol. 2003, 77, 6376–6384. [Google Scholar] [CrossRef] [PubMed]
- Rashkova, S.; Karam, S.E.; Pardue, M.L. Element-specific localization of Drosophila retrotransposon Gag proteins occurs in both nucleus and cytoplasm. Proc. Natl. Acad. Sci. USA 2002, 99, 3621–3626. [Google Scholar] [CrossRef] [PubMed]
- Casacuberta, E.; Marin, F.A.; Pardue, M.L. Intracellular targeting of telomeric retrotransposon Gag proteins of distantly related Drosophila species. Proc. Natl. Acad. Sci. USA 2007, 104, 8391–8396. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Beaucher, M.; Cheng, Y.; Rong, Y.S. Coordination of transposon expression with DNA replication in the targeting of telomeric retrotransposons in Drosophila. EMBO J. 2014, 33, 1148–1158. [Google Scholar] [CrossRef] [PubMed]
- López-Panadès, E.; Casacuberta, E. NAP-1, Nucleosome assembly protein 1, a histone chaperone involved in Drosophila telomeres. Insect Biochem. Mol. Biol. 2016, 70, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Kapitonov, V.V.; Jurka, J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat. Rev. Genet. 2008, 9, 411–412. [Google Scholar] [CrossRef] [PubMed]
- Beaucher, M.; Zheng, X.F.; Amariei, F.; Rong, Y.S. Multiple pathways suppress telomere addition to DNA breaks in the Drosophila germline. Genetics 2012, 191, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Shpiz, S.; Kwon, D.; Uneva, A.; Kim, M.; Klenov, M.; Rozovsky, Y.; Georgiev, P.; Savitsky, M.; Kalmykova, A. Characterization of Drosophila telomeric retroelement TAHRE: Transcription, transpositions, and RNAi-based regulation of expression. Mol. Biol. Evol. 2007, 24, 2535–2545. [Google Scholar] [CrossRef] [PubMed]
- Casacuberta, E.; Pardue, M.L. HeT-A elements in Drosophila virilis: Retrotransposon telomeres are conserved across the Drosophila genus. Proc. Natl. Acad. Sci. USA 2003, 100, 14091–14096. [Google Scholar] [CrossRef] [PubMed]
- Casacuberta, E.; Pardue, M.L. Coevolution of the telomeric retrotransposons across Drosophila species. Genetics 2002, 161, 1113–1124. [Google Scholar] [PubMed]
- Villasante, A.; Abad, J.P.; Planello, R.; Méndez-Lago, M.; Celniker, S.E.; de Pablos, B. Drosophila telomeric retrotransposons derived from an ancestral element that was recruited to replace telomerase. Genome Res. 2007, 17, 1909–1918. [Google Scholar] [CrossRef] [PubMed]
- Raffa, G.D.; Siriaco, G.; Cugusi, S.; Ciapponi, L.; Cenci, G.; Wojcik, E.; Gatti, M. The Drosophila modigliani (moi) gene encodes a HOAP-interacting protein required for telomere protection. Proc. Natl. Acad. Sci. USA 2009, 106, 2271–2276. [Google Scholar] [CrossRef] [PubMed]
- Raffa, G.D.; Raimondo, D.; Sorino, C.; Cugusi, S.; Cenci, G.; Cacchione, S.; Gatti, M.; Ciapponi, L. Verrocchio, a Drosophila OB fold-containing protein, is a component of the terminin telomere-capping complex. Genes Dev. 2010, 24, 1596–1601. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Walser, J.C.; Beaucher, M.L.; Morciano, P.; Wesolowska, N.; Chen, J.; Rong, Y.S. HipHop interacts with HOAP and HP1 to protect Drosophila telomeres in a sequence-independent manner. EMBO J. 2010, 29, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Jakubczak, J.L.; Burke, W.D.; Eickbush, T.H. Retrotransposable elements R1 and R2 interrupt the rRNA genes of most insects. Proc. Natl. Acad. Sci. USA 1991, 88, 3295–3299. [Google Scholar] [CrossRef] [PubMed]
- Kojima, K.K.; Fujiwara, H. Long-term inheritance of the 28S rDNA-specific retrotransposon R2. Mol. Biol. Evol. 2005, 22, 2157–2165. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, N.; Lutz-Prigge, S.; Moran, J.V. Genomic deletions created upon LINE-1 retrotransposition. Cell 2002, 110, 315–325. [Google Scholar] [CrossRef]
- Luan, D.D.; Korman, M.H.; Jakubczak, J.L.; Eickbush, T.H. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: A mechanism for non-LTR retrotransposition. Cell 1993, 72, 595–605. [Google Scholar] [CrossRef]
- Fujiwara, H. Site-specific non-LTR retrotransposons. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Gilbert, N.; Ooi, S.L.; Lawler, J.F.; Ostertag, E.M.; Kazazian, H.H.; Boeke, J.D.; Moran, J.V. Human L1 retrotransposition: Cis preference versus trans complementation. Mol. Cell Biol. 2001, 21, 1429–1439. [Google Scholar] [CrossRef] [PubMed]
- Mason, J.M.; Frydrychova, R.C.; Biessmann, H. Drosophila telomeres: An exception providing new insights. Bioessays 2008, 30, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Savitsky, M.; Kravchuk, O.; Melnikova, L.; Georgiev, P. Heterochromatin protein 1 is involved in control of telomere elongation in Drosophila melanogaster. Mol. Cell Biol. 2002, 22, 3204–3218. [Google Scholar] [CrossRef] [PubMed]
- Cicconi, A.; Micheli, E.; Verni, F.; Jackson, A.; Gradilla, A.C.; Cipressa, F.; Raimondo, D.; Bosso, G.; Wakefield, J.G.; Ciapponi, L.; et al. The Drosophila telomere-capping protein Verrocchio binds single-stranded DNA and protects telomeres from DNA damage response. Nucleic Acids Res. 2017, 45, 3068–3085. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, L.; Tang, X.; Bhardwaj, S.R.; Ji, J.; Rong, Y.S. MTV, an ssDNA Protecting Complex Essential for Transposon-Based Telomere Maintenance in Drosophila. PLoS Genet. 2016, 12, e1006435. [Google Scholar] [CrossRef] [PubMed]
- DeZwaan, D.C.; Freeman, B.C. The conserved Est1 protein stimulates telomerase DNA extension activity. Proc. Natl. Acad. Sci. USA 2009, 106, 17337–17342. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Shtessel, L.; Brady, M.M.; Ahmed, S. Caenorhabditis elegans POT-2 telomere protein represses a mode of alternative lengthening of telomeres with normal telomere lengths. Proc. Natl. Acad. Sci. USA 2012, 109, 7805–7810. [Google Scholar] [CrossRef] [PubMed]
- Silva-Sousa, R.; Varela, M.D.; Casacuberta, E. The putzig partners DREF,TRF2 and Ken are invoved in the regulation of the Drosophila telomere retrotransposons, HeT-A and TART. Mob. DNA 2013, 4–18. [Google Scholar] [CrossRef] [Green Version]
- Silva-Sousa, R.; Casacuberta, E. The JIL-1 kinase affects telomere expression in the different telomere domains of Drosophila. PLoS ONE 2013, 8, e81543. [Google Scholar] [CrossRef] [PubMed]
- Osanai-Futahashi, M.; Fujiwara, H. Coevolution of telomeric repeats and telomeric repeat-specific non-LTR retrotransposons in insects. Mol. Biol. Evol. 2011, 28, 2983–2986. [Google Scholar] [CrossRef] [PubMed]
- Arkhipova, I.R.; Yushenova, I.A.; Rodriguez, F. Giant reverse transcriptase-encoding transposable elements at telomeres. Mol. Biol. Evol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Osanai, M.; Kojima, K.K.; Futahashi, R.; Yaguchi, S.; Fujiwara, H. Identification and characterization of the telomerase reverse transcriptase of Bombyx mori (silkworm) and Tribolium castaneum (flour beetle). Gene 2006, 376, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Arkhipova, I.R.; Morrison, H.G. Three retrotransposon families in the genome of Giardia lamblia: Two telomeric, one dead. Proc. Natl. Acad. Sci. USA 2001, 98, 14497–14502. [Google Scholar] [CrossRef] [PubMed]
- Sultana, T.; Zamborlini, A.; Cristofari, G.; Lesage, P. Integration site selection by retroviruses and transposable elements in eukaryotes. Nat. Rev. Genet. 2017, 18, 292–308. [Google Scholar] [CrossRef] [PubMed]
- Gai, X.; Voytas, D.F. A single amino acid change in the yeast retrotransposon Ty5 abolishes targeting to silent chromatin. Mol. Cell 1998, 1, 1051–1055. [Google Scholar] [CrossRef]
- Xie, W.; Gai, X.; Zhu, Y.; Zappulla, D.C.; Sternglanz, R.; Voytas, D.F. Targeting of the yeast Ty5 retrotransposon to silent chromatin is mediated by interactions between integrase and Sir4p. Mol. Cell Biol. 2001, 21, 6606–6614. [Google Scholar] [CrossRef] [PubMed]
- McClintock, B. The significance of responses of the genome to challenge. Science 1984, 226, 792–801. [Google Scholar] [CrossRef] [PubMed]
- Evgen’ev, M.B.; Arkhipova, I.R. Penelope-like elements—A new class of retroelements: Distribution, function and possible evolutionary significance. Cytogenet. Genome Res. 2005, 110, 510–521. [Google Scholar] [CrossRef] [PubMed]
- Gladyshev, E.A.; Arkhipova, I.R. Telomere-associated endonuclease-deficient Penelope-like retroelements in diverse eukaryotes. Proc. Natl. Acad. Sci. USA 2007, 104, 9352–9357. [Google Scholar] [CrossRef] [PubMed]
- Arkhipova, I.R.; Yushenova, I.A.; Rodriguez, F. Endonuclease-containing Penelope retrotransposons in the bdelloid rotifer Adineta vaga exhibit unusual structural features and play a role in expansion of host gene families. Mob. DNA 2013, 4, 19. [Google Scholar] [CrossRef] [PubMed]
- Morrish, T.A.; Bekbolysnov, D.; Velliquette, D.; Morgan, M.; Ross, B.; Wang, Y.; Chaney, B.; McQuigg, J.; Fager, N.; Maine, I.P. Multiple Mechanisms Contribute to Telomere Maintenance. J. Cancer Biol. Res. 2013, 1, 1012. [Google Scholar] [PubMed]
- Kheimar, A.; Previdelli, R.L.; Wight, D.J.; Kaufer, B.B. Telomeres and Telomerase: Role in Marek’s Disease Virus Pathogenesis, Integration and Tumorigenesis. Viruses 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Arbuckle, J.H.; Medveczky, P.G. The molecular biology of human herpesvirus-6 latency and telomere integration. Microbes Infect. 2011, 13, 731–741. [Google Scholar] [CrossRef] [PubMed]
- Osterrieder, N.; Wallaschek, N.; Kaufer, B.B. Herpesvirus Genome Integration into Telomeric Repeats of Host Cell Chromosomes. Annu. Rev. Virol. 2014, 1, 215–235. [Google Scholar] [CrossRef] [PubMed]
- Wallaschek, N.; Sanyal, A.; Pirzer, F.; Gravel, A.; Mori, Y.; Flamand, L.; Kaufer, B.B. The Telomeric Repeats of Human Herpesvirus 6A (HHV-6A) Are Required for Efficient Virus Integration. PLoS Pathog. 2016, 12, e1005666. [Google Scholar] [CrossRef] [PubMed]
- Cenci, G.; Ciapponi, L.; Gatti, M. The mechanism of telomere protection: A comparison between Drosophila and humans. Chromosoma 2005, 114, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Diede, S.J.; Gottschling, D.E. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell 1999, 99, 723–733. [Google Scholar] [CrossRef]
- Marcand, S.; Brevet, V.; Mann, C.; Gilson, E. Cell cycle restriction of telomere elongation. Curr. Biol. 2000, 10, 487–490. [Google Scholar] [CrossRef]
- Wellinger, R.J.; Zakian, V.A. Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: Beginning to end. Genetics 2012, 191, 1073–1105. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Sfeir, A.J.; Zou, Y.; Buseman, C.M.; Chow, T.T.; Shay, J.W.; Wright, W.E. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell 2009, 138, 463–475. [Google Scholar] [CrossRef] [PubMed]
- Price, C.M.; Boltz, K.A.; Chaiken, M.F.; Stewart, J.A.; Beilstein, M.A.; Shippen, D.E. Evolution of CST function in telomere maintenance. Cell Cycle 2010, 9, 3157–3165. [Google Scholar] [CrossRef] [PubMed]
- Morgunova, V.; Akulenko, N.; Radion, E.; Olovnikov, I.; Abramov, Y.; Olenina, L.V.; Shpiz, S.; Kopytova, D.V.; Georgieva, S.G.; Kalmykova, A. Telomeric repeat silencing in germ cells is essential for early development in Drosophila. Nucleic Acids Res. 2015, 43, 8762–8773. [Google Scholar] [CrossRef] [PubMed]
- Scheibe, M.; Arnoult, N.; Kappei, D.; Buchholz, F.; Decottignies, A.; Butter, F.; Mann, M. Quantitative interaction screen of telomeric repeat-containing RNA reveals novel TERRA regulators. Genome Res. 2013, 23, 2149–2157. [Google Scholar] [CrossRef] [PubMed]
- Arkhipova, I.R.; Pyatkov, K.I.; Meselson, M.; Evgen’ev, M.B. Retroelements containing introns in diverse invertebrate taxa. Nat. Genet. 2003, 33, 123–124. [Google Scholar] [CrossRef] [PubMed]
- Curcio, M.J.; Belfort, M. The beginning of the end: Links between ancient retroelements and modern telomerases. Proc. Natl. Acad. Sci. USA 2007, 104, 9107–9108. [Google Scholar] [CrossRef] [PubMed]
- Wells, R.A.; Germino, G.G.; Krishna, S.; Buckle, V.J.; Reeders, S.T. Telomere-related sequences at interstitial sites in the human genome. Genomics 1990, 8, 699–704. [Google Scholar] [CrossRef]
© 2017 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Casacuberta, E. Drosophila: Retrotransposons Making up Telomeres. Viruses 2017, 9, 192. https://doi.org/10.3390/v9070192
Casacuberta E. Drosophila: Retrotransposons Making up Telomeres. Viruses. 2017; 9(7):192. https://doi.org/10.3390/v9070192
Chicago/Turabian StyleCasacuberta, Elena. 2017. "Drosophila: Retrotransposons Making up Telomeres" Viruses 9, no. 7: 192. https://doi.org/10.3390/v9070192



