Comprehensive Characterization of Serum MicroRNA Profile in Response to the Emerging Avian Influenza A (H7N9) Virus Infection in Humans
Abstract
:1. Introduction
2. Results and Discussion
2.1. Sample Characteristics
| Sample Characteristic | TaqMan Low-Density Array | Validation Study | ||
|---|---|---|---|---|
| Controls | Patients | Controls | Patients | |
| Number | 10 | 10 | 36 | 21 |
| Sex (Male/Female) | 6/4 | 6/4 | 19/17 | 13/8 |
| Age (Years, mean ± SD) | 52.1 ± 17.2 | 54.0 ± 19.4 | 48.6 ± 11.6 | 51.9 ± 18.7 |
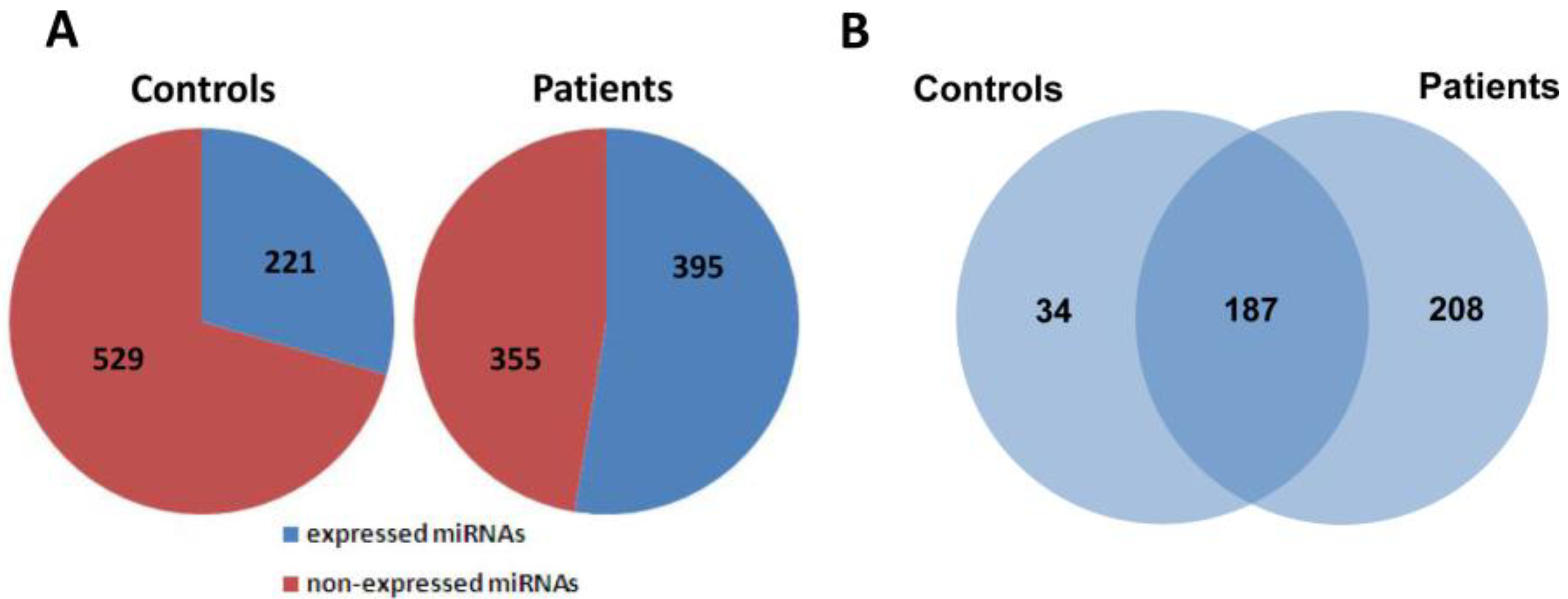
2.2. Global Analysis of Serum MiRNA Expression Profiles
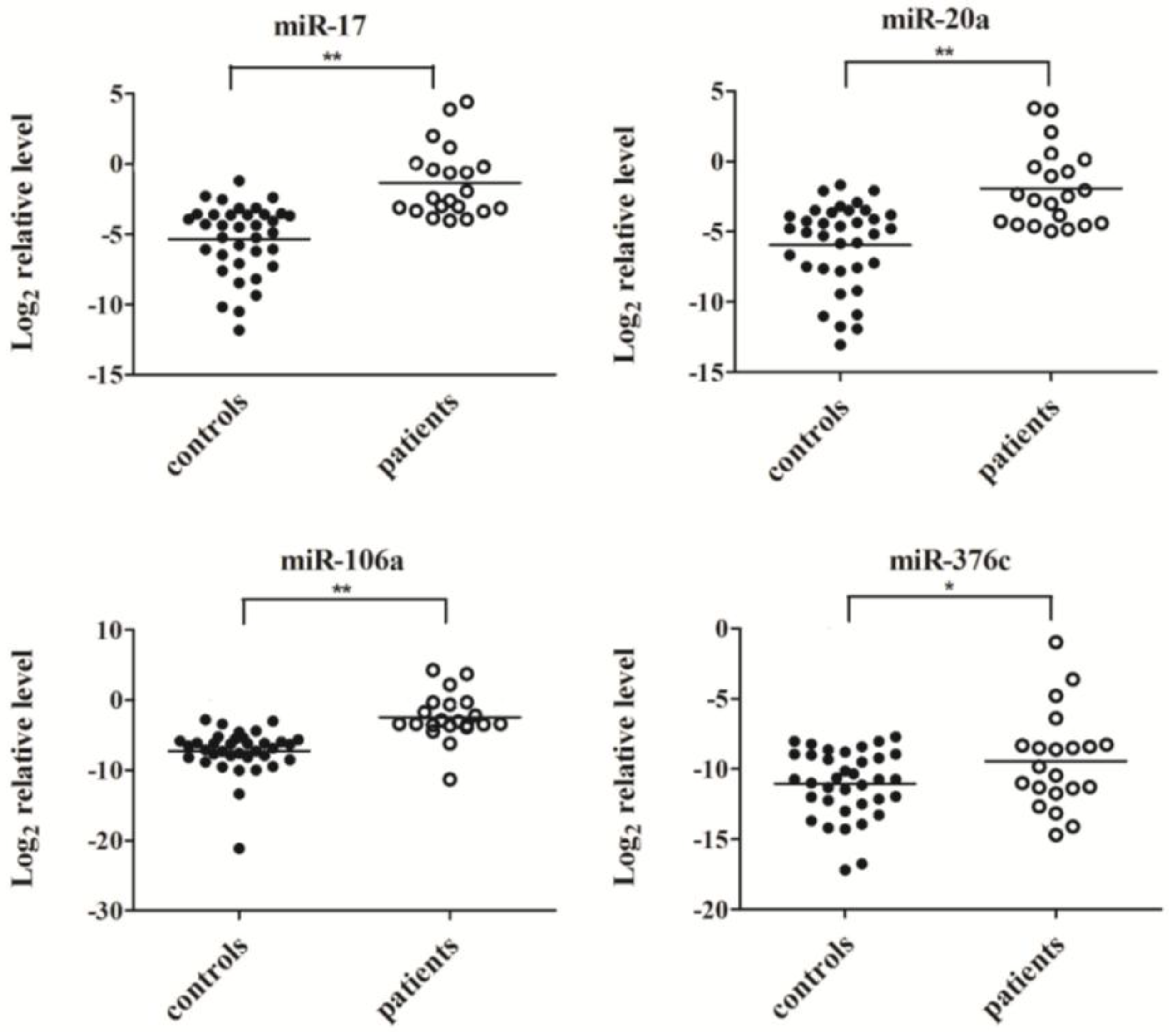
2.3. Confirmation of MiRNA Expression by Real Time RT-PCR
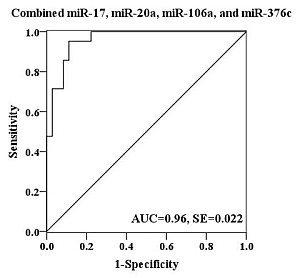
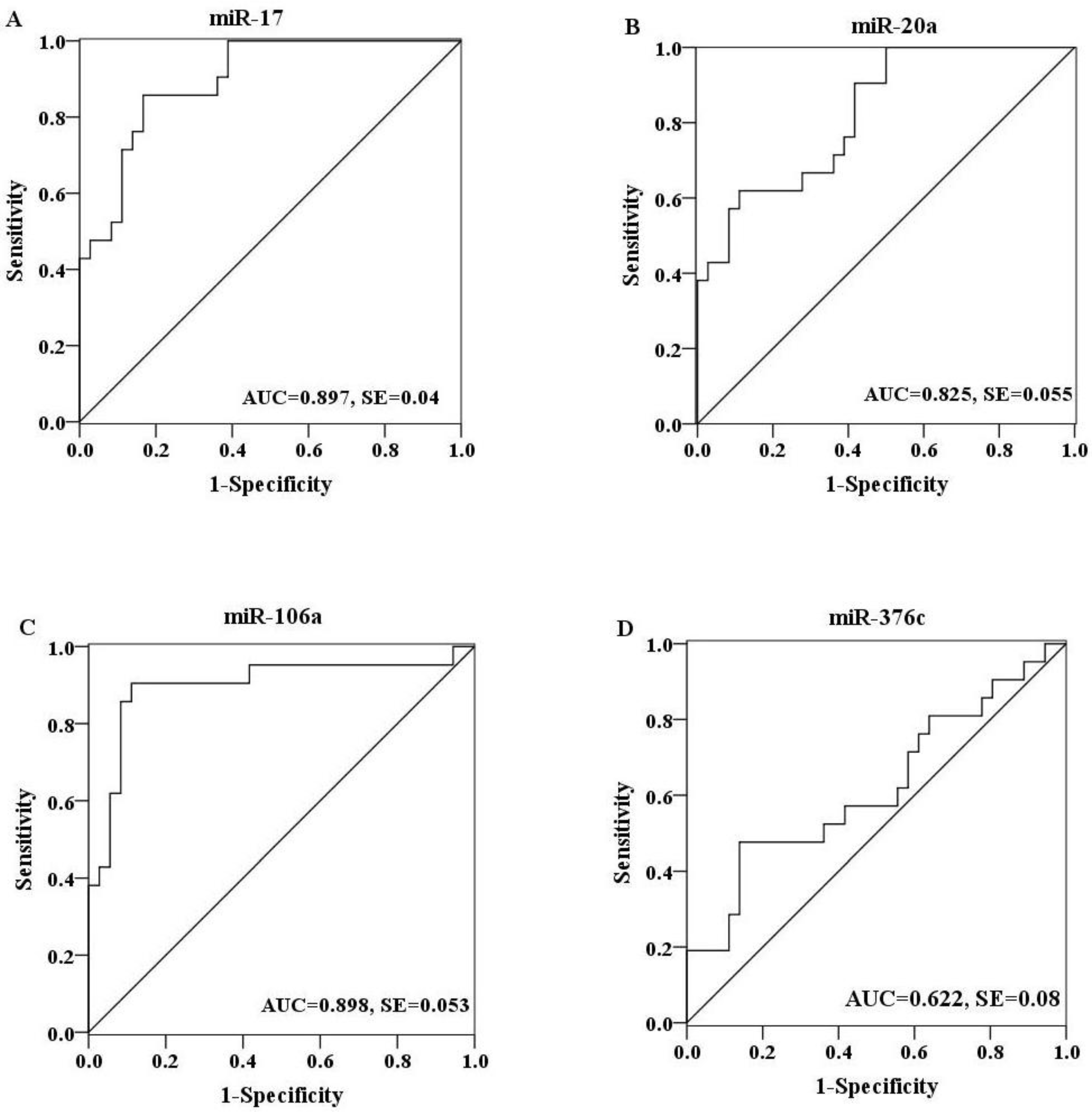
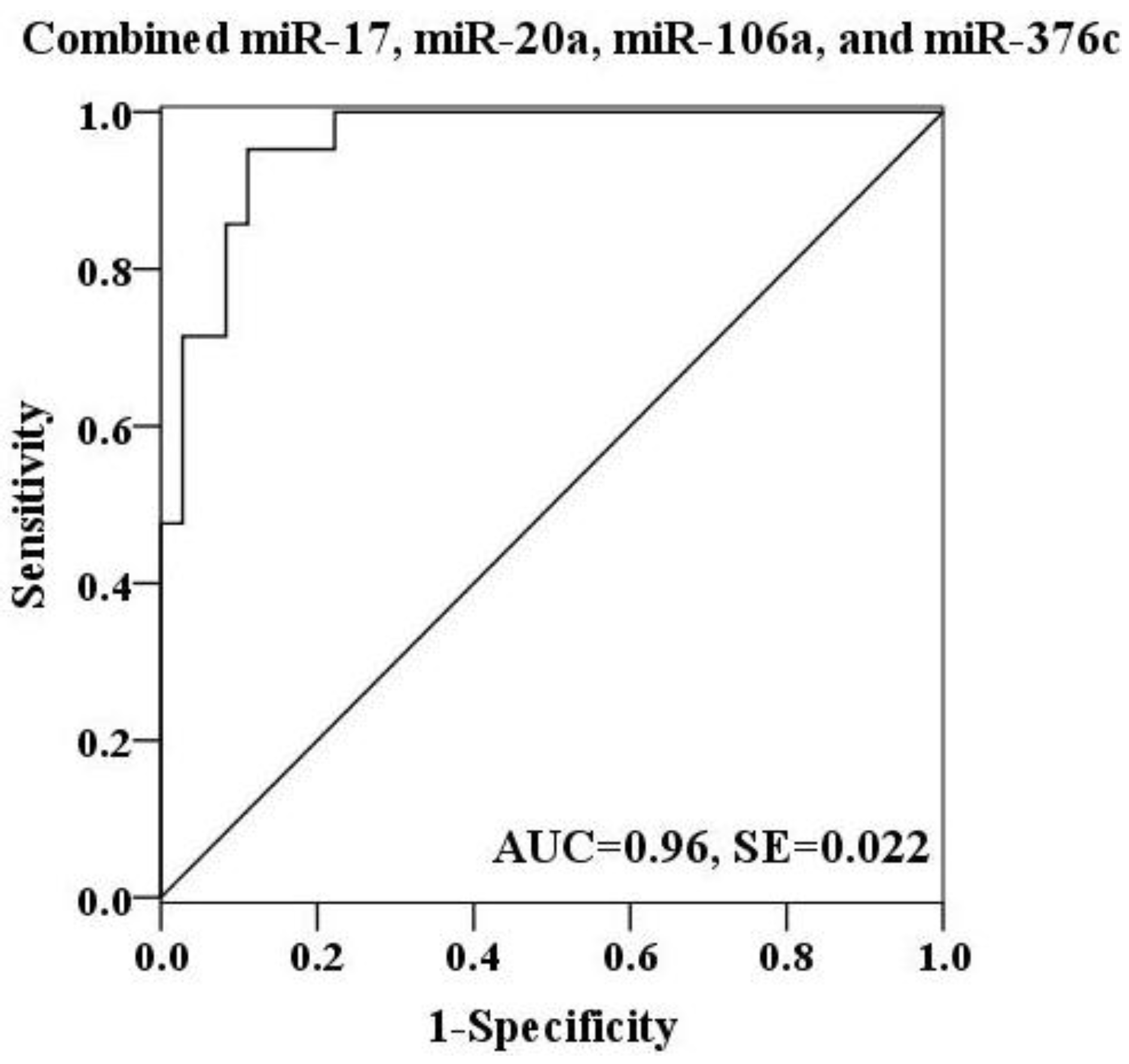
2.4. Evaluation of the Diagnostic Potential of Serum miRNAs for H7N9 Virus Infection
3. Experimental
3.1. Sample Collection
3.2. RNA Extraction
3.3. miRNA Expression Profile Examination Using TaqMan Low Density Arrays
3.4. Quantification of Candidate miRNAs by Real Time RT-PCR
3.5. Data Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References and Notes
- Gao, R.; Cao, B.; Hu, Y.; Feng, Z.; Wang, D.; Hu, W.; Chen, J.; Jie, Z.; Qiu, H.; Xu, K.; et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 2013, 368, 1888–1897. [Google Scholar] [CrossRef]
- Chen, Y.; Liang, W.; Yang, S.; Wu, N.; Gao, H.; Sheng, J.; Yao, H.; Wo, J.; Fang, Q.; Cui, D.; et al. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: Clinical analysis and characterisation of viral genome. Lancet 2013, 381, 1916–1925. [Google Scholar] [CrossRef]
- Li, Q.; Zhou, L.; Zhou, M.; Chen, Z.; Li, F.; Wu, H.; Xiang, N.; Chen, E.; Tang, F.; Wang, D.; et al. Preliminary report: Epidemiology of the avian influenza A (H7N9) outbreak in China. N. Engl. J. Med. 2013, 370, 520–532. [Google Scholar]
- World Health Organization. Number of confirmed human cases of avian influenza A (H7N9) reported to WHO, as of 25 October 2013. Available online: http://www.who.int/influenza/human_animal_interface/influenza_h7n9/10u_ReportWebH7N9Number.pdf (accessed on 25 October 2013).
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef]
- Bushati, N.; Cohen, S.M. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007, 23, 175–205. [Google Scholar] [CrossRef]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef]
- Lawrie, C.H.; Gal, S.; Dunlop, H.M.; Pushkaran, B.; Liggins, A.P.; Pulford, K.; Banham, A.H.; Pezzella, F.; Boultwood, J.; Wainscoat, J.S.; et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008, 141, 672–675. [Google Scholar] [CrossRef]
- Wu, Q.; Wang, C.; Lu, Z.; Guo, L.; Ge, Q. Analysis of serum genome-wide microRNAs for breast cancer detection. Clin. Chim. Acta 2012, 413, 1058–1065. [Google Scholar] [CrossRef]
- Hofsli, E.; Sjursen, W.; Prestvik, W.S.; Johansen, J.; Rye, M.; Trano, G.; Wasmuth, H.H.; Hatlevoll, I.; Thommesen, L. Identification of serum microRNA profiles in colon cancer. Br. J. Cancer 2013, 108, 1712–1719. [Google Scholar] [CrossRef]
- Fleischhacker, S.N.; Bauersachs, S.; Wehner, A.; Hartmann, K.; Weber, K. Differential expression of circulating microRNAs in diabetic and healthy lean cats. Vet. J. 2013, 197, 688–693. [Google Scholar] [CrossRef]
- Iborra, M.; Bernuzzi, F.; Correale, C.; Vetrano, S.; Fiorino, G.; Beltran, B.; Marabita, F.; Locati, M.; Spinelli, A.; Nos, P.; et al. Identification of serum and tissue micro-RNA expression profiles in different stages of inflammatory bowel disease. Clin. Exp. Immunol. 2013, 173, 250–258. [Google Scholar] [CrossRef]
- Hu, G.; Drescher, K.M.; Chen, X.M. Exosomal miRNAs: Biological properties and therapeutic potential. Front. Genet. 2012, 3. [Google Scholar] [CrossRef]
- Dorval, V.; Nelson, P.T.; Hebert, S.S. Circulating microRNAs in Alzheimer’s disease: The search for novel biomarkers. Front. Mol. Neurosci. 2013, 6. [Google Scholar] [CrossRef]
- Pescador, N.; Perez-Barba, M.; Ibarra, J.M.; Corbaton, A.; Martinez-Larrad, M.T.; Serrano-Rios, M. Serum circulating microRNA profiling for identification of potential type 2 diabetes and obesity biomarkers. PLoS One 2013, 8, e77251. [Google Scholar]
- Sayed, A.S.; Xia, K.; Yang, T.L.; Peng, J. Circulating microRNAs: A potential role in diagnosis and prognosis of acute myocardial infarction. Dis. Markers 2013, 35, 561–566. [Google Scholar] [CrossRef]
- Roth, C.; Kasimir-Bauer, S.; Pantel, K.; Schwarzenbach, H. Screening for circulating nucleic acids and caspase activity in the peripheral blood as potential diagnostic tools in lung cancer. Mol. Oncol. 2011, 5, 281–291. [Google Scholar] [CrossRef]
- Cuk, K.; Zucknick, M.; Heil, J.; Madhavan, D.; Schott, S.; Turchinovich, A.; Arlt, D.; Rath, M.; Sohn, C.; Benner, A.; et al. Circulating microRNAs in plasma as early detection markers for breast cancer. Int. J. Cancer 2012, 132, 1602–1612. [Google Scholar]
- Chan, M.; Liaw, C.S.; Ji, S.M.; Tan, H.H.; Wong, C.Y.; Thike, A.A.; Tan, P.H.; Ho, G.H.; Lee, A.S. Identification of circulating microRNA signatures for breast cancer detection. Clin. Cancer Res. 2013, 19, 4477–4487. [Google Scholar]
- Cui, L.; Qi, Y.; Li, H.; Ge, Y.; Zhao, K.; Qi, X.; Guo, X.; Shi, Z.; Zhou, M.; Zhu, B.; et al. Serum microRNA expression profile distinguishes enterovirus 71 and coxsackievirus 16 infections in patients with hand-foot-and-mouth disease. PLoS One 2011, 6, e27071. [Google Scholar]
- Shrivastava, S.; Petrone, J.; Steele, R.; Lauer, G.M.; di Bisceglie, A.M.; Ray, R.B. Up-regulation of circulating miR-20a is correlated with hepatitis C virus-mediated liver disease progression. Hepatology 2013, 58, 863–871. [Google Scholar] [CrossRef]
- Chanteux, H.; Guisset, A.C.; Pilette, C.; Sibille, Y. LPS induces IL-10 production by human alveolar macrophages via MAPKinases- and Sp1-dependent mechanisms. Respir. Res. 2007, 8. [Google Scholar] [CrossRef]
- World Health Organization. Real-time RT-PCR protocol for the detection of Avian Influenza A(H7N9) virus. Available online: http://www.who.int/influenza/gisrs_laboratory/cnic_realtime_rt_pcr_protocol_a_h7n9.pdf (accessed on 1 December 2013).
- Qi, Y.; Cui, L.; Ge, Y.; Shi, Z.; Zhao, K.; Guo, X.; Yang, D.; Yu, H.; Shan, Y.; Zhou, M.; et al. Altered serum microRNAs as biomarkers for the early diagnosis of pulmonary tuberculosis infection. BMC Infect. Dis. 2012, 12. [Google Scholar] [CrossRef]
- SDS Relative Quantification Software, version 2.3; Applied Biosystems: Foster City, CA, USA, 2008.
- SPSS software, version 19.0; SPSS Inc: Chicago, IL, USA, 2010.
- Redell, J.B.; Moore, A.N.; Ward, N.H., 3rd; Hergenroeder, G.W.; Dash, P.K. Human traumatic brain injury alters plasma microRNA levels. J. Neurotrauma 2010, 27, 2147–2156. [Google Scholar] [CrossRef]
- Bakre, A.; Mitchell, P.; Coleman, J.K.; Jones, L.P.; Saavedra, G.; Teng, M.; Tompkins, S.M.; Tripp, R.A. Respiratory syncytial virus modifies microRNAs regulating host genes that affect virus replication. J. Gen. Virol. 2012, 93, 2346–2356. [Google Scholar] [CrossRef]
- Ma, Y.J.; Yang, J.; Fan, X.L.; Zhao, H.B.; Hu, W.; Li, Z.P.; Yu, G.C.; Ding, X.R.; Wang, J.Z.; Bo, X.C.; et al. Cellular microRNA let-7c inhibits M1 protein expression of the H1N1 influenza A virus in infected human lung epithelial cells. J. Cell Mol. Med. 2012, 16, 2539–2546. [Google Scholar] [CrossRef]
- Onnis, A.; Navari, M.; Antonicelli, G.; Morettini, F.; Mannucci, S.; de Falco, G.; Vigorito, E.; Leoncini, L. Epstein-Barr nuclear antigen 1 induces expression of the cellular microRNA hsa-miR-127 and impairing B-cell differentiation in EBV-infected memory B cells. New insights into the pathogenesis of Burkitt lymphoma. Blood Cancer J. 2012, 2, e84. [Google Scholar]
- Song, L.; Liu, H.; Gao, S.; Jiang, W.; Huang, W. Cellular microRNAs inhibit replication of the H1N1 influenza A virus in infected cells. J. Virol. 2010, 84, 8849–8860. [Google Scholar] [CrossRef]
- Terrier, O.; Textoris, J.; Carron, C.; Marcel, V.; Bourdon, J.C.; Rosa-Calatrava, M. Host microRNA molecular signatures associated with human H1N1 and H3N2 influenza A viruses reveal an unanticipated antiviral activity for miR-146a. J. Gen. Virol. 2013, 94, 985–995. [Google Scholar] [CrossRef]
- Vasilescu, C.; Rossi, S.; Shimizu, M.; Tudor, S.; Veronese, A.; Ferracin, M.; Nicoloso, M.S.; Barbarotto, E.; Popa, M.; Stanciulea, O.; et al. MicroRNA fingerprints identify miR-150 as a plasma prognostic marker in patients with sepsis. PLoS One 2009, 4, e7405. [Google Scholar] [CrossRef]
- Ji, F.; Yang, B.; Peng, X.; Ding, H.; You, H.; Tien, P. Circulating microRNAs in hepatitis B virus-infected patients. J. Viral. Hepat. 2011, 18, e242–e251. [Google Scholar] [CrossRef]
- Li, L.M.; Hu, Z.B.; Zhou, Z.X.; Chen, X.; Liu, F.Y.; Zhang, J.F.; Shen, H.B.; Zhang, C.Y.; Zen, K. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. 2010, 70, 9798–9807. [Google Scholar] [CrossRef]
- Xu, J.; Wu, C.; Che, X.; Wang, L.; Yu, D.; Zhang, T.; Huang, L.; Li, H.; Tan, W.; Wang, C.; et al. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol. Carcinog. 2011, 50, 136–142. [Google Scholar] [CrossRef]
- Tambyah, P.A.; Sepramaniam, S.; Mohamed Ali, J.; Chai, S.C.; Swaminathan, P.; Armugam, A.; Jeyaseelan, K. microRNAs in circulation are altered in response to influenza A virus infection in humans. PLoS One 2013, 8, e76811. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Ng, E.K.; Chong, W.W.; Jin, H.; Lam, E.K.; Shin, V.Y.; Yu, J.; Poon, T.C.; Ng, S.S.; Sung, J.J. Differential expression of microRNAs in plasma of patients with colorectal cancer: A potential marker for colorectal cancer screening. Gut 2009, 58, 1375–1381. [Google Scholar] [CrossRef] [Green Version]
- Huang, Z.; Huang, D.; Ni, S.; Peng, Z.; Sheng, W.; Du, X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int. J. Cancer 2010, 127, 118–126. [Google Scholar] [CrossRef]
- Chen, Y.; Li, L.; Zhou, Z.; Wang, N.; Zhang, C.Y.; Zen, K. A pilot study of serum microRNA signatures as a novel biomarker for occult hepatitis B virus infection. Med. Microbiol. Immunol. 2012, 201, 389–395. [Google Scholar] [CrossRef]
Appendix
| miRNA | △△Ct | 2−△△Ct |
|---|---|---|
| hsa-miR-1290 | −10.05 | 1063.66 |
| hsa-miR-1275 | −9.36 | 655.36 |
| hsa-miR-1260 | −9.25 | 609.09 |
| hsa-miR-574-3p | −4.49 | 22.48 |
| hsa-miR-454 | −4.45 | 21.93 |
| hsa-miR-148a | −4.38 | 20.85 |
| hsa-miR-539 | −4.36 | 20.57 |
| hsa-miR-223 | −4.27 | 19.29 |
| hsa-miR-142-5p | −4.27 | 19.26 |
| hsa-miR-485-3p | −4.21 | 18.56 |
| hsa-miR-548c-5p | −4.13 | 17.56 |
| hsa-miR-17 | −4.12 | 17.41 |
| hsa-miR-484 | −4.10 | 17.17 |
| hsa-miR-652 | −4.09 | 17.01 |
| hsa-miR-660 | −4.07 | 16.78 |
| hsa-miR-20b | −4.06 | 16.66 |
| hsa-miR-511 | −4.05 | 16.61 |
| hsa-miR-26b | −4.01 | 16.15 |
| hsa-miR-210 | −4.01 | 16.15 |
| hsa-miR-489 | −3.98 | 15.78 |
| hsa-miR-22* | −3.98 | 15.74 |
| hsa-miR-15a* | −3.97 | 15.70 |
| hsa-miR-106a | −3.92 | 15.15 |
| hsa-miR-331-5p | −3.91 | 15.04 |
| hsa-miR-194 | −3.88 | 14.73 |
| hsa-miR-139-5p | −3.85 | 14.38 |
| hsa-miR-193a-5p | −3.84 | 14.37 |
| hsa-miR-29a | −3.80 | 13.94 |
| hsa-miR-24 | −3.75 | 13.43 |
| hsa-miR-140-5p | −3.71 | 13.12 |
| hsa-miR-28-3p | −3.69 | 12.91 |
| hsa-miR-151-3p | −3.67 | 12.73 |
| hsa-miR-192 | −3.66 | 12.62 |
| hsa-miR-190b | −3.60 | 12.15 |
| hsa-miR-143 | −3.59 | 12.04 |
| hsa-miR-425 | −3.58 | 11.96 |
| hsa-miR-101 | −3.55 | 11.70 |
| hsa-miR-146b-5p | −3.52 | 11.45 |
| hsa-miR-26a | −3.47 | 11.06 |
| hsa-miR-425* | −3.46 | 11.02 |
| hsa-miR-25 | −3.45 | 10.92 |
| hsa-miR-186 | −3.44 | 10.86 |
| hsa-miR-191 | −3.42 | 10.72 |
| hsa-miR-9* | −3.42 | 10.72 |
| hsa-miR-494 | −3.41 | 10.65 |
| hsa-miR-323-3p | −3.41 | 10.64 |
| hsa-miR-34a | −3.27 | 9.65 |
| hsa-miR-125a-5p | −3.26 | 9.58 |
| hsa-miR-152 | −3.25 | 9.51 |
| hsa-miR-19b | −3.24 | 9.45 |
| hsa-miR-532-5p | −3.23 | 9.41 |
| hsa-miR-324-5p | −3.21 | 9.25 |
| hsa-miR-106b | −3.19 | 9.12 |
| hsa-miR-886-5p | −3.19 | 9.11 |
| hsa-miR-744 | −3.14 | 8.83 |
| hsa-miR-451 | −3.13 | 8.75 |
| hsa-miR-130a | −3.13 | 8.73 |
| hsa-miR-200c | −3.11 | 8.63 |
| hsa-miR-625* | −3.11 | 8.61 |
| hsa-miR-532-3p | −3.10 | 8.56 |
| hsa-miR-720 | −3.08 | 8.44 |
| hsa-miR-338-5p | −3.05 | 8.30 |
| hsa-miR-324-3p | −3.05 | 8.29 |
| hsa-miR-203 | −3.05 | 8.27 |
| hsa-miR-30a-3p | −3.01 | 8.07 |
| hsa-miR-345 | −3.01 | 8.04 |
| hsa-miR-27a | −2.92 | 7.57 |
| hsa-miR-30e-3p | −2.88 | 7.37 |
| hsa-miR-664 | −2.86 | 7.28 |
| hsa-miR-340 | −2.77 | 6.84 |
| hsa-miR-495 | −2.76 | 6.79 |
| hsa-miR-21 | −2.75 | 6.74 |
| hsa-miR-335 | −2.72 | 6.58 |
| hsa-miR-181a | −2.71 | 6.53 |
| hsa-miR-218 | −2.62 | 6.16 |
| hsa-miR-223* | −2.58 | 5.99 |
| hsa-miR-93* | −2.57 | 5.95 |
| hsa-miR-151-5p | −2.52 | 5.74 |
| hsa-miR-27b | −2.51 | 5.70 |
| hsa-miR-199a-5p | −2.47 | 5.56 |
| hsa-miR-505 | −2.47 | 5.53 |
| hsa-miR-93 | −2.38 | 5.21 |
| hsa-miR-144* | −2.34 | 5.06 |
| hsa-let-7g | −2.34 | 5.06 |
| hsa-miR-130b | −2.32 | 5.00 |
| hsa-miR-128 | −2.23 | 4.70 |
| hsa-miR-132 | −2.22 | 4.66 |
| hsa-miR-331-3p | −2.10 | 4.29 |
| hsa-miR-375 | −2.07 | 4.20 |
| hsa-miR-125b | −2.02 | 4.06 |
| hsa-miR-148b | −2.00 | 4.00 |
| hsa-miR-19b-1* | −2.00 | 3.99 |
| hsa-miR-122 | −1.90 | 3.73 |
| hsa-miR-486-5p | −1.86 | 3.64 |
| hsa-miR-409-3p | −1.85 | 3.60 |
| hsa-miR-142-3p | −1.81 | 3.50 |
| hsa-miR-150 | −1.80 | 3.48 |
| hsa-let-7b | −1.76 | 3.38 |
| hsa-miR-541* | −1.73 | 3.32 |
| hsa-miR-221 | −1.62 | 3.08 |
| hsa-miR-649 | −1.61 | 3.06 |
| hsa-miR-92a | −1.54 | 2.92 |
| hsa-miR-518f | −1.33 | 2.51 |
| has-miR-155 | −1.28 | 2.43 |
| hsa-miR-301a | −1.12 | 2.17 |
| hsa-miR-486-3p | −1.07 | 2.10 |
| hsa-miR-10a | −1.05 | 2.07 |
| hsa-miR-642 | 1.09 | 0.47 |
| hsa-miR-654-3p | 1.22 | 0.43 |
| hsa-let-7e | 1.28 | 0.41 |
| hsa-miR-548c-3p | 1.34 | 0.39 |
| hsa-miR-29b | 1.41 | 0.38 |
| hsa-miR-548p | 1.77 | 0.29 |
| hsa-miR-1243 | 9.65 | 0.00 |
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Zhu, Z.; Qi, Y.; Ge, A.; Zhu, Y.; Xu, K.; Ji, H.; Shi, Z.; Cui, L.; Zhou, M. Comprehensive Characterization of Serum MicroRNA Profile in Response to the Emerging Avian Influenza A (H7N9) Virus Infection in Humans. Viruses 2014, 6, 1525-1539. https://doi.org/10.3390/v6041525
Zhu Z, Qi Y, Ge A, Zhu Y, Xu K, Ji H, Shi Z, Cui L, Zhou M. Comprehensive Characterization of Serum MicroRNA Profile in Response to the Emerging Avian Influenza A (H7N9) Virus Infection in Humans. Viruses. 2014; 6(4):1525-1539. https://doi.org/10.3390/v6041525
Chicago/Turabian StyleZhu, Zheng, Yuhua Qi, Aihua Ge, Yefei Zhu, Ke Xu, Hong Ji, Zhiyang Shi, Lunbiao Cui, and Minghao Zhou. 2014. "Comprehensive Characterization of Serum MicroRNA Profile in Response to the Emerging Avian Influenza A (H7N9) Virus Infection in Humans" Viruses 6, no. 4: 1525-1539. https://doi.org/10.3390/v6041525