PepGMV Rep-Protein Expression in Mammalian Cells
Abstract
:1. Introduction
2. Results and Discussion
2.1. Expression Vector pTracer-SV40 and Ligation with PepGMV Rep-Protein Gene
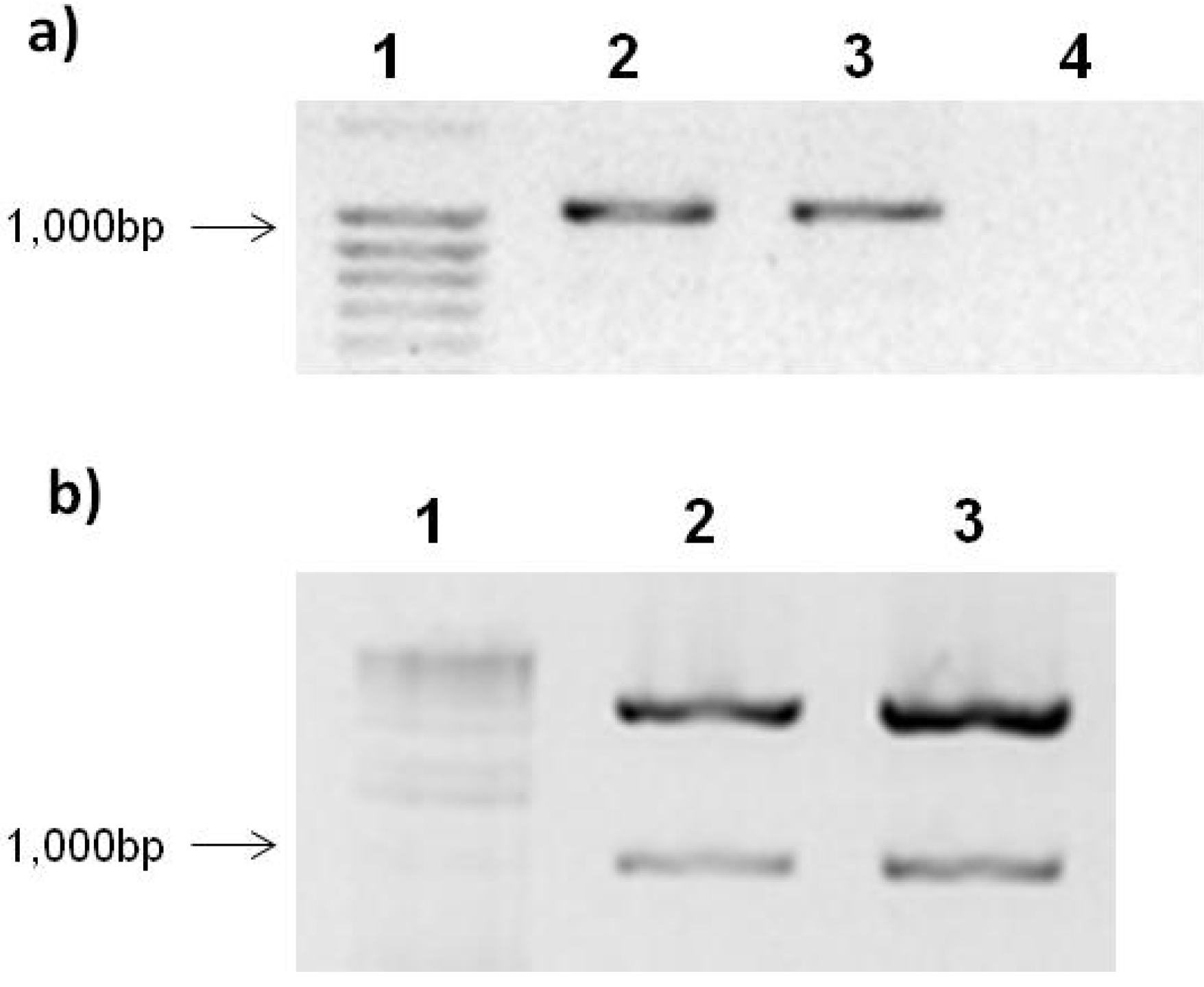
2.2. PepGMV Cell Transfection
| Oligonuceotide name | Forward | Reverse | Fragment size |
|---|---|---|---|
| RepPK | GGTACCACCATGCCACTACCACCAAAATC | ACTAGTTTAGCTATCCTGTGCTGTGCT | 1,047 bp |
| RepPA | GGTACCACCActGCACTACCACCAAAATC | ACTAGTTTAGCTATCCTGTGCTGTGCT | 1,047 bp |
| JM23 | TGGTGTAGGACTCCAGCAGAGTC | 288 bp | |
| JM24 | TAGGCCCACACCTTGGTGACCAA | ||
| β actin | AGGTATCCTGACCCTGAAGTACCCC | GGCCACACGCAGCTCATTGTA | 106 bp |
| GAPDH | ATTGTTGCCATCAACGACCCC | CAAGCTTCCCATTCTCGGCC | 116 bp |
| RepPepGMV | CAAAGCTGGTGATCCGAAAACG | GTTAAACGAGGATAATGGATAAGG | 120 bp |
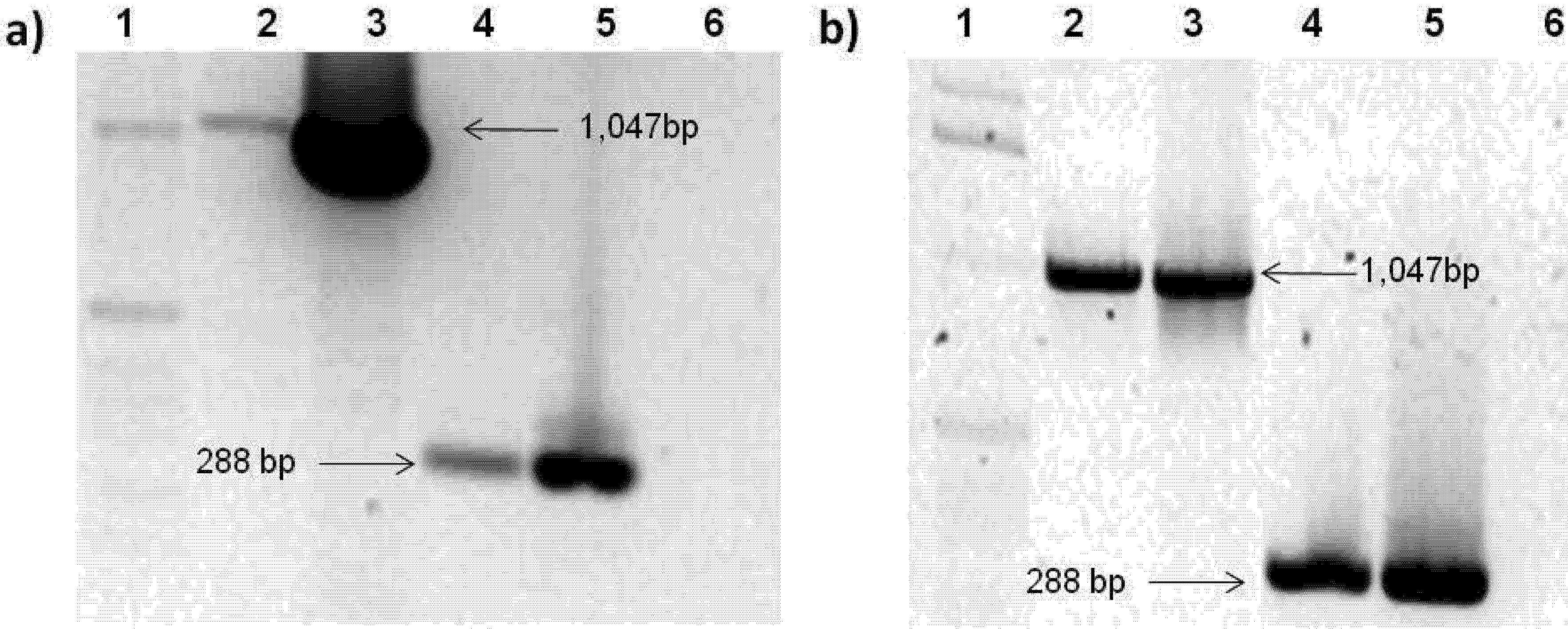

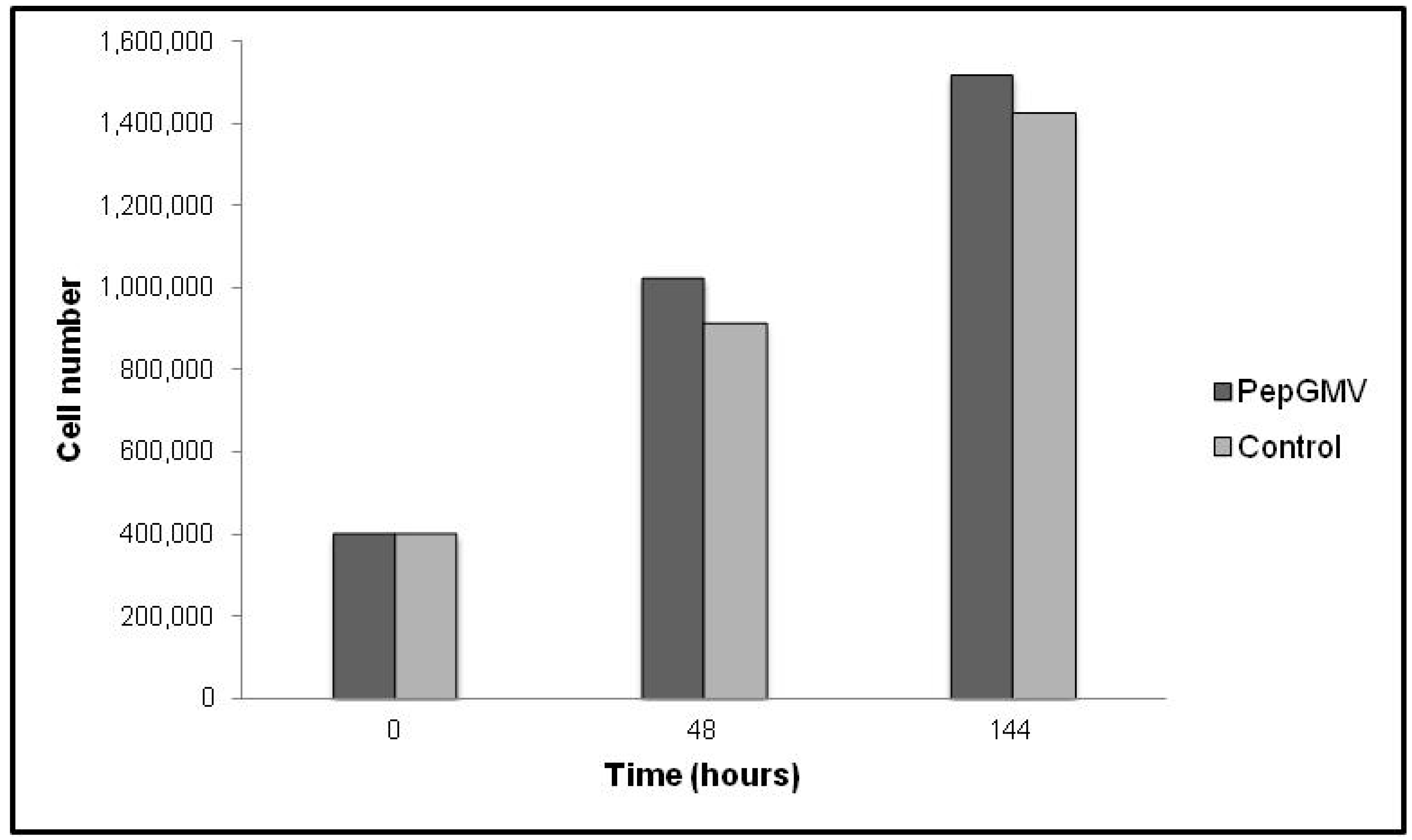
2.3. Rep-Gene Expression
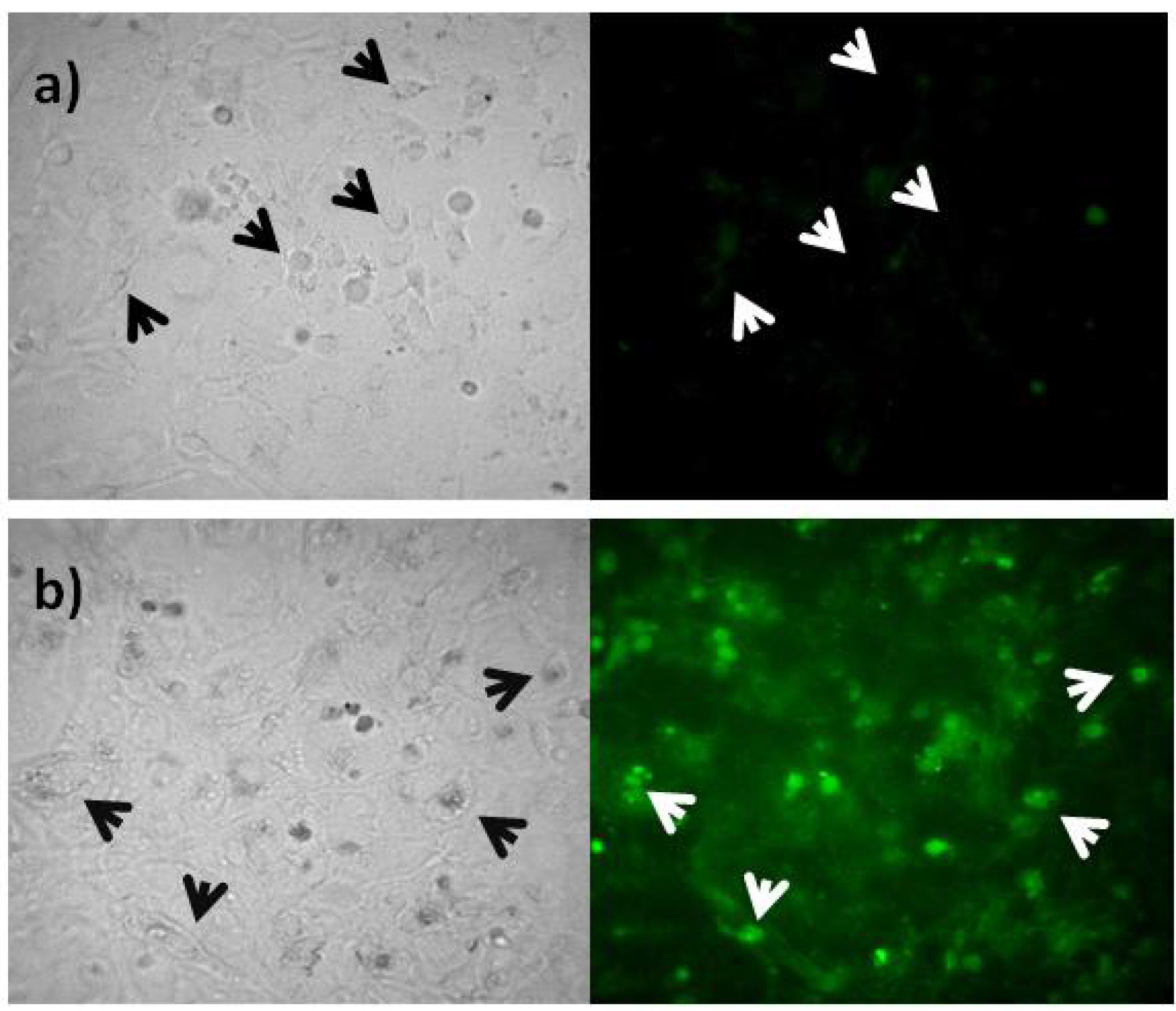

3. Experimental Section
3.1. Viral Clone and Rep-Gene PCR Amplification
3.2. Construction of pTracer™-SV40:Rep Expression Vector
3.3. Cell Culture and Transfection
3.4. Rep Expression
4. Conclusions
Acknowledgments
Conflict of Interest
References and Notes
- Gutierrez, C.; Ramirez-Parra, E.; Castellano, M.M.; Sanz-Burgos, A.P.; Luque, A.; Missich, R. Geminivirus DNA replication and cell cycle interactions. Vet. Microbiol. 2004, 98, 111–119. [Google Scholar] [CrossRef]
- Kittelmann, K.; Rau, P.; Gronenborn, B.; Jeske, H. Plant geminivirus Rep protein induces rereplication in fission yeast. J. Virol. 2009, 83, 6769–6778. [Google Scholar] [CrossRef]
- Hanley-Bowdoin, L.; Settlage, S.B.; Robertson, D. Reprogramming plant gene expression: A prerequisite to geminivirus DNA replication. Mol. Plant Pathol. 2004, 5, 149–156. [Google Scholar] [CrossRef]
- Nash, T.E.; Dallas, M.B.; Reyes, M.I.; Buhrman, G.K.; Ascencio-Ibañez, J.T.; Hanley-Bowdoin, L. Functional analysis of a novel motif conserved across geminivirus Rep proteins. J. Virol. 2011, 85, 1182–1192. [Google Scholar]
- Yadava, P.; Suyal, G.; Mukherjee, S.K. Begomovirus DNA replication and pathogenicity. Curr. Sci. 2010, 98, 360–368. [Google Scholar]
- Carrillo-Tripp, J.; Lozoya-Gloria, E.; Rivera-Bustamante, R.F. Symptom remission and specific resistance of pepper plants after infection by Pepper golden mosaic virus. Virology 2007, 97, 51–59. [Google Scholar]
- Ascencio-Ibáñez, J.T.; Sozzani, R.; Lee, T.J.; Chu, T.M.; Wolfinger, R.D.; Cella, R.; Hanley Bowdoin, L. Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 2008, 148, 436–454. [Google Scholar] [CrossRef]
- Torres-Pacheco, I.; Guevara-González, R.G.; Ruíz-Medrano, R.; Rivera-Bustamante, R.F. Los geminivirus como modelos de estudio de ciclo celular y de la apoptosis en plantas (in Spanish). Revista Mexicana de Fitolpatología 1996, 14, 88–96. [Google Scholar]
- Herwing, S.; Strauss, M. The retinoblastoma protein: A master regulator of cell cycle, differentiation and apoptosis. Eur. J. Biochem. 1997, 246, 581–601. [Google Scholar]
- Sidle, A.; Palaty, C.; Dirks, P.; Wiggan, O.; Kiess, M.; Gill, R.M.; Wong, A.K.; Hamel, P.A. Activity of the retinoblastoma family proteins, pRB, p107, and p130, during cellular proliferation and differentiation. Crit. Rev. Biochem. Molec. Biol. 1996, 31, 237–271. [Google Scholar] [CrossRef]
- Burkhart, D.L.; Sage, J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nature Rev. Cancer 2008, 8, 671–682. [Google Scholar] [CrossRef]
- Desvoyes, B.; Ramirez-Parra, E.; Xie, Q.; Chua, N.H.; Gutierrez, C. Cell type-specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant Physiol. 2006, 140, 67–80. [Google Scholar]
- Nagar, S.; Pedersen, T.J.; Carrick, K.M.; Hanley-Bowdoin, L.; Robertson, D. A geminivirus induces expression of a host DNA synthesis protein in terminally differentiated plant cells. Plant Cell 1995, 7, 705–719. [Google Scholar]
- Martin, D.P.; Biagini, P.; Lefeuvre, P.; Golden, M.; Roumagnac, P.; Varsani, A. Recombination in eukaryotic single stranded DNA viruses. Viruses 2011, 3, 1699–1738. [Google Scholar] [CrossRef] [Green Version]
- Laufs, J.; Jupin, I.; David, C.; Schumacher, S.; Heyraud-Nitschke, F.; Gronenbom, B. Geminivirus replication: Genetic and biochemical characterization of Rep protein function, a review. Biochimie 1995, 77, 765–773. [Google Scholar] [CrossRef]
- Chapa-Oliver, A.; Guevara-Gonzalez, R.G.; González-Chavira, M.M.; Ocampo-Velazquez, A.A.; Feregrino-Pérez, A.A.; Mejía-Teniente, L.; Herrera-Ruíz, G.; Torres-Pacheco, I. Analogies between geminivirus and oncovirus: Cell cycle regulation. Afr. J. Biotechnol. 2011, 10, 11327–11332. [Google Scholar]
- Doonan, J.H.; Sablowski, R. Walls around tumors—Why do plants do not develop cancer. Nat. Rev. Canc. 2010, 10, 794–802. [Google Scholar] [CrossRef]
- Kong, L.; Orozco, B.M.; Roe, J.L.; Nagar, S.; Ou, S.; Feiler, H.S.; Durfee, T.; Miller, A.B.; Gruissem, W.; Robertson, D.; Hanley-Bowdoin, L. A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. EMBO J. 2000, 19, 3485–3495. [Google Scholar] [CrossRef]
- Castillo, A.; Collinet, D.; Deret, S.; Kashoggi, A.; Bejarano, E.R. Dual interaction of plant PCNA with geminivirus replication accessory protein (Ren) and viral replication protein (Rep). Virology 2003, 312, 381–394. [Google Scholar] [CrossRef]
- Jordan, M.; Schallhorn, A.; Wurm, F.M. Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res. 1996, 24, 596–601. [Google Scholar] [CrossRef]
- Sugden, K.; Pariante, C.M.; McGuffin, P.; Aitchison, K.J.; D’Souza, U.M. Housekeeping gene expression is affected by antidepressant treatment in a mouse fibroblast cell line. J. Psychopharmacol. 2010, 24, 1253–1259. [Google Scholar] [CrossRef]
- Bocchetta, M.; Eliasz, S.; de Marco, M.A.; Rudzinski, J.; Zhang, L.; Carbone, M. The SV40 large T antigen-p53 complexes bind and actívate the insulin-like growth factor-I promoter stimulating cell growth. Cancer Res. 2008, 68, 1022–1028. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Chapa-Oliver, A.M.; Mejía-Teniente, L.; García-Gasca, T.; Guevara-Gonzalez, R.G.; Torres-Pacheco, I. PepGMV Rep-Protein Expression in Mammalian Cells. Viruses 2012, 4, 1792-1801. https://doi.org/10.3390/v4091792
Chapa-Oliver AM, Mejía-Teniente L, García-Gasca T, Guevara-Gonzalez RG, Torres-Pacheco I. PepGMV Rep-Protein Expression in Mammalian Cells. Viruses. 2012; 4(9):1792-1801. https://doi.org/10.3390/v4091792
Chicago/Turabian StyleChapa-Oliver, Angela María, Laura Mejía-Teniente, Teresa García-Gasca, Ramon Gerardo Guevara-Gonzalez, and Irineo Torres-Pacheco. 2012. "PepGMV Rep-Protein Expression in Mammalian Cells" Viruses 4, no. 9: 1792-1801. https://doi.org/10.3390/v4091792








