Diversity of Dicotyledenous-Infecting Geminiviruses and Their Associated DNA Molecules in Southern Africa, Including the South-West Indian Ocean Islands
Abstract
:1. Introduction
2. Diversity of Dicotyledenous-Infecting Begomoviruses in SADC Countries and South-West Indian Ocean (SWIO) Islands
2.1. Cassava (Manihot esculenta Crantz)
2.1.1. General Introduction
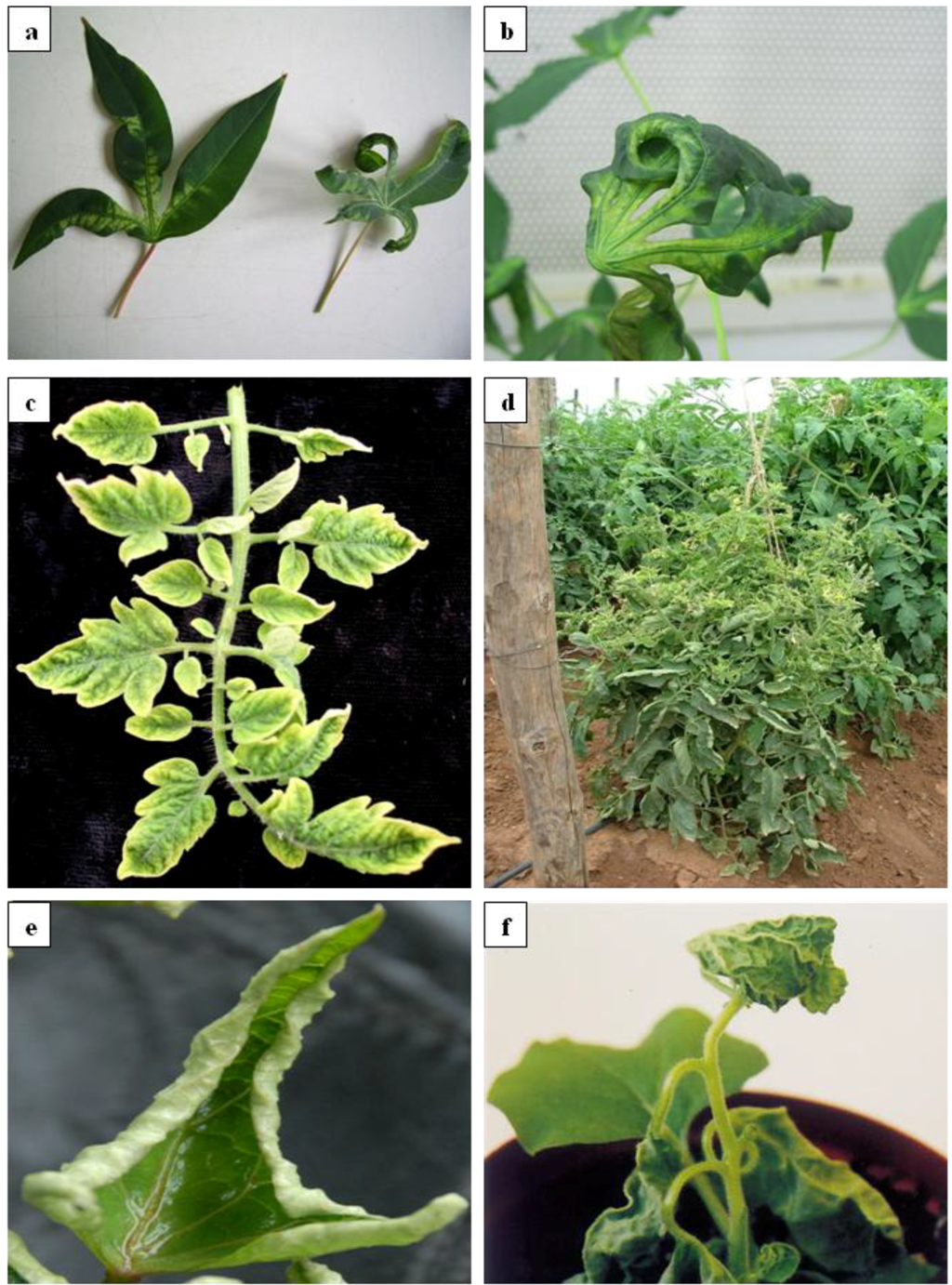
2.1.3. Transmission and Diversity of Begomoviruses in South Africa and Mozambique
2.1.4. Recombination
2.1.5. Defective Interfering DNAs Associated with CMBs
2.2. Tomato
2.2.1. General Introduction
2.2.2. Geographic Diversity of Tomato Begomovirus Species and Variants
| Geminivirus | Accession Number | Host | Location | Reference |
|---|---|---|---|---|
| African cassava mosaic virus | ||||
| African cassava mosaic virus-[Tanzania:2001] (ACMV-[TZ:01]) | AY795982 | Cassava | Tanzania | [83] |
| African cassava mosaic virus-[Tanzania:2001] (ACMV-[TZ:01]) | AY795982 | Cassava | Tanzania | [83] |
| African cassava mosaic virus-[Angola:2008] (ACMV-[An:08]) | FJ807631 | Cassava | Angola | Unpublished |
| African cassava mosaic virus-[Angola:AOS:2009] (ACMV-[An:09]) | GU580897 | Cassava | Angola | Unpublished |
| African cassava mosaic virus-[Congo:2008] (ACMV-[DRC:08]) | FN435277 | Cassava | Congo | [108] |
| African cassava mosaic virus-[Congo:2008] (ACMV-[DRC:08]) | FN435275 | Cassava | Congo | [108] |
| African cassava mosaic virus-[Congo:2008] (ACMV-[DRC:08]) | FN435273 | Cassava | Congo | [108] |
| African cassava mosaic virus-[Congo:2008] (ACMV-[DRC:08]) | FN435271 | Cassava | Congo | [108] |
| African cassava mosaic virus-[Congo:2008] (ACMV-[DRC:08]) | FN435276 | Cassava | Congo | [108] |
| African cassava mosaic virus-[Congo:2008] (ACMV-[DRC:08]) | FN435274 | Cassava | Congo | [108] |
| African cassava mosaic virus-[Congo:2008] (ACMV-[DRC:08]) | FN435272 | Cassava | Congo | [108] |
| African cassava mosaic virus-[Congo:2008] (ACMV-[DRC:08]) | FN668378 | Cassava | Congo | Unpublished |
| African cassava mosaic virus-[Congo:2008] (ACMV-[DRC:08]) | FN668379 | Cassava | Congo | Unpublished |
| Bean yellow dwarf virus | ||||
| Bean yellow dwarf virus-[South Africa:Mpumalanga:1994] (BeYDV-[ZA:Mpu:94]) | Y11023 | Bean | South Africa | [72] |
| Cassava mosaic Madagascar virus | ||||
| Cassava mosaic Madagascar virus-[Madagascar:Toliary:2006] (CMMGV-[MG:Tol:06]) | HE617299 | Madagascar | [107] | |
| Cotton leaf curl Gezira virus | ||||
| Cotton leaf curl Gezira virus-[Madagascar: Fort Dauphin:2001] (CLCuGV-Be[MG:For:01]) | AM701767 | Bean | Madagascar | [63] |
| East African cassava mosaic virus | ||||
| East African cassava mosaic virus-[Congo:2008] (EACMV-[DRC:08]) | FN435281 | Cassava | Congo | [108] |
| East African cassava mosaic virus-[Congo:2008] (EACMV-[DRC:08]) | FN435279 | Cassava | Congo | [108] |
| East African cassava mosaic virus-[Congo:2008] (EACMV-[DRC:08]) | FN435280 | Cassava | Congo | [108] |
| East African cassava mosaic virus-[Congo:2008] (EACMV-[DRC:08]) | FN435278 | Cassava | Congo | [108] |
| East African cassava mosaic-Kenya [Tanzania:Dar Es Salaam:1996] (EACMV-KE[TZ:Dar:96]) | Z83256 | Cassava | Tanzania | [73] |
| East African cassava mosaic-Kenya [Tanzania:M] (EACMV-KE:[TZ:M]) | AY795986 | Cassava | Tanzania | [83] |
| East African cassava mosaic-Kenya [Tanzania:T] (EACMV-Ke[TZ:T]) | AY795985 | Cassava | Tanzania | [83] |
| East African cassava mosaic-Tanzania [Tanzania:YV] (EACMV-[TZ:YV]) | AY795987 | Cassava | Tanzania | [83] |
| East African cassava mosaic virus-Uganda-[Tanzania:10] (EACMV-UG[TZ:10]) | AY795988 | Cassava | Tanzania | [83] |
| East African cassava mosaic Cameroon virus | ||||
| East African cassava mosaic Cameroon virus-Tanzania[Tanzania:7:2001](EACMCV-[TZ:7:01]) | AY795984 | Cassava | Tanzania | [74] |
| East African cassava mosaic Malawi virus | ||||
| East African cassava mosaic Malawi virus-[Malawi:MH:1996] (EACMMV-[MW:MH:96]) | AJ006459 | Cassava | Malawi | [73] |
| East African cassava mosaic Malawi virus-[Malawi:K:1996] (EACMMV-[MW:K:96]) | AJ006460 | Cassava | Malawi | [73] |
| East African cassava mosaic Zanzibar virus | ||||
| East African cassava mosaic Zanzibar virus-[Tanzania:Uguja:1998] (EACMZV-[TZ:Ugu:98]) | AF422174 | Cassava | Zanzibar | [74] |
| South African cassava mosaic virus | ||||
| South African cassava mosaic virus-South Africa-[South Africa:99] (SACMV-[ZA:99]) | AF155806 | Cassava | South Africa | [76] |
| South African cassava mosaic virus-[Madagascar:12] (SACMV-[MG:12]) | AJ422132 | Cassava | Madagascar | [79] |
| South African cassava mosaic virus-[Zimbabwe:Muzarabani] (SACMV-[ZW:Muz]) | AJ575560 | Cassava | Zimbabwe | [80] |
| Sweet potato mosaic associated virus | ||||
| Sweet potato mosaic associated virus (SPMaV-[ZA:WP:2011]) | JQ621843 | Sweet potato | South Africa | [64] |
| Sweet potato leaf curl Sao Paulo virus | ||||
| Sweet potato leaf curl Sao Paulo virus (SPLCSPV-[ZA;WP:2011]) | JQ621844 | Sweet potato | South Africa | [64] |
| Tobacco leaf curl Comoros virus | ||||
| Tobacco leaf curl Comoros virus-[Grande Comore:Foubouni99:2005] (TbLCKMV-[GC:Fou99:05]) | AM701762 | Tobacco | Comoros | [63] |
| Tobacco leaf curl Comoros virus-[Grande Comore:Simboussa18:2004] (TbLCKMV-[GC:Sim18:04]) | AM701760 | Tobacco | Comoros | [63] |
| Tobacco leaf curl Zimbabwe virus | ||||
| Tobacco leaf curl Zimbabwe virus-[Zimbabwe:2001] (TbLCZV-[ZW:01]) | AF350330 | Tobacco | Zimbabwe | [78] |
| Tobacco leaf curl Zimbabwe virus-[Grande Comore:Foumboudziouni95:2005] (TbLCZV-[GC:Sim18:05]) | AM701756 | Tobacco | Comoros archipelago; Grande Comore | [63] |
| Tomato curly stunt virus | ||||
| Tomato curly stunt virus-[South Africa:Onderberg:1998](ToCSV-[ZA:Ond:98]) | AF261885 | Tomato | South Africa | [86] |
| Tomato curly stunt Lanseria virus* | ||||
| Tomato curly stunt Lanseria virus-[South Africa:Lanseria:2008] (ToCSLV-[ZA:Lan:08]) | Access # | Tomato | South Africa | [145] |
| Tomato curly stunt Noordoewer virus* | ||||
| Tomato curly stunt Noordoewer virus-[South Africa:Noordoewer06:2008](ToCSNV-[ZA:Nwr06:08]) | Access # | Tomato | South Africa | [145] |
| Tomato curly stunt Mooketsi virus* | ||||
| Tomato curly stunt Mooketsi virus-[South Africa:Mooketsi:2007] (ToCSMV-[ZA:Mks:07]) | Access # | Tomato | South Africa | [145] |
| Tomato leaf curl Antsiranana virus* | ||||
| Tomato leaf curl Antsiranana virus-[Madagascar:Antsalaka6:2001] (ToLCAntV-[MG:Mia6:01]) | AM701766 | Tomato | Madagascar | [63] |
| Tomato leaf curl Antsiranana virus-[Madagascar:Miandrivazo1:2001] (ToLCTolV-[MG:Mia1:01]) | AM701767 | Tomato | Madagascar | [63] |
| Tomato leaf curl Arusha virus | ||||
| Tomato leaf curl Arusha virus-[Tanzania:Tengelu:2005] (ToLCArV-[TZ:Ten:05]) | DQ519575 | Tomato | Tanzania | [84] |
| Tomato leaf curl Anjouan virus | ||||
| Tomato leaf curl Anjouan virus-[Anjouan:Ouani3:2004] (ToLCAnjV-[Anj:Oua3:04]) | AM701758 | Tomato | Comoros archipelago; Anjouan | [63] |
| Tomato leaf curl Comoros virus* | ||||
| Tomato leaf curl Comoros virus-Mayotte:Dembeni:2003] (ToLCKMV-[YT:Dem:03]) | AJ865341 | Tomato | Comoros | [63] |
| Tomato leaf curl Diana virus* | ||||
| Tomato leaf curl Diana virus - [Madagascar:Namakely5:2001] (ToLCDiaV-[MG:Nam5:01]) | AM701765 | Tomato | Madagascar | [82] |
| Tomato leaf curl Madagascar virus | ||||
| Tomato leaf curl Madagascar virus-Androy [Madagascar:Toliary:2001] (ToLCMGV-And[MG:Tol:01]) | AJ865339 | Tomato | Madagascar | [82] |
| Tomato leaf curl Madagascar virus-Menabe [Madagascar:Morondova:2001] (ToLCMGV-Men[MG:Mor:01]) | AJ865338 | Tomato | Madagascar | [82] |
| Tomato leaf curl Mayotte virus | ||||
| Tomato leaf curl Mayotte virus-[Mayotte:Kahani:2003] (ToLCYTV-[YT:Kah:03]) | AJ865340 | Tomato | Comoros archipelago; Mayotte | [82] |
| Tomato leaf curl Moheli virus | ||||
| Tomato leaf curl Moheli virus-[Comoros:Fomboni163:2005] (ToLCMohV-[KM:Fom163:05]) | AM701763 | Tomato | Comoros archipelago; Grande Comore | [63] |
| Tomato leaf curl Namakely virus* | ||||
| Tomato leaf curl Namakely virus-[Comoros:Dimadjou:2001] (ToLCNaV:Dim:01) | AM701761 | Tomato | Comoros archipelago; Dimadjou | [63] |
| ToLCNamV-[Madagascar:Namakely:2001] ([MG:Nam:01]) | AM701764 | Tomato | Madagascar | [63] |
| Tomato leaf curl Seychelles virus | ||||
| Tomato leaf curl Seychelles virus-[Mahe:Val d’Endor77:2004] (ToLCSV-[Mah:VE77:04]) | AM491778 | Tomato | Seychelles | [71] |
| Tomato leaf curl Tanzania virus-[Tanzania:1994]) (TYLCTZV-[TZ:94]) | U734981523 bp | Tomato | Tanzania | [71] |
| Tomato leaf curl Toliara virus* | ||||
| Tomato leaf curl Toliara virus-[Madagascar:Miandrivazo2:2001] (ToLCTolV-[MG:Mia2:01]) | AM701768 | Tomato | Madagascar | [63] |
| Tomato leaf curl Tanzania virus | ||||
| Tomato leaf curl Tanzania virus (ToLCTZV) | U73498 | Tomato | Tanzania | [71] |
| Tomato yellow leaf curl virus | ||||
| Tomato yellow leaf curl virus | HM448447 | Tomato | Mauritius | [89] |
| Defective subgenomic DNAs | ||||
| East African cassava mosaic virus associated defective DNA-A (1,525 nt) | Cassava | Tanzania | [65] | |
| TbLCZV-[ZW] associated ‘HG’ DI (1,341 nt) | Tobacco | Zimbabwe | [78] | |
| TbLCZV-[ZW] associated ‘mild’ DI (1,421 nt) | Tobacco | Zimbabwe | [78] | |
| SACMV-[ZA] associated defective DNA-B (1,389 nt) | Cassava | South Africa | [66] | |
2.2.3. Evolutionary Relationships between SADC and SWIO Tomato-Infecting Begomoviruses
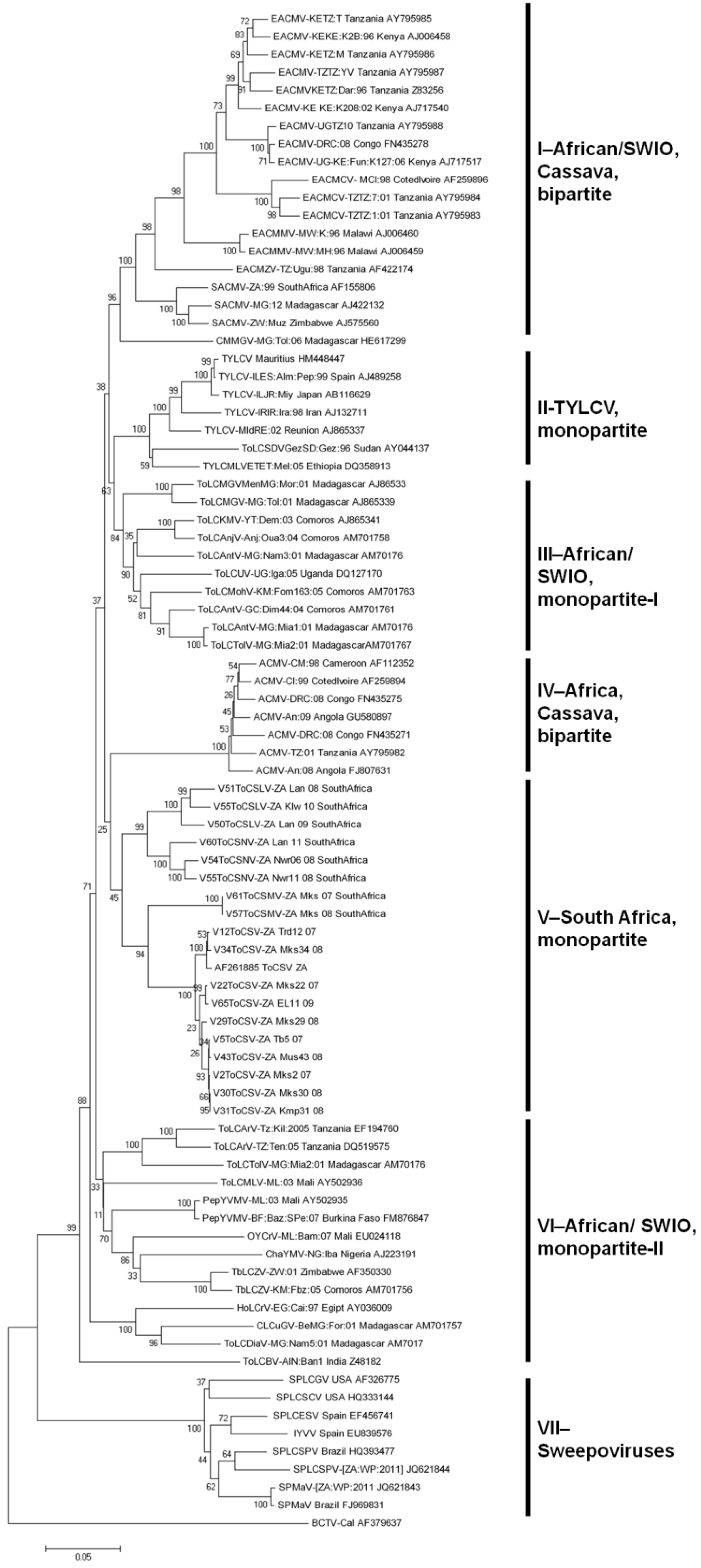
2.2.4. Emergence and Transmission
2.2.5. Management and Control of Tomato-Infecting Begomoviruses
2.3. Sweet Potato
2.3.1. General Introduction
2.3.2. Recent Detection of Sweepoviruses in South Africa
2.4. Tobacco
2.4.1. General Introduction
2.4.2. Geographic Diversity of Tobacco Begomovirus Species
2.4.3. Atypical Defective DNA Molecules Associated with TbLCZV from Zimbabwe
2.5. Bean
3. Conclusions
References and Notes
- Polston, J.E.; Anderson, P.K. The emergence of whitefly-transmitted geminiviruses in tomato in the Western Hemisphere. Plant Dis. 1997, 81, 1358–1369. [Google Scholar] [CrossRef]
- Morales, F.J.; Anderson, P.K. The emergence and dissemination of whitefly-transmitted geminiviruses in Latin America—Brief review. Arch. Virol. 2001, 146, 415–441. [Google Scholar] [CrossRef]
- Varma, A.; Malathi, V.G. Emerging geminivirus problems: A serious threat to crop production. Ann. Appl. Biol. 2003, 142, 145–164. [Google Scholar] [CrossRef]
- Mansoor, S.; Briddon, R.W.; Zafar, Y.; Stanley, J. Geminivirus disease complexes: An emerging threat. Trends Plant Sci. 2003, 8, 128–134. [Google Scholar] [CrossRef]
- Legg, J.P.; Fauquet, C.M. Cassava mosaic geminiviruses in Africa. Plant Mol. Biol. 2004, 56, 585–599. [Google Scholar] [CrossRef]
- Hanssen, I.M.; Lapidot, M.; Thomma, B.P.H.J. Emerging viral diseases of tomato crops. Mol. Plant Microbe In. 2010, 23, 539–548. [Google Scholar] [CrossRef]
- Navas-Castillo, J.; Fiallo-Olive, E.; Sanchez-Campos, S. Emerging virus diseases transmitted by whiteflies. Annu. Rev. Phytopathol. 2011, 49, 15.1–15.30. [Google Scholar]
- Harrison, B.D. Advances in geminivirus research. Annu. Rev. Phytopathol. 1985, 23, 55–82. [Google Scholar] [CrossRef]
- Fauquet, C.M.; Briddon, R.W.; Brown, J.K.; Moriones, E.; Stanley, J.; Zerbini, M.; Zhou, X. Geminivirus strain demarcation and nomenclature. Arch. Virol. 2008, 153, 783–821. [Google Scholar] [CrossRef]
- Brown, J.K.; Fauquet, C.M.; Briddon, R.W.; Zerbini, M.; Moriones, E.; Navas-Castillo, J. Family Geminiviridae. In Virus Taxonomy. Ninth Report of the International Committee on Taxonomy of Viruses, 1st ed; Elsevier-Academic: Amsterdam, The Netherlands, 2011; pp. 351–373. [Google Scholar]
- Hamilton, W.D.O.; Bisaro, D.M.; Coutts, R.H.A.; Buck, K.W. Demonstration of the bipartite nature of the genome of a single-stranded DNA plant virus by infection with the cloned DNA components. Nucleic Acids Res. 1983, 11, 7387–7396. [Google Scholar] [CrossRef]
- Briddon, R.W.; Mansoor, S.; Bedford, I.D.; Pinner, M.S; Markham, P.G. Clones of cotton leaf curl geminivirus induce symptoms atypical of cotton leaf curl disease. Virus Genes 2000, 20, 19–26. [Google Scholar] [CrossRef]
- Saunders, K.; Bedford, I.D.; Briddon, R.W.; Markham, P.G.; Wong, S.M.; Stanley, J. A unique virus complex causes Ageratum yellow vein disease. Proc. Natl. Acad. Sci. U. S. A. 2000, 97, 6890–6895. [Google Scholar]
- Zhou, X.P.; Xie, Y.; Tao, X.R.; Zhang, Z.K.; Li, Z.H.; Fauquet, C.M. Characterization of DNA beta associated with begomoviruses in China and evidence for co-evolution with their cognate viral DNA-A. J. Gen. Virol. 2003, 84, 237–247. [Google Scholar] [CrossRef]
- Kheyr-Pour, A.; Bendahmane, M.; Matzeit, V.; Accotto, G.P.; Crespi, S.; Gronenborn, B. Tomato yellow leaf curl virus from Sardinia is a whitefly-transmitted monopartite geminivirus. Nucleic Acids Res. 1991, 19, 6763–6769. [Google Scholar] [CrossRef]
- Navot, N.; Pichersky, E.; Zeidan, M.; Zamir, D.; Czosnek, H. Tomato yellow leaf curl virus—A whitefly-transmitted geminivirus with a single genomic component. Virology 1991, 185, 151–161. [Google Scholar] [CrossRef]
- Dry, I.B.; Rigden, J.E.; Krake, L.R.; Mullineaux, P.M.; Rezaian, M.A. Nucleotide-sequence and genome organization of tomato leaf curl geminivirus. J. Gen. Virol. 1993, 74, 147–151. [Google Scholar] [CrossRef]
- Brown, J.K.; Frohlich, D.R.; Rosell, R.C. The sweet potato/silverleaf whiteflies: Biotypes of Bemisia tabaci or a species complex? Annu. Rev. Entomol. 1995, 40, 511–534. [Google Scholar] [CrossRef]
- Ghanim, M.; Czosnek, H. Tomato leaf curl virus (TYLCV-Is) is transmitted among whitefly in a sex-related manner. J. Virol. 2000, 74, 4738–4745. [Google Scholar] [CrossRef]
- Moriones, E.; Navas-Castillo, J. Tomato yellow leaf curl virus, an emerging virus complex causing epidemics worldwide. Virus Res. 2000, 71, 123–134. [Google Scholar] [CrossRef]
- Czosnek, H.; Ghanim, M.; Ghanim, M. The circulative pathway of begomovirsues in the whitefly vector Bemisia tabaci-insights from studies with Tomato yellow leaf curl virus. Ann. Appl. Biol. 2002, 140, 215–231. [Google Scholar] [CrossRef]
- Brown, J.K. Phylogenetic biology of the Bemisia tabaci sibling species group. In Bionomics and Management of a Global Pest; Stansly, P.A., Naranjo, S.E., Eds.; Springer: Amsterdam, The Netherlands, 2010; pp. 31–67. [Google Scholar]
- Brown, J.K.; Bird, J. Whitefly-transmitted geminiviruses and associated disorders in the America and the Caribbean basin. Plant Dis. 1992, 76, 220–225. [Google Scholar] [CrossRef]
- Anderson, P.K.; Cunningham, A.A.; Patel, N.G.; Morales, F.J.; Epstein, P.R.; Daszak, P. Emerging infectious diseases of plants: Pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol. 2004, 19, 535–544. [Google Scholar] [CrossRef]
- Seal, S.E.; van den Bosch, F.; Jeger, M.J. Factors influencing begomovirus evolution and their increasing global significance: Implications for sustainable control. Crit. Rev. Plant Sci. 2006, 25, 23–46. [Google Scholar] [CrossRef]
- Jeske, H. Geminiviruses. In Torque Teno Virus: The Still Elusive Human Pathogens, 1st ed; Springer: Berlin, Germany, 2009; Volume 331, pp. 185–226. [Google Scholar]
- Rybicki, E.P.; Pietersen, G. Plant virus disease problems in the developing world. Adv. Virus Res. 1999, 53, 127–175. [Google Scholar] [CrossRef]
- Seal, S.E.; Jeger, M.J.; van den Bosch, F. Begomovirus evolution and disease management. Adv. Virus Res. 2006, 67, 297–316. [Google Scholar] [CrossRef]
- Padidam, M.; Sawyer, S.; Fauquet, C.M. Possible emergence of new geminiviruses by frequent recombination. Virology 1999, 265, 218–225. [Google Scholar] [CrossRef]
- Martin, D.P.; Williamson, C.; Posada, D. RDP2: Recombination detection and analysis from sequence alignments. Bioinformatics 2005, 21, 260–262. [Google Scholar] [CrossRef]
- Rojas, M.R.; Hagen, C.; Lucas, W.J.; Gilbertson, R.L. Exploiting chinks in the plant’s armor: Evolution and emergence of geminiviruses. Annu. Rev. Phytopathol. 2005, 43, 361–394. [Google Scholar] [CrossRef]
- Briddon, R.W.; Stanley, J. Subviral agents associated with plant single-stranded DNA viruses. Virology 2006, 344, 198–210. [Google Scholar] [CrossRef]
- Duffy, S.; Holmes, E.C. Phylogenetic evidence for rapid rates of molecular evolution in the single-stranded DNA begomovirus. Tomato Yellow Leaf curl virus. J. Virol. 2008, 82, 957–965. [Google Scholar] [CrossRef]
- Martin, D.P.; Biagini, P.; Lefeuvre, P.; Golden, M.; Roumagnac, P.; Varsani, A. Recombination in eukaryotic single stranded DNA. Viruses 2011, 3, 1669–1738. [Google Scholar]
- Bedford, I.D.; Briddon, R.W.; Brown, J.K.; Rosell, R.C.; Markham, P.G. Geminivirus-transmission and biological characterization of Bemisia tabaci (Gennadius) biotypes from different geographic regions. Ann. Appl. Biol. 1994, 125, 311–325. [Google Scholar] [CrossRef]
- Frohlich, D.R.; Torres-Jerez, I.; Bedford, I.D.; Markham, P.G.; Brown, J.K. A phylogeographical analysis of the Bemisia tabaci species complex based on mitochondrial DNA markers. Mol. Ecol. 1999, 8, 1683–1691. [Google Scholar] [CrossRef]
- Gill, R.; Brown, J.K. Systematics of Bemisia and Bemisia Relatives: Can molecular techniques solve the Bemisia tabaci complex conundrum—A Taxonomist’s viewpoint. In Bionomics and Management of a Global Pest, 1st ed; Springer: Amsterdam, The Netherlands, 2010; pp. 5–29. [Google Scholar]
- De Barro, P.J.; Liu, S.S.; Boykin, L.M.; Dinsdale, A.B. Bemisia tabaci: A statement of species status. Annu. Rev. Entomol. 2011, 56, 1–19. [Google Scholar] [CrossRef]
- Berry, S.D.; Fondong, V.N.; Rey, M.E.C.; Rogan, D.; Fauquet, C.; Brown, J.K. Molecular evidence for five distinct Bemisia tabaci (Homoptera: Aleyrodidae) geographic haplotypes associated with cassava plants in sub-Saharan Africa. Ann. Entomol. Soc. Am. 2004, 97, 852–859. [Google Scholar] [CrossRef]
- De La Rua, P.; Simon, B.; Cifuentes, D.; Martinez-Mora, C.; Cenis, J.L. New insights into the mitochondrial phylogeny of the whitefly Bemisia tabaci (Hemiptera Aleyrodidae) in Mediterranean basin. J. Zool. Syst. Evol. Res. 2006, 44, 25–33. [Google Scholar] [CrossRef]
- Boykin, L.M.; Shatters, R.G.; Rosell, R.C.; McKenzie, C.L.; Bagnall, R.A.; de Barro, P.J.; Frohlich, D.R. Global relationships of Bemisia tabaci (Hemiptera: Aleyrodidae) revealed using Bayesian analysis of mitochondrial COI DNA sequences. Mol. Phylogenet. Evol. 2007, 44, 1306–1319. [Google Scholar] [CrossRef]
- Dinsdale, L.; Cook, C.; Riginos, Y.; Buckley, M.; de Barro, P.J. Refined global analysis of Bemisia tabaci (Hemiptera: Sternorrhyncha: Aleyrodoidea: Aleyrodidae) mitochondrial cytochrome oxidase I to identify species level genetic boundaries. Ann. Entomol. Soc. Am. 2010, 103, 196–208. [Google Scholar] [CrossRef]
- Esterhuizen, L.L.; Mabasa, K.G.; van Heerden, S.W.; Czosnek, H.; Brown, K.; van Heerden, H.; Rey, M.E.C. Genetic identification of members of the Bemisia tabaci cryptic species complex from South Africa reveals native and introduced haplotypes. J. Appl. Entomol. 2012. [Google Scholar]
- Legg, J.P.; French, R.; Rogan, D.; Okao-Okuja, G.; Brown, J.K. A distinct Bemisia tabaci (Gennadius) (Hemiptera: Sternorrhyncha: Aleyrodidae) genotype cluster is associated with the epidemic of severe cassava mosaic virus disease in Uganda. Mol. Ecol. 2002, 11, 1219–1229. [Google Scholar] [CrossRef]
- Abdullahi, I.; Winterm, S.; Atirim, G.I.; Thottappillym, G. Molecular characterization of whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae) populations infesting cassava. B. Entomol. Res. 2003, 93, 97–106. [Google Scholar]
- Maruthi, M.N.; Colvin, J.; Thwaites, R.M.; Banks, G.K.; Gibson, G.; Seal, S. Reproductive incompatibility and cytochrome oxidase I gene sequence variability amongst host-adapted and geographically separate Bemisia tabaci populations (Hemiptera: Aleyrodidae). Syst. Entomol. 2004, 29, 560–568. [Google Scholar] [CrossRef]
- Brown, J.K.; Idris, A.M. Genetic differentiation of whitefly (Bemisia tabaci) mitochondrial cytochrome oxidase I, and phylogeographic concordance with the coat protein of the plant virus genus Begomoviridae. Ann. Entomol. Soc. Am. 2005, 95, 827–837. [Google Scholar]
- Sseruwagi, P.; Legg, J.P.; Maruthi, M.N.; Colvin, J.; Rey, M.E.C.; Brown, J.K. Genetic diversity of Bemisia tabaci (Gennadius)(Hemiptera: Aleyrodidae) populations and presence of the B biotype and non-B biotype that can induce silverleaf symptoms in squash, in Uganda. Ann. Appl. Biol. 2005, 147, 253–265. [Google Scholar] [CrossRef]
- Sseruwagi, P.; Maruthi, M.N.; Colvin, J.; Rey, M.E.C.; Brown, J.K.; Legg, J.P. Colonization of non-cassava plant species by whiteflies (Bemisia tabaci) in Uganda. Entomol. Exp. Appl. 2006, 119, 145–153. [Google Scholar] [CrossRef]
- Carabalí, A.; Bellotti, A.C.; Montoya-Lerma, J. Biological parameters of Bemisia tabaci (Gennadius) biotype B (Hemiptera: Aleyrodidae) on Jatropha gossypiifolia, commercial (Manihot esculenta) and wild cassava (Manihot flabellifolia and M. carthaginensis) (Euphorbiaceae). Neotrop. Entomol. 2010, 39, 562–567. [Google Scholar] [CrossRef]
- Thompson, W.M.O. A new host plant species for the cassava biotype of Bemisia tabaci (Gennadius) (Hom., Aleyrodidae). J. Appl. Entomol. 2003, 127, 374–376. [Google Scholar] [CrossRef]
- De Barro, P.J.; Trueman, J.W.; Frohlich, D.R. Bemisia argentifolii is a race of B. tabaci (Hemiptera: Aleyrodidae): The molecular genetic differentiation of B. tabaci populations around the world. B. Entomol. Res. 2005, 95, 193–203. [Google Scholar] [CrossRef]
- Legg, J.P. Host-associated strains within Ugandan populations of the whitefly Bemisia tabaci (Genn.) (Hom, Aleyrodidae). J. Appl. Entomol. 1996, 120, 523–527. [Google Scholar] [CrossRef]
- Polston, J.E.; Chellemi, D.O.; Schuster, D.J.; McGovern, R.J.; Stansly, P.A. Spatial and temporal dynamics of tomato mottle geminivirus and Bemisia tabaci (Genn) in Florida tomato fields. Plant Dis. 1996, 80, 1022–1028. [Google Scholar] [CrossRef]
- Ribeiro, S.G.; de Avila, A.C.; Bezerra, I.C.; Fernandes, J.J.; Faria, J.C.; Lima, M.F.; Gilbertson, R.L.; Maciel Zambolim, E.; Zerbini, F.M. Widespread occurrence of tomato geminiviruses in Brazil, associated with the new biotype of the whitefly vector. Plant Dis. 1998, 82, 830. [Google Scholar]
- Nawaz-Ul-Rehman, M.S.; Fauquet, C.M. Evolution of geminiviruses and their satellites. FEBS Lett. 2009, 583, 1825–1832. [Google Scholar] [CrossRef]
- South African Development Community. Available online: http://en.wikipedia (accessed on 26 June 2012).
- SADC. South African Development Community. Measures to address Food Security in the SADC Region. Background Paper (SADC Secretariat) (2009). Available online: http://www.sadc.int/fanr/ (accessed on 7 June 2012).
- Harrison, B.D.; Zhou, X.; Otim-Nape, G.W.; Liu, Y.; Robinson, D.J. Role of a novel type of double infection in the geminivirus-induced epidemic of severe cassava mosaic in Uganda. Ann. Appl. Biol. 1997, 131, 437–448. [Google Scholar] [CrossRef]
- Zhou, X.P.; Liu, Y.L.; Calvert, L.; Munoz, C.; Otim-Nape, G.W.; Robinson, D.J.; Harrison, B.D. Evidence that DNA-A of a geminivirus associated with severe cassava mosaic disease in Uganda has arisen by interspecific recombination. J. Gen. Virol. 1997, 78, 2101–2111. [Google Scholar]
- Fondong, V.N.; Pita, J.S.; Rey, M.E.C.; de Kochko, A.; Beachy, R.N.; Fauquet, C.M. Evidence of synergism between African cassava mosaic virus and a new double-recombinant geminivirus infecting cassava in Cameroon. J. Gen. Virol. 2000, 81, 287–297. [Google Scholar]
- Pita, J.S.; Fondong, V.N.; Sangare, A.; Kokora, R.N.N.; Fauquet, C.M. Genomic and biological diversity of the African cassava geminiviruses. Euphytica 2001, 120, 115–125. [Google Scholar] [CrossRef]
- Lefeuvre, P.; Martin, D.P.; Hoareau, M.; Naze, F.; Delatte, H.; Thierry, M.; Varsani, A.; Becker, N.; Reynaud, B.; Lett, J.M. Begomovirus ‘melting pot’ in the south-west Indian Ocean islands: Molecular diversity and evolution through recombination. J. Gen. Virol. 2007, 88, 3458–3468. [Google Scholar] [CrossRef]
- Esterhuizen, L.L.; van Heerden, S.W.; Rey, M.E.C.; van Heerden, H. Genetic identification of two sweet potato infecting begomoviruses in South Africa. Arch. Virol. 2012. [Google Scholar]
- Ndunguru, J.; Legg, J.P.; Fofana, I.B.F.; Aveling, T.A.S.; Thompson, G.; Fauquet, C.M. Identification of a defective molecule derived from DNA-A of the bipartite begomovirus of East African cassava mosaic virus. Plant Pathol. 2006, 55, 2–10. [Google Scholar] [CrossRef]
- Choge, I.; Paximadis, M.; Rey, M.E.C. A 1389bp defective molecule associated with South African cassava mosaic virus in South Africa. In Proceedings of the 3rd International Geminivirus Symposium, John Innes Center, Norwich, UK, 24–28 July 2001. abstract 90.
- Thresh, J.M.; Cooter, R.J. Strategies for controlling cassava mosaic disease in Africa. Plant Pathol. 2005, 54, 587–614. [Google Scholar] [CrossRef]
- Storey, H.H. Virus diseases of East African plants. VI-A progress report on studies of the disease of cassava. East Afr. Agr. Forest J. 1936, 2, 34–39. [Google Scholar]
- Story, H.H.; Nichols, R.F.W. Studies of the mosaic diseases of cassava. Ann. Appl. Biol. 1938, 25, 790–806. [Google Scholar] [CrossRef]
- Bock, K.R.; Woods, R.D. Etiology of African cassava mosaic disease. Plant Dis. 1983, 67, 994–995. [Google Scholar] [CrossRef]
- Chiang, B.T.; Nakhla, M.K.; Maxwell, D.P. A new geminivirus associated with a leaf curl disease of tomato in Tanzania. Plant Dis. 1997, 81, 111. [Google Scholar]
- Liu, L.; van Tonder, T.; Pietersen, G.; Davies, J.W.; Stanley, J. Molecular characterization of a subgroup I geminivirus from a legume in South Africa. J. Gen. Virol. 1997, 78, 2113–2117. [Google Scholar]
- Ogbe, F.O.; Legg, J.; Raya, M.D.; Muimba-Kankalongo, A.; Theu, M.P.; Kaitisha, G.; Phiri, N.A.; Chalwe, A. Diagnostic survey of cassava mosaic viruses in Tanzania, Malawi and Zambia. Roots 1997, 4, 12–15. [Google Scholar]
- Zhou, X.; Robinson, D.J.; Harrison, B.D. Types of variation in DNA-A among isolates of East African cassava mosaic virus from Kenya, Malawi and Tanzania. J. Gen. Virol. 1998, 79, 2835–2840. [Google Scholar]
- Paximadis, M.; Idris, A.M.; Torres-Jerez, I.; Villarreal, A.; Rey, M.E.C.; Brown, J.K. Characterization of tobacco geminiviruses in the old and new world. Arch. Virol. 1999, 144, 703–717. [Google Scholar] [CrossRef]
- Berrie, L.C.; Rybicki, E.P.; Rey, M.E.C. Complete nucleotide sequence and host range of South African cassava mosaic virus: Further evidence for recombination amongst begomoviruses. J. Gen. Virol. 2001, 82, 53–58. [Google Scholar]
- Berry, S.; Rey, M.E.C. Molecular evidence for the existence of biodiverse begomovirus populations in cassava in southern Africa. Arch. Virol. 2011, 146, 1795–1802. [Google Scholar] [CrossRef]
- Paximadis, M.; Rey, M.E.C. Genome organization of Tobacco leaf curl Zimbabwe virus, a new, distinct monopartite begomovirus associated with subgenomic defective DNA molecules. J. Gen. Virol. 2001, 82, 3091–3097. [Google Scholar]
- Ranomenjanahary, S.; Rabindran, R.; Robinson, D.J. Occurrence of three distinct begomoviruses in cassava in Madagascar. Ann. Appl. Biol. 2002, 140, 315–318. [Google Scholar] [CrossRef]
- Briddon, R.W.; Robertson, I.; Markham, P.G.; Stanley, J. Occurrence of South African cassava mosaic virus (SACMV) in Zimbabwe. Plant Pathol. 2004, 53, 233. [Google Scholar] [CrossRef]
- Delatte, H.; Holota, H.; Naze, F.; Peterschmitt, M.; Reynaud, B.; Lett, J.M. The presence of both recombinant and non-recombinant strains of Tomato yellow leaf curl virus on tomato in Réunion Island. Plant Pathol. 2005, 54, 262. [Google Scholar] [CrossRef]
- Delatte, H.; Martin, D.P.; Naze, F.; Golbach, R.W.; Reynaud, B.; Peterschmitt, M; Lett, J.M. South West Indian Ocean islands tomato begmovirus populations represent a new major monopartite begomovirus group. J. Gen. Virol. 2005, 86, 1533–1542. [Google Scholar] [CrossRef]
- Ndunguru, J.; Legg, J.P.; Aveling, T.A.; Thompson, G.; Fauquet, C.M. Molecular biodiversity of cassava begomoviruses in Tanzania: Evolution of cassava geminiviruses in Africa and evidence for East Africa being a center of diversity of cassava geminiviruses. Virol. J. 2005, 2, 21. [Google Scholar] [CrossRef]
- Shih, S.L.; Tsai, W.S.; Green, S.K.; Lee, L.M. Molecular characterization of a distinct begomovirus associated with tomato leaf curl diseases in Arusha of Tanzania. Plant Dis. 2006, 90, 1550. [Google Scholar]
- Domola, M.J. Survey and characterization of sweet potato viruses in South Africa. M.Sc. thesis, University of Pretoria, Pretoria, South Africa, December 2003. [Google Scholar]
- Pietersen, G.; Idris, A.M.; Kruger, K.; Brown, J.K. Characterization of Tomato curly stunt virus: A new tomato-infecting begomovirus from South Africa. Plant Pathol. 2008, 57, 809–818. [Google Scholar] [CrossRef]
- Abraham, N. Characterization of a subgenomic molecule associated with South African cassava mosaic virus. M.Sc. thesis, University of the Witwatersrand, Johannesburg, South Africa, March 2012. [Google Scholar]
- Esterhuizen, L.L.; van Heerden, S.W.; Rey, M.E.C.; van Heerden, H. Epidemiology and molecular characterization of Tomato curly stunt virus and its insect vector Bemisia tabaci in South Africa. In Abstract of the 6th international geminivirus symposium, Guanajuato, Mexico, 7-12 November 2010.
- Lobin, K.; Druffel, K.L.; Pappu, H.R.; Benimadhu, S.P. First report of Tomato yellow leaf curl virus in Mauritius. Plant Dis. 2010, 94, 1261. [Google Scholar]
- Thierry, M.; Lefeuvre, P.; Hoareau, M.; Perefarres, F.; Delatte, H.; Reynaud, B.; Martin, D.M.; Lett, J.M. Differential disease phenotype of begomoviruses associated with tobacco leaf curl disease in Comoros. Arch. Virol. 2012, 157, 545–550. [Google Scholar] [CrossRef]
- FAOSTAT. Food and Agriculture Organization of the United Nations. Available online: http://faostat.fao.org/ (accessed on 21 April 2012).
- Woodward, B.R.; Allemann, J.; O'Regan, B.P. Tissue culture of cassava: A South African perspective. African Journal of Root and Tuber Crops 1997, 2, 243–245. [Google Scholar]
- Trench, T.N.; Martin, M.M. An assessment of cassava African mosaic disease in South Africa and Swaziland. South African Journal of Plant and Soil 1985, 2, 169–170. [Google Scholar]
- Haggblade, S.; Zulu, B. The recent cassava surge in Zambia and Malawi. In Proceedings of the Went, IFPRI, NEPAD, CTA Conference Success in African Agriculture, Pretoria, South Africa, 1–3 December 2003.
- Burns, A.; Gleadow, R.; Cliff, J.; Zacarias, T.; Cavagnaro, A. Cassava: The drought, war and famine crop in a changing world. Sustainability 2010, 2, 3572–3607. [Google Scholar] [CrossRef]
- Hillocks, R.J. Cassava in Africa. In Cassava: Biology, Production and Utilization, 1st ed; CABI Publishing: Wallingford, UK, 2002; pp. 41–54. [Google Scholar]
- Balat, M.; Balat, H. Recent trends in global production and utilization of bio-ethanol fuel. Appl. Energ. 2009, 86, 2273–2282. [Google Scholar] [CrossRef]
- Warburg, O. Die Kulturpflanzen Usambaras (in Dutch). Miteilung Deutsch Schutzgeb 1894, 7, 131. [Google Scholar]
- Fauquet, C.M.; Fargette, D. African cassava mosaic virus: Etiology, epidemiology, and control. Plant Dis. 1990, 74, 404–411. [Google Scholar] [CrossRef]
- Bock, K.R.; Guthrie, E.J. Transmission of African cassava mosaic by mechanical inoculation. Plant Dis. Rep. 1978, 62, 580–581. [Google Scholar]
- Bock, K.R.; Woods, R.D. Etiology of African cassava mosaic disease. Plant Dis. 1983, 67, 994–995. [Google Scholar] [CrossRef]
- Legg, J.P.; Owor, B.; Sseruwagi, P.; Ndunguru, J. Cassava mosaic virus disease in east and central Africa: Epidemiology and management of a regional pandemic. Adv. Virus Res. 2006, 67, 355–418. [Google Scholar] [CrossRef]
- Fauquet, C.M.; Stanley, J. Geminivirus classification and nomenclature: Progress and problems. Ann. Appl. Biol. 2003, 142, 165–189. [Google Scholar] [CrossRef]
- Swanson, M.M.; Harrison, B.D. Properties, relationships and distribution of cassava mosaic geminiviruses. Trop. Sci. 1994, 34, 15–25. [Google Scholar]
- Patil, B.L.; Fauquet, C.M. Cassava mosaic geminiviruses: Actual knowledge and perspectives. Mol. Plant Pathol. 2009, 10, 685–701. [Google Scholar]
- Lava Kumar, P.; Akinbade, S.A.; Dixon, A.G.O.; Mahungu, N.M.; Mutunda, M.P.; Kiala, D.; Londa, L.; Legg, J.P. First report of the occurrence of East African cassava mosaic virus-Uganda (EACMV-UG) in Angola. Plant Pathol. 2009, 58, 402. [Google Scholar]
- Harimalala, M.A.; Lefeuvre, P.; de Bruyn, A.; Tiendrebeogo, F.; Hoareau, M.; Villemot, J.; Ranomenjanahary, S.; Andrianjaka, A.; Reynaud, B.; Lett, J-M. A novel cassava-infecting begomovirus from Madagascar: Cassava mosaic Madagascar virus. Arch. Virol. 2012. [Google Scholar]
- Monde, G.; Walangululu, J.; Winter, S.; Bragard, C. Dual infection by cassava begomoviruses in two leguminous species (Fabaceae) in Yangambi, Northeastern Democratic Republic of Congo. Arch. Virol. 2010, 155, 1865–1869. [Google Scholar] [CrossRef]
- Neuenschwander, P.; Hughes, J.; Ogbe, F.; Ngatse, J.M.; Legg, J.P. Occurrence of the Uganda variant of East African Cassava Mosaic Virus (EACMV-Ug) in Western Democratic Republic of Congo and the Congo Republic defines the westernmost extent of the CMD pandemic in East/Central Africa. Plant Pathol. 2002, 51, 385. [Google Scholar] [CrossRef]
- Tata-Hangy, K.; Legg, J.R.; Hanna, J.R.; Toko, M.; Lema, K.M.A.; Dixon, A.; Mahungu, N.M. Incidence and distribution of cassava diseases and pests in the Democratic Republic of Congo. In Proceedings of the 91st ISTRC-AB Symposium, Mombasa, Kenya, 31 October–5 November 2004; pp. 614–622.
- Tata-Hangy, K.; Koffi-Tete, M.; Obonyo, R.; Okao-Okuja, G.; Asiimwe, P.; Legg, J.P. Monitoring and diagnostic survey of cassava mosaic virus disease (CMD) in Eastern Democratic Congo. Available online: http://www.iita.org/reports (accessed on 13 September 2012).
- Bisimwa, E.; Walangululu, J.; Bragard, C. Occurrence and Distribution of cassava mosaic begomovirus related to agro-ecosystems in the Sud-kivu Province, Democratic Republic of Congo. Asian J. Plant Pathol. 2012, 6, 1–12. [Google Scholar] [CrossRef]
- Berrie, L.C.; Palmer, K.E.; Rybicki, E.P.; Hiyadat, S.H.; Maxwell, D.P.; Rey, M.E.C. A new isolate of African cassava mosaic virus in South Africa. African Journal of Root and Tuber Crops 1997, 2, 49–52. [Google Scholar]
- Mabasa, K.G. Epidemiology of Cassava moaic disease and molecular characterization of cassava mosaic viruses and their associated whitefly (Bemisia tabaci) vector in South Africa. M.Sc. thesis, University of the Witwatersrand, Johannesburg, South Africa, 28 February 2008. [Google Scholar]
- Cossa, N. Epidemiology of Cassava Mosaic Disease in Mozambique. M.Sc. thesis, University of the Witwatersrand, Johannesburg, South Africa, 28 February 2010. [Google Scholar]
- Briddon, R.W.; Bull, S.E.; Mansoor, S.; Amin, I; Markham, P.G. Universal primers for the PCR-mediated amplification of DNA β. Mol. Biotech. 2002, 20, 315–318. [Google Scholar] [CrossRef]
- Maruthi, M.N.; Colvin, J.; Seal, S.; Gibson, G.; Cooper, J. Co-adaptation between cassava mosaic geminiviruses and their local vector populations. Virus Res. 2002, 86, 71–85. [Google Scholar] [CrossRef]
- Tiendrébéogo, F.; Lefeuvre, P.; Hoareau, M.; Harimalala, M.A.; de Bruyn, A.; Villemot, J.; Traoré, V.S.E.; Konaté, G.; Traoré, A.S.; Barro, N.; et al. Evolution of African cassava mosaic virus by recombination between bipartite and monopartite begomoviruses. Virol. J. 2012, 9, 67. [Google Scholar] [CrossRef] [Green Version]
- Stanley, J.; Townsend, R. Characterization of DNA forms associated with cassava latent virus infection. Nucleic Acids Res. 1985, 13, 2189–2206. [Google Scholar] [CrossRef]
- Stenger, D.C.; Stevenson, M.C.; Hormuzdi, S.G.; Bisaro, D.M. A number of subgenomic DNAs are produced following agroinoculation of plants with beet curly top virus. J. Gen. Virol. 1992, 73, 237–242. [Google Scholar] [CrossRef]
- Patil, B.L.; Dasgupta, I. Defective interfering DNAs of plant viruses. Crit. Rev. Plant Sci. 2006, 25, 47–64. [Google Scholar] [CrossRef]
- Stanley, J.; Saunders, K.; Pinner, M.S.; Wong, S.M. Novel defective interfering DNAs associated with Ageratum Yellow Vein Geminivirus infection of Ageratum conyzoides. Virology 1997, 239, 87–96. [Google Scholar] [CrossRef]
- Stanley, J.; Frischmuth, T.; Ellwood, S. Defective viral DNA ameliorates symptoms of geminivirus infection in transgenic plants. Genetics 1990, 87, 6291–6295. [Google Scholar]
- Stanley, J. Subviral DNAs associated with geminivirus disease complexes. Vet. Microbiol. 2004, 98, 121–129. [Google Scholar] [CrossRef]
- Patil, B.L.; Dutt, N.; Briddon, R.W.; Bull, S.E.; Rothenstein, D.; Borah, B.K.; Dasgupta, I.; Stanley, J.; Jeske, H. Deletion and recombination events between the DNA-A and DNA-B components of Indian cassava-infecting geminiviruses generate defective molecules in Nicotiana benthamiana. Virus Res. 2007, 124, 59–67. [Google Scholar] [CrossRef]
- Kuhn, S.L. The characterization of a SACMV subgenomic DNA molecule for use in symptom amelioration of SACMV infected cassava. M.Sc. thesis, University of the Witwatersrand, Johannesburg, South Africa, 22 December 2003. [Google Scholar]
- Whitham, S.; Wang, Y. Roles for host factors in plant viral pathogenicity. Curr. Opin. Plant Biol. 2004, 7, 365–71. [Google Scholar] [CrossRef]
- Goodin, M.M.; Zaitlin, M.; Naidu, R.A.; Lommel, S.A. Nicotiana benthamiana: It’s history and future as a model for plant-pathogen interactions. Mol. Plant Microbe Interact. 2008, 21, 1015–1026. [Google Scholar] [CrossRef]
- Townsend, R.; Watts, J.; Stanley, J. Synthesis of viral DNA forms in Nicotiana plumbaginifolia protoplasts inoculated with cassava latent virus (CLV): Evidence for the independent replication of one component of the CLV genome. Nucleic Acids Res. 1986, 14, 1253–1265. [Google Scholar] [CrossRef]
- Foolad, M.R. Genome mapping and molecular breeding of tomato. Int. J. Plant Genom. 2007, 64358, 1–52. [Google Scholar]
- Diez, M.J.; Nuez, F. Tomato. In Handbook of Plant Breeding: Vegetables II; Springer: New York, NY, USA, 2008; pp. 249–323. [Google Scholar]
- Labate, J.A.; Grandillo, S.; Fulton, T.; Munos, S.; Caicedo, A.L.; Peralta, I.; Ji, Chetelat, R.T.; Scott, J.W.; Gonzalo, M.J.; et al. Tomato. In Genome Mapping and Molecular Breeding in Plants, Vegetables; Kole, C.R., Ed.; Springer-Verlag: Berlin, Heidelberg, Germany, 2007; pp. 1–125. [Google Scholar]
- Lapidot, M.; Ben-Joseph, R.; Cohen, L.; Machbash, Z.; Levy, D. Development of a scale for evaluation of Tomato yellow leaf curl virus resistance level in tomato plants. Phytopathology 2006, 96, 1404–1408. [Google Scholar] [CrossRef]
- Levy, D.; Lapidot, M. Effect of plant age at inoculation on expression of genetic resistance to tomato yellow leaf curl virus. Arch. Virol. 2008, 153, 171–179. [Google Scholar] [CrossRef]
- Lefeuvre, P.; Martin, D.P.; Harkins, G.; Lemey, P.; Gray, A.J.A.; Meredith, S.; Lakay, F.; Monjane, A.; Lett, J.-M.; Varsani, A.; Haydarnejads, J. The spread of tomato yellow leaf curl virus from the Middle East to the world. PLoS Pathog. 2010, 6, e1001164. [Google Scholar]
- Duffy, S.; Holmes, E.C. Multiple introductions of the Old World Begomovirus Tomato yellow leaf curl virus into the New World. Appl. Environ. Microbiol. 2007, 73, 7114–7117. [Google Scholar] [CrossRef]
- Czosnek, H.; Navot, N.; Laterrot, H. Geographical distribution of tomato yellow leaf curl virus. A first survey using a specificc DNA probe. Phytopatol. Medit. 1990, 29, 1–6. [Google Scholar]
- Czosnek, H.; Laterrot, H. A worldwide survey of tomato yellow leaf curl viruses. Arch. Virol. 1997, 142, 1391–1406. [Google Scholar] [CrossRef]
- Thindwa, H.; Khonje, P. Whiteflies as vectors of cassava and sweetpotato in Africa: Malawi. In Whiteflies and Whitefly-Borne Viruses in the Tropics: Building a Knowledge Base for Global Action, 1st ed; Centro Internacional de Agricultura Tropical: Cali, Colombia, 2005; pp. 150–156. [Google Scholar]
- Harrison, B.D.; Robinson, D.J. Natural genomic and antigenic variation in whitefly-transmitted geminiviruses (begomoviruses). Annu. Rev. Phytopathol. 1999, 37, 369–398. [Google Scholar] [CrossRef]
- Kashina, B.D.; Mabagala, R.B.; Mpunami, A.A. Molecular characterization of isolates of Tomato yellow leaf curl from Tanzania. Archiv. Phytopath. Pflanz. 2002, 35, 255–267. [Google Scholar] [CrossRef]
- Peterschmitt, M.; Granier, M.; Mekdoud, R.; Dalmon, A.; Vayssieres, J.F.; Gambin, O.; Reynaud, B. First report of Tomato yellow leaf curl virus in Réunion. Plant Dis. 1999, 83, 303. [Google Scholar]
- Delatte, H.; Reynaud, B.; Lett, J.M.; Peterschmitt, M.; Granier, M.; Ravololonandrianina, J.; Goldbach, W.R. First molecular identification of a begomovirus isolated from tomato in Madagascar. Plant Dis. 2002, 86, 1404. [Google Scholar]
- Pietersen, G.; Idris, A.M.; Krüger, K.; Brown, J.K. Tomato curly stunt virus, a new begomovirus of tomato within the Tomato yellow leaf curl virus—Is cluster in South Africa. Plant Dis. 2000, 84, 810. [Google Scholar]
- Esterhuizen, L.L. A study of the South African Tomato curly stunt virus pathosystem: Epidemiology, molecular diversity and resistance. Ph.D. thesis, University of Johannesburg, Johanesburg, South Africa, May 2012. [Google Scholar]
- Polston, J.E.; Chellemi, D.O.; Schuster, D.J.; McGovern, R.J.; Stansly, P.A. Spatial and temporal dynamics of tomato mottle geminivirus and Bemisia tabaci (Genn) in Florida tomato fields. Plant Dis. 1996, 80, 1022–1028. [Google Scholar] [CrossRef]
- Morales, F.J. History and current distribution of begomoviruses in Latin America. Adv. Virus Res. 2006, 67, 127–162. [Google Scholar] [CrossRef]
- Jones, R.A.C. Plant virus emergence and evolution: Origins, new encounter scenarios, factors driving emergence, effects of changing world conditions and prospects for control. Virus Res. 2009, 141, 113–130. [Google Scholar] [CrossRef]
- Delatte, H.; Holota, H.; Warren, B.H.; Becker, N.; Thierry, M.; Reynaud, B. Genetic diversity, geographical range and origin of Bemisia tabaci (Hemiptera: Aleyrodidae) Indian Ocean Ms. B. Entomol. Res. 2011, 101, 1–11. [Google Scholar] [CrossRef]
- Nuaila, V.N. Epidemiology of Tomato curly stunt disease in Mozambique. M.Sc. thesis, University of the Witwatersrand, Johannesburg, South Africa.
- Pietersen, G.; Smith, M.F. Tomato yellow leaf curl virus resistant tomatoes show resistance to Tomato curly stunt virus. Plant Dis. 2002, 86, 528–534. [Google Scholar] [CrossRef]
- Kays, S.J. Sweet potato production worldwide: Assessment, trends, and the future. Acta Hortic. 2005, 670, 19–25. [Google Scholar]
- Kreuze, J.F. Molecular studies on the sweet potato virus disease and it two causal agents. Ph.D. thesis, Swedish University of Agricultural Sciences, Sweden, January 2002. [Google Scholar]
- Valverde, R.A.; Clark, C.A.; Valkonen, J.P.T. Viruses and virus disease complexes of sweetpotato. Plant Viruses 2007, 1, 116–126. [Google Scholar]
- Tesfaye, T.; Feyissa, T.; Abraham, A. Survey and serological detection of sweet potato (Ipomoea batatas (L.) Lam) viruses in Ethiopia. J. Appl. Biosciences 2011, 41, 2746–2756. [Google Scholar]
- Gibson, R.W.; Mwanga, R.O.M.; Kasule, S.; Mpembe, I.; Carey, E.E. Apparent absence of viruses in most symptomless field-grown sweet potato in Uganda. Ann. Appl. Biol. 1997, 130, 481–490. [Google Scholar] [CrossRef]
- Mukasa, S.B.; Rubaihayo, P.R.; Valkonen, J.P.T. Incidence of viruses and viruslike diseases of sweetpotato in Uganda. Plant Dis. 2003, 87, 329–335. [Google Scholar] [CrossRef]
- Aritua, V.; Adipala, E. Characteristics and diversity in sweetpotato-infecting viruses in Africa. Acta Hort. 2006, 703, 175–180. [Google Scholar]
- Ateka, E.M.; Njeru, R.W.; Kibaru, A.G.; Kimenju, J.W.; Barg, E.; Gibson, R.W.; Vetten, H.J. Identification and distribution of viruses infecting sweet potato in Kenya. Ann. Appl. Biol. 2004, 144, 371–379. [Google Scholar] [CrossRef]
- Fauquet, C.M.; Stanley, J. Revising the way we conceive and name viruses below the species level: A review of geminivirus taxonomy calls for new standardized isolate descriptors. Arch. Virol. 2005, 150, 2151–2179. [Google Scholar] [CrossRef]
- Lozano, G.; Trenado, H.P.; Valverde, R.A.; Navas-Castillo, J. Novel begomovirus species of recombinant nature in sweet potato (Ipomoea batatas) and Ipomoea indica: taxonomic and phylogenetic implications. J. Gen. Virol. 2009, 90, 2550–2562. [Google Scholar] [CrossRef]
- Paprotka, T.; Boiteux, L.S.; Fonseca, M.E.; Resende, R.O.; Jeske, H.; Faria, J.C.; Ribeiro, S.G. Genomic diversity of sweet potato geminiviruses in a Brazilian germplasm bank. Virus Res. 2010, 149, 224–233. [Google Scholar] [CrossRef]
- Albuquerque, L.C.; Inoue-Nagata, A.K.; Pinheiro, B.; Ribeiro, S.G.; Resende, R.O.; Moriones, E.; Navas-Castillo, J. A novel monopartite begomovirus infecting sweet potato in Brazil. Arch. Virol. 2011, 156, 1291–129. [Google Scholar] [CrossRef] [Green Version]
- Miano, D.W.; LaBonte, D.R.; Clark, C.A.; Valverde, R.A.; Hoy, M.W.; Hurtt, S.; Li, R. First report of a begomovirus infecting sweet potato in Kenya. Plant Dis. 2006, 90, 832. [Google Scholar]
- Wasswa, P.; Otto, B.; Maruthi, M.N.; Mukasa, S.B.; Monger, W.; Gibson, R.W. First identification of a sweet potato begomovirus (swepovirus) in Uganda: Characterization, detection and distribution. Plant Pathol. 2011, 60, 1030–1039. [Google Scholar] [CrossRef]
- Banks, G.K.; Bedford, I.D.; Beitia, F.J.; Rodriguez-Cerezo, E.; Markham, P.G. A novel geminivirus of Ipomoea indica (Convolvulacae) from Southern Spain. Plant Dis. 1999, 83, 486. [Google Scholar]
- Ling, K.S.; Jackson, D.M.; Harrison, H.; Simmons, A.M.; Pesic-VanEsbroeck, Z. Field evaluation of yield effects on the U.S.A. heirloom sweet potato cultivars infected by Sweet potato leaf curl virus. Crop Protect. 2010, 29, 757–765. [Google Scholar] [CrossRef]
- Briddon, R.W.; Bull, S.E.; Bedford, I.D. Occurrence of Sweet potato leaf curl virus in Sicily. Plant Pathol. 2006, 55, 286–286. [Google Scholar]
- Fuentes, S.; Salazar, L.F. First report of Sweet potato leaf curl virus in Peru. Plant Dis. 2003, 87, 98. [Google Scholar]
- Lotrakul, P.; Valverde, R.A.; Clark, C.A.; Sim, J.; de La Torre, R. Detection of a geminivirus infecting sweet potato in the United States. Plant Dis. 1998, 85, 1253–1257. [Google Scholar]
- Luan, Y.S.; Zhang, J.; Liu, D.M.; Li, W.L. Molecular characterization of Sweet potato leaf curl virus isolate from China (SPLCV-CN) and its phylogenetic relationship with other members of the Geminiviridae. Virus Genes 2007, 35, 379–385. [Google Scholar] [CrossRef]
- Prasanth, G.; Hegde, V. Occurrence of Sweet potato feathery mottle virus and Sweet potato leaf curl Georgia virus on sweet potato in India. Plant Dis. 2008, 92, 311. [Google Scholar]
- Trenado, H.P.; Orilio, A.F.; Marquez-Martin, B.; Moriones, E.; Navas-Castillo, J. Sweepoviruses cause disease in swet potato and related Ipomoea spp.: Fulfilling Koch’s Postulates for a divergent group in the Genus Begomovirus. PLoS One 2011, 6, e27329. [Google Scholar]
- FAO. Food and Agricultural Organization of the United Nations, Rome. Available online: http://www.fao.org/DOCREP/006/Y4956E/Y4956E00.htm (accessed on 20 June 2012).
- Lucas, G.B. Diseases of Tobacco, 1st ed; Harold E. Parker and Sons: Raleigh, NC, USA, 1975. [Google Scholar]
- Storey, H.H. A new virus disease of the tobacco plant. Nature 1931, 128, 187–188. [Google Scholar] [CrossRef]
- Storey, H.H. Leaf curl diseases of tobacco in southern Rhodesia. Rhod. Agr. J. 1932, 29, 186–192. [Google Scholar]
- Lucas, G.B. Diseases of Tobacco; Scarecrow Press, Inc.: New York, NY, USA, 1958. [Google Scholar]
- Osaki, T.; Inouye, T. Tobacco leaf curl virus. CMI/AAB Descriptions of Plant Viruses 1981, No. 232.
- Pal, B.P.; Tandon, R.K. Types of tobacco leaf curl in Northern India. Indian J. Agr. Sci. 1937, 7, 363–393. [Google Scholar]
- McLean, A.P.D. Some leaf curl diseases in South Africa. Science Bulletin Department of Agriculture South Africa 1940, 225, 1–45. [Google Scholar]
- Hill, B.G. Occurrence of Bemisia tabaci (Genn.) in the field and its relationship to the leaf curl disease of tobacco. S. Afr. J. Agr. Sci. 1968, 11, 583–593. [Google Scholar]
- Paximadis, M.; Rey, M.E.C. Aetiology of tobacco leaf curl in southern Africa. Ann. Appl. Biol. 1997, 131, 449–457. [Google Scholar] [CrossRef]
- Moran, Y.M.; Ramos, P.L.; Dominguez, M.; Fuentes, A.D.; Sanchez, Y.; Crespo, A. Tobacco leaf curl Cuba virus, a new begomovirus infecting tobacco (Nicotiana tabacum) in Cuba. Plant Pathol. 2006, 55, 570. [Google Scholar]
- Shimizu, S.; Ikegami, M. Complete nucleotide sequence and the genome organization of Tobacco leaf curl from Japan. Microbiol. Immunol. 1999, 43, 989–992. [Google Scholar]
- Xie, Y.; Jiang, T.; Zhou, X.P. Agroinoculation shows Tobacco leaf curl Yunnan virus is a monopartite begomovirus. Eur. J. Plant Pathol. 2006, 115, 369–375. [Google Scholar] [CrossRef]
- Xie, Y.; Peijun, W.; Liu, P.; Gong, H.; Zhou, X.P. Characterization of alphasatellites associated with monopartite begomovirus/betasatellite complexes in Yunnan, China. Virol. J. 2010, 7, 178. [Google Scholar] [CrossRef]
- Briddon, R.W.; Bull, S.E.; Mansoor, S.; Amin, L.; Markham, P.G. Universal primers for the PCR-mediated amplification of DNA: A molecule associated with some monopartite begomoviruses. Mol. Biotechnol. 2002, 20, 315–318. [Google Scholar] [CrossRef]
- Gutierrez, C. Geminivirus DNA replication. Cell. Mol. Life Sci. 1999, 56, 313–329. [Google Scholar] [CrossRef]
- Valand, G.B.; Muniyappa, V. Epidemiology of tobacco leaf curl virus in India. Ann. Appl. Biol. 1992, 120, 257–267. [Google Scholar] [CrossRef]
- Spence, N.J.; Walkey, D.G.A. Variation for pathogenicity among isolates of bean common mosaic virus in Africa and a reinterpretation of the genetic relationship between cultivars of Phaseolus vulgaris and pathotypes of BCMV. Plant Pathol. 1995, 44, 527–546. [Google Scholar] [CrossRef]
- Howarth, A.J.; Caton, J.; Bossert, M.; Goodman, M. Nucelotide sequence of a bean golden mosaic virus and a model for gene regulation in geminiviruses. Proc. Natl. Acad. Sci. U. S. A. 1985, 82, 3572–3576. [Google Scholar] [CrossRef]
- Hidayat, S.H.; Gilbertson, R.L.; Hanson, S.F.; Morales, F.J.; Ahlquist, P.; Maxwell, D.P. Complete nucleotide sequences of the infectious cloned DNAs of bean dwarf mosaic geminivirus. Phytopathology 1993, 83, 181–187. [Google Scholar] [CrossRef]
- Morris, B.A.M.; Richardson, K.A.; Haley, A.; Zhan, X.; Thomas, E. The nucleotide sequence of the infectious cloned DNA components of tobacco yellow dwarf virus reveals features geminiviruses infecting monocotyledenous plants. Virology 1992, 187, 633–642. [Google Scholar] [CrossRef]
- Alabi, J.O.; Ogbe, F.; Bandyopadhyay, R.; Lava Kumar, P. Alternative hosts of African cassava mosaic virus and East African cassava Camerron virus in Nigeria. Arch. Virol. 2008, 153, 1743–1747. [Google Scholar] [CrossRef]
- Kitamura, K.; Murayama, A.; Ikegami, M. Evidence for recombination among isolates of Tobacco leaf curl Japan virus and Honeysuckle yellow vein mosaic virus. Arch. Virol. 2004, 149, 1221–1229. [Google Scholar] [CrossRef]
- Briddon, R.W.; Patil, B.L.; Bagewadi, B.; Nawaz-ul-Rehman, M.S.; Fauquet, C.M. Distinct evolutionary histories of the DNA-A and DNA-B components of bipartite begomoviruses. BMC Evol. Biol. 2010, 97. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Rey, M.E.C.; Ndunguru, J.; Berrie, L.C.; Paximadis, M.; Berry, S.; Cossa, N.; Nuaila, V.N.; Mabasa, K.G.; Abraham, N.; Rybicki, E.P.; et al. Diversity of Dicotyledenous-Infecting Geminiviruses and Their Associated DNA Molecules in Southern Africa, Including the South-West Indian Ocean Islands. Viruses 2012, 4, 1753-1791. https://doi.org/10.3390/v4091753
Rey MEC, Ndunguru J, Berrie LC, Paximadis M, Berry S, Cossa N, Nuaila VN, Mabasa KG, Abraham N, Rybicki EP, et al. Diversity of Dicotyledenous-Infecting Geminiviruses and Their Associated DNA Molecules in Southern Africa, Including the South-West Indian Ocean Islands. Viruses. 2012; 4(9):1753-1791. https://doi.org/10.3390/v4091753
Chicago/Turabian StyleRey, Marie E. C., Joseph Ndunguru, Leigh C. Berrie, Maria Paximadis, Shaun Berry, Nurbibi Cossa, Valter N. Nuaila, Kenneth G. Mabasa, Natasha Abraham, Edward P. Rybicki, and et al. 2012. "Diversity of Dicotyledenous-Infecting Geminiviruses and Their Associated DNA Molecules in Southern Africa, Including the South-West Indian Ocean Islands" Viruses 4, no. 9: 1753-1791. https://doi.org/10.3390/v4091753





