Glycosphingolipids as Receptors for Non-Enveloped Viruses
Abstract
:1. Introduction
2. Discussion
2.1. Structure and function of glycosphingolipids
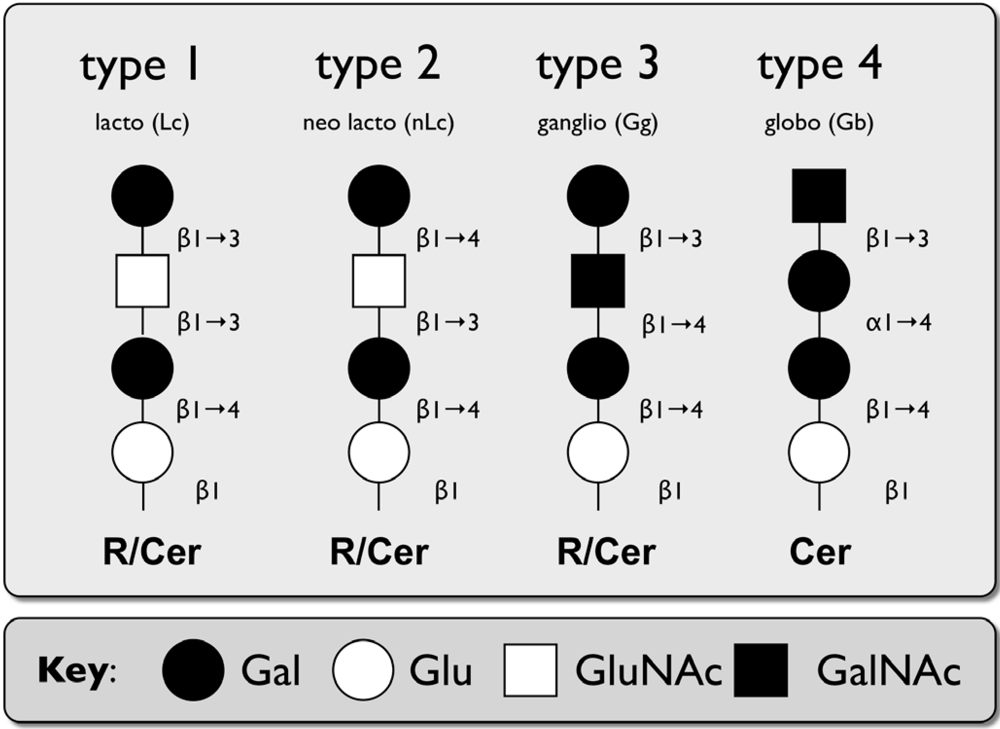
2.2. Caliciviridae
2.2.1. Noroviruses (NoV)
2.2.1.1. Human noroviruses (HuNoV)
| Virus | Glycosphingolipid Receptors | Other Receptors | Reference |
|---|---|---|---|
| Caliciviridae | |||
| Norovirus | |||
| Human Norovirus(HuNoV) | Histo-blood group antigens (HBGA) on type 1, 2, and 3 GSLs | Heparan sulfate,sialic acid on SLex | [44,61,62,63,64] |
| Murine Norovirus(MNV) | Terminal α2,3-linked sialic acid on GD1a and GT1b | [70] and this article | |
| Bovine Norovirus(BoNoV) | αGal of HBGA | [71] | |
| Lagovirus | |||
| Rabbit Hemorrhagic Disease Virus (RHDV) | A and H type 2 HBGA | [72] | |
| Reoviridae | |||
| Rotavirus | Integrins α2β1, α4β1, αvβ3, αxβ2, hsc70 | [73] | |
| Neuraminidase-sensitive | |||
| Porcine Rotavirus: OSU strain | Sialic acid on GM3 | [74,75] | |
| Porcine Rotavirus: CRW-8 strain | Terminal and internal α2,3-linked sialic acid on GD1a | [76] | |
| Simian Rotavirus: SA11 strain | Sialyl-galactose on NeuGcGM3, sialylneolactotetraosylceramide, GM2, and GD1a | [77] | |
| Bovine Rotavirus:NCVD strain | Sialyl-galactose (NeuGc/NeuAcα3-Galβ) on NeuGcGM3, sialylneolactotetraosylceramide, GM2, and GD1a | [77] | |
| Rhesus Rotavirus: RRV strain | N-acetyl neuraminic acid | [78] | |
| Neuraminidase-insensitive | |||
| Bovine Rotavirus:UK strain | Sialyl-galactose (NeuAc) on NeuGcGM3, GM1, GD1a, GM2, sialylneolactotetraosylceramide | [77] | |
| Human Rotavirus:KU, MO, DS-1, Wa strains | GM1 | [76,79] | |
| Polyomaviridae | |||
| Murine Polyomavirus (MPyV) | Terminal α2,3-linked sialic acid on GD1a and GT1b | Integrin α4β1 | [80,81,82,83] |
| Simian Virus 40 (SV40) | GM1 | Class I MHC | [80,84,85] |
| BK Virus (BKV) | α2,8-linked disialic acid on GD1b and GT1b | Unknown glycoprotein | [86,87] |
| JC Virus (JCV) | Terminal α2,3-linked sialic acid on GT1b | Serotonin receptor 5HT2aR; Terminal α2,6-linked sialic acid on an unknown glycoprotein | [88,89,90,91] |
| Merkel Cell Polyomavirus (MCPyV) | Terminal α2,3-linked sialic acid and α2,8-linked disialic acid on GT1b | [92] | |
| Parvoviridae | |||
| Erythrovirus | |||
| Human Parvovirus B19 | HexNAcβ1,3Gal on globoside Gb4 (P antigen), SSEA-3, SSEA-4, and nLc4 | Integrin α5β1, autoantigen Ku80 | [93,94,95,96] |
| Simian Parvovirus | Globoside and Forssmann antigen | [9797] | |
| Dependovirus | |||
| Bovine Adeno-associated Virus (BAAV) | Unknown ganglioside | [98] |
| Name | Structure | Residue (R) |
|---|---|---|
| Type 1 | Galβ1-3, GlcNAcβ1-R | N-,O-glycoproteins, GSLs of the lactoseries (Lc) |
| Type 2 | Galβ1-4, GlcNAcβ1-R | N-,O-glycoproteins, GSLs of the neolactoseries (nLc) |
| Type 3 | Galβ1-3, GalNAcα1-R | O-glycoproteins (core 1), GSLs of the ganglioseries (Gg) |
| Type 4 | Galβ1-3, GalNAcβ1-R | GSLs of the globoseries (Gb) |
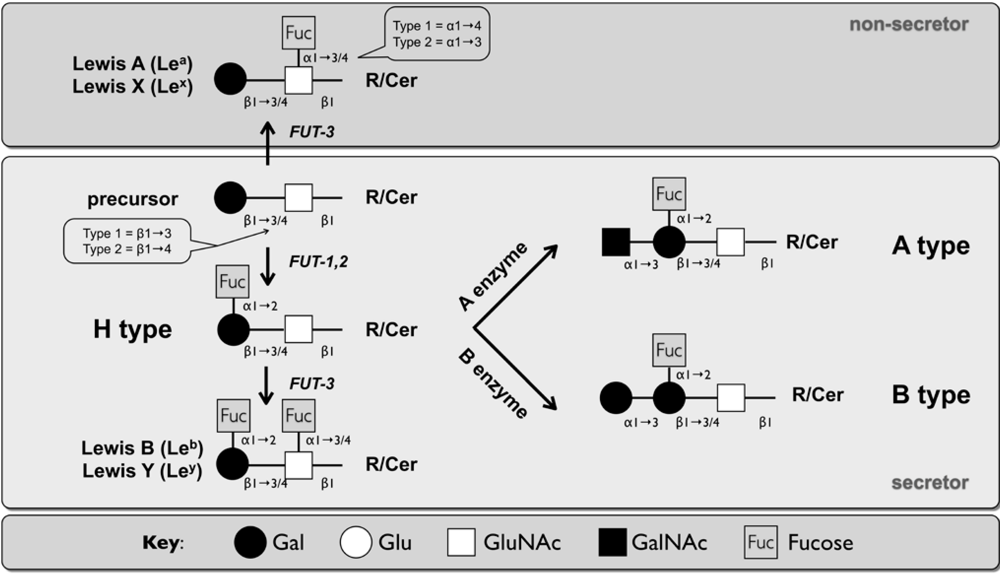

2.2.1.2. Murine Norovirus

2.2.1.3. Bovine Norovirus (BoNoV)
2.2.2. Lagovirus
2.3. Rotaviruses
2.3.1. Neuraminidase-sensitive strains
2.3.1.1 Porcine rotaviruses (OSU, CRW strains)
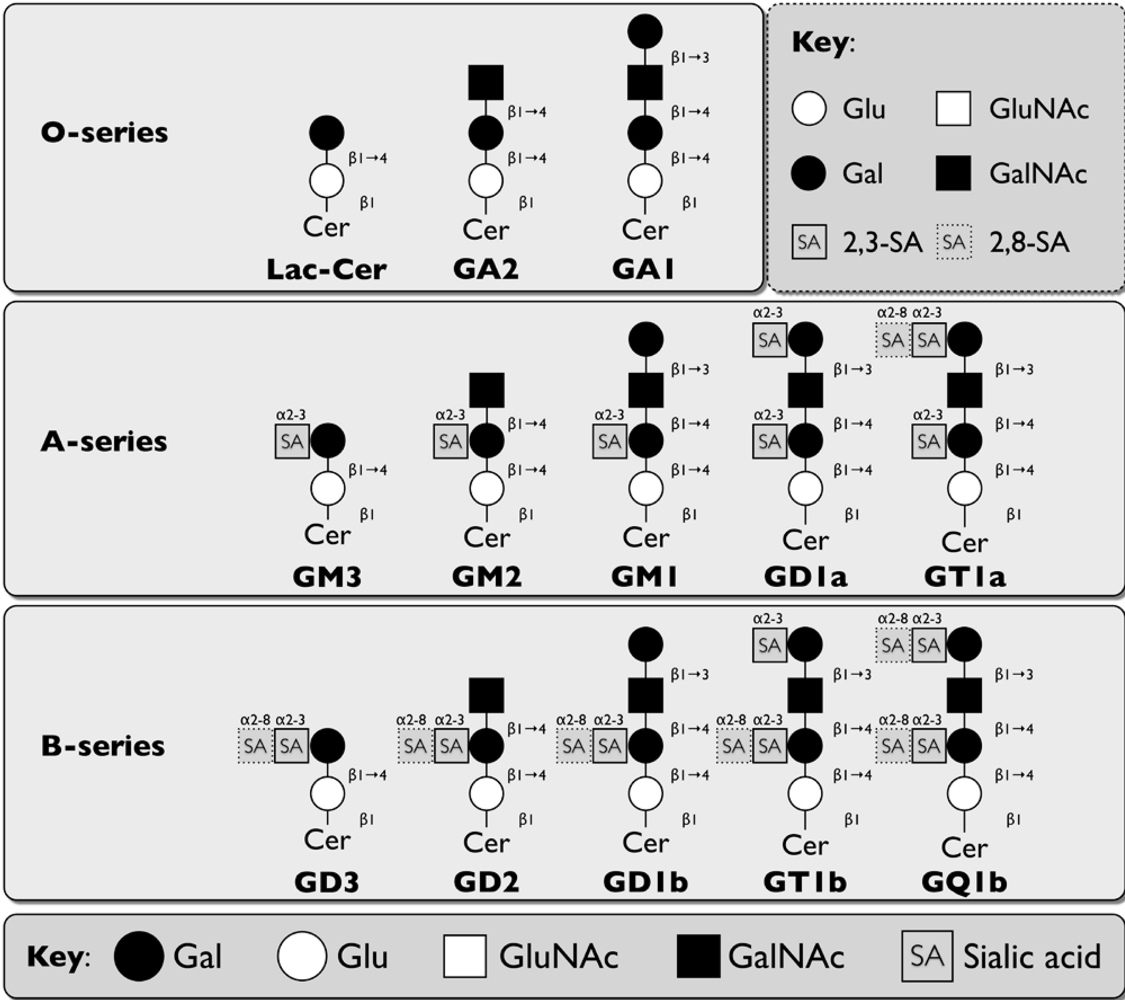
2.3.1.2. Simian rotavirus (SA11 strain)
2.3.1.3. Bovine rotavirus (NCVD, UK strain)
2.3.1.4. Rhesus rotavirus (RRV strain)
2.3.2. Neuraminidase-insensitive human rotaviruses (KUN, MO, DS-1, Wa strains)
2.4. Polyomaviridae
2.4.1. Murine Polyomavirus (MPyV)
2.4.2. Simian Virus 40 (SV40)
2.4.3. BK virus (BKV) and JC virus (JCV)
2.4.4. Merkel Cell Polyomavirus (MCPyV)
2.5. Parvoviridae
2.5.1. Erythroviruses
2.5.1.1. Human parvovirus B19

2.5.1.2. Simian parvovirus (SPV)
2.5.2. Dependoviruses
3. Conclusions
Acknowledgments
References
- Helenius, A. Virus entry and uncoating. In Fields Virology, 5th; Knipe, D.M., Howley, P.M., Eds.; Wolters Kluwer: Philadelphia, 2007. [Google Scholar]
- Olofsson, S.; Bergstrom, T. Glycoconjugate glycans as viral receptors. Ann. Med. 2005, 37, 154–172. [Google Scholar] [CrossRef] [PubMed]
- Iwamori, M. A new turning point in glycosphingolipid research. Hum. Cell 2005, 18, 117–133. [Google Scholar]
- Prinetti, A.; Loberto, N.; Chigorno, V.; Sonnino, S. Glycosphingolipid behaviour in complex membranes. Biochim. Biophys. Acta 2009, 1788, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Hakomori, S. Structure, organization, and function of glycosphingolipids in membrane. Curr. Opin. Hematol. 2003, 10, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Chester, M.A. IUPAC-IUB Joint Commission on Biochemical Nomenclature (JCBN). Nomenclature of glycolipids--recommendations 1997. Eur. J. Biochem. 1998, 257, 293–298. [Google Scholar] [PubMed]
- Spitalnik, P.F.; Spitalnik, S.L. The P blood group system: biochemical, serological, and clinical aspects. Transfus. Med. Rev. 1995, 9, 110–122. [Google Scholar] [CrossRef] [PubMed]
- Hakomori, S. Traveling for the glycosphingolipid path. Glycoconj. J. 2000, 17, 627–647. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, K.A. Animal glycosphingolipids as membrane attachment sites for bacteria. Annu. Rev. Biochem. 1989, 58, 309–350. [Google Scholar] [CrossRef] [PubMed]
- Green, K.Y. Caliciviridae. In Fields Virology, 5th; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, 2007. [Google Scholar]
- Farkas, T.; Sestak, K.; Wei, C.; Jiang, X. Characterization of a rhesus monkey calicivirus representing a new genus of Caliciviridae. J. Virol. 2008, 82, 5408–5416. [Google Scholar] [CrossRef] [PubMed]
- Oliver, S.L.; Asobayire, E.; Dastjerdi, A.M.; Bridger, J.C. Genomic characterization of the unclassified bovine enteric virus Newbury agent-1 (Newbury1) endorses a new genus in the family Caliciviridae. Virology 2006, 350, 240–250. [Google Scholar] [CrossRef] [PubMed]
- Sosnovtsev, S.V.; Green, K.Y. Identification and genomic mapping of the ORF3 and VPg proteins in feline calicivirus virions. Virology 2000, 277, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Thackray, L.B.; Wobus, C.E.; Chachu, K.A.; Liu, B.; Alegre, E.R.; Henderson, K.S.; Kelley, S.T.; Virgin IV, H.W. Murine noroviruses comprising a single genogroup exhibit biological diversity despite limited sequence divergence. J. Virol. 2007, 81, 10460–10473. [Google Scholar] [CrossRef] [PubMed]
- Prasad, B.V.; Hardy, M.E.; Dokland, T.; Bella, J.; Rossmann, M.G.; Estes, M.K. X-ray crystallographic structure of the Norwalk virus capsid. Science 1999, 286, 287–290. [Google Scholar] [CrossRef] [PubMed]
- Prasad, B.V.; Hardy, M.E.; Jiang, X.; Estes, M.K. Structure of Norwalk virus. Arch. Virol. Suppl. 1996, 12, 237–242. [Google Scholar] [PubMed]
- Prasad, B.V.; Rothnagel, R.; Jiang, X.; Estes, M.K. Three-dimensional structure of baculovirus-expressed Norwalk virus capsids. J. Virol. 1994, 68, 5117–5125. [Google Scholar] [PubMed]
- Katpally, U.; Wobus, C.E.; Dryden, K.; Virgin IV, H.W.; Smith, T.J. Structure of antibody-neutralized murine norovirus and unexpected differences from viruslike particles. J. Virol. 2008, 82, 2079–2088. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Neill, J.D.; Noel, J.S.; Hutson, A.M.; Glass, R.I.; Estes, M.K.; Prasad, B.V. Inter- and intragenus structural variations in caliciviruses and their functional implications. J. Virol. 2004, 78, 6469–6479. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Neill, J.D.; Estes, M.K.; Prasad, B.V. X-ray structure of a native calicivirus: structural insights into antigenic diversity and host specificity. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 8048–8053. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Hegde, R.S.; Jiang, X. The P domain of norovirus capsid protein forms dimer and binds to histo-blood group antigen receptors. J. Virol. 2004, 78, 6233–6242. [Google Scholar] [CrossRef] [PubMed]
- Bertolotti-Ciarlet, A.; White, L.J.; Chen, R.; Prasad, B.V.; Estes, M.K. Structural requirements for the assembly of Norwalk virus-like particles. J. Virol. 2002, 76, 4044–4055. [Google Scholar] [CrossRef] [PubMed]
- Katpally, U.; Voss, N.R.; Cavazza, T.; Taube, S.; Rubin, J.R.; Young, V.L.; Stuckey, J.; Ward, V.K.; Virgin, H.W.t.; Wobus, C.E.; Smith, T.J. High-resolution cryo-electron microscopy structures of MNV-1 and RHDV reveals marked flexibility in the receptor binding domains. J. Virol. 2010. [Google Scholar]
- Taube, S.; Rubin, J.R.; Katpally, U.; Smith, T.J.; Kendall, A.; Stuckey, J.A.; Wobus, C.E. High Resolution X-Ray Structure and Functional Analysis of the Murine Norovirus (MNV)-1 Capsid Protein Protruding (P) Domain. J. Virol. 2010. [Google Scholar]
- Bu, W.; Mamedova, A.; Tan, M.; Xia, M.; Jiang, X.; Hegde, R.S. Structural basis for the receptor binding specificity of Norwalk virus. J. Virol. 2008, 82, 5340–5347. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.M.; Hutson, A.M.; Estes, M.K.; Prasad, B.V. Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 9175–9180. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Lou, Z.; Tan, M.; Chen, Y.; Liu, Y.; Zhang, Z.; Zhang, X.C.; Jiang, X.; Li, X.; Rao, Z. Structural basis for the recognition of blood group trisaccharides by norovirus. J. Virol. 2007, 81, 5949–5957. [Google Scholar] [CrossRef] [PubMed]
- Santi, L.; Batchelor, L.; Huang, Z.; Hjelm, B.; Kilbourne, J.; Arntzen, C.J.; Chen, Q.; Mason, H.S. An efficient plant viral expression system generating orally immunogenic Norwalk virus-like particles. Vaccine 2008, 26, 1846–1854. [Google Scholar] [CrossRef] [PubMed]
- Green, K.Y.; Lew, J.F.; Jiang, X.; Kapikian, A.Z.; Estes, M.K. Comparison of the reactivities of baculovirus-expressed recombinant Norwalk virus capsid antigen with those of the native Norwalk virus antigen in serologic assays and some epidemiologic observations. J. Clin. Microbiol. 1993, 31, 2185–2191. [Google Scholar] [PubMed]
- Taube, S.; Kurth, A.; Schreier, E. Generation of recombinant norovirus-like particles (VLP) in the human endothelial kidney cell line 293T. Arch. Virol. 2005, 150, 1425–1431. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Wang, M.; Graham, D.Y.; Estes, M.K. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J. Virol. 1992, 66, 6527–6532. [Google Scholar] [PubMed]
- Harrington, P.R.; Yount, B.; Johnston, R.E.; Davis, N.; Moe, C.; Baric, R.S. Systemic, mucosal, and heterotypic immune induction in mice inoculated with Venezuelan equine encephalitis replicons expressing Norwalk virus-like particles. J. Virol. 2002, 76, 730–742. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Buehner, N.A.; Hutson, A.M.; Estes, M.K.; Mason, H.S. Tomato is a highly effective vehicle for expression and oral immunization with Norwalk virus capsid protein. Plant Biotechnol. J. 2006, 4, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Wobus, C.E.; Karst, S.M.; Thackray, L.B.; Chang, K.O.; Sosnovtsev, S.V.; Belliot, G.; Krug, A.; Mackenzie, J.M.; Green, K.Y.; Virgin, H.W. Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2004, 2, e432. [Google Scholar] [CrossRef] [PubMed]
- Parwani, A.V.; Flynn, W.T.; Gadfield, K.L.; Saif, L.J. Serial propagation of porcine enteric calicivirus in a continuous cell line. Effect of medium supplementation with intestinal contents or enzymes. Arch. Virol. 1991, 120, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Bidawid, S.; Malik, N.; Adegbunrin, O.; Sattar, S.A.; Farber, J.M. A feline kidney cell line-based plaque assay for feline calicivirus, a surrogate for Norwalk virus. J. Virol. Methods 2003, 107, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Stuart, A.D.; Brown, T.D. Alpha2,6-linked sialic acid acts as a receptor for Feline calicivirus. J. Gen. Virol. 2007, 88, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Ossiboff, R.J.; Parker, J.S. Identification of regions and residues in feline junctional adhesion molecule required for feline calicivirus binding and infection. J. Virol. 2007, 81, 13608–13621. [Google Scholar] [CrossRef] [PubMed]
- Makino, A.; Shimojima, M.; Miyazawa, T.; Kato, K.; Tohya, Y.; Akashi, H. Junctional adhesion molecule 1 is a functional receptor for feline calicivirus. J. Virol. 2006, 80, 4482–4490. [Google Scholar] [CrossRef] [PubMed]
- Stuart, A.D.; Brown, T.D. Entry of feline calicivirus is dependent on clathrin-mediated endocytosis and acidification in endosomes. J. Virol. 2006, 80, 7500–7509. [Google Scholar] [CrossRef] [PubMed]
- Gerondopoulos, A.; Jackson, T.; Monaghan, P.; Doyle, N.; Roberts, L.O. Murine norovirus-1 cell entry is mediated through a non-clathrin, non-caveolae, dynamin and cholesterol dependent pathway. J. Gen. Virol. 2010. [Google Scholar]
- Perry, J.W.; Taube, S.; Wobus, C.E. Murine norovirus-1 entry into permissive macrophages and dendritic cells is pH-independent. Virus Res. 2009, 143, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Perry, J.; Wobus, C.E. Endocytosis of Murine Norovirus 1 into Murine Macrophages is dependent on dynamin II and cholesterol. J. Virol. 2010, 84. [Google Scholar]
- Estes, M.K.; Prasad, B.V.; Atmar, R.L. Noroviruses everywhere: has something changed? Curr. Opin. Infect. Dis. 2006, 19, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Widdowson, M.A.; Monroe, S.S.; Glass, R.I. Are noroviruses emerging? Emerg. Infect. Dis. 2005, 11, 735–737. [Google Scholar] [PubMed]
- Koopmans, M.; Duizer, E. Foodborne viruses: an emerging problem. Int. J. Food Microbiol. 2004, 90, 23–41. [Google Scholar] [CrossRef] [PubMed]
- Skovgaard, N. New trends in emerging pathogens. Int. J. Food Microbiol. 2007, 120, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Karst, S.M.; Wobus, C.E.; Lay, M.; Davidson, J.; Virgin IV, H.W. STAT1-dependent innate immunity to a Norwalk-like virus. Science 2003, 299, 1575–1578. [Google Scholar] [CrossRef] [PubMed]
- Martella, V.; Lorusso, E.; Decaro, N.; Elia, G.; Radogna, A.; D'Abramo, M.; Desario, C.; Cavalli, A.; Corrente, M.; Camero, M.; Germinario, C.A.; Banyai, K.; Di Martino, B.; Marsilio, F.; Carmichael, L.E.; Buonavoglia, C. Detection and molecular characterization of a canine norovirus. Emerg. Infect. Dis. 2008, 14, 1306–1308. [Google Scholar] [CrossRef] [PubMed]
- van Der Poel, W.H.; Vinje, J.; van Der Heide, R.; Herrera, M.I.; Vivo, A.; Koopmans, M.P. Norwalk-like calicivirus genes in farm animals. Emerg. Infect. Dis. 2000, 6, 36–41. [Google Scholar] [PubMed]
- Wise, A.G.; Monroe, S.S.; Hanson, L.E.; Grooms, D.L.; Sockett, D.; Maes, R.K. Molecular characterization of noroviruses detected in diarrheic stools of Michigan and Wisconsin dairy calves: circulation of two distinct subgroups. Virus Res. 2004, 100, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; Williamson, W.; Hewitt, J.; Lin, S.; Rivera-Aban, M.; Ball, A.; Scholes, P.; Savill, M.; Greening, G.E. Molecular detection of norovirus in sheep and pigs in New Zealand farms. Vet. Microbiol. 2008. [Google Scholar]
- Zheng, D.P.; Ando, T.; Fankhauser, R.L.; Beard, R.S.; Glass, R.I.; Monroe, S.S. Norovirus classification and proposed strain nomenclature. Virology 2006, 346, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, E.F.; Lindesmith, L.C.; Lobue, A.D.; Baric, R.S. Viral shape-shifting: norovirus evasion of the human immune system. Nat. Rev. Microbiol. 2010, 8, 231–241. [Google Scholar] [CrossRef]
- Lopman, B.; Vennema, H.; Kohli, E.; Pothier, P.; Sanchez, A.; Negredo, A.; Buesa, J.; Schreier, E.; Reacher, M.; Brown, D.; Gray, J.; Iturriza, M.; Gallimore, C.; Bottiger, B.; Hedlund, K.O.; Torven, M.; von Bonsdorff, C.H.; Maunula, L.; Poljsak-Prijatelj, M.; Zimsek, J.; Reuter, G.; Szucs, G.; Melegh, B.; Svennson, L.; van Duijnhoven, Y.; Koopmans, M. Increase in viral gastroenteritis outbreaks in Europe and epidemic spread of new norovirus variant. Lancet 2004, 363, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Widdowson, M.A.; Cramer, E.H.; Hadley, L.; Bresee, J.S.; Beard, R.S.; Bulens, S.N.; Charles, M.; Chege, W.; Isakbaeva, E.; Wright, J.G.; Mintz, E.; Forney, D.; Massey, J.; Glass, R.I.; Monroe, S.S. Outbreaks of acute gastroenteritis on cruise ships and on land: identification of a predominant circulating strain of norovirus--United States, 2002. J. Infect. Dis. 2004, 190, 27–36. [Google Scholar] [CrossRef] [PubMed]
- White, P.A.; Hansman, G.S.; Li, A.; Dable, J.; Isaacs, M.; Ferson, M.; McIver, C.J.; Rawlinson, W.D. Norwalk-like virus 95/96-US strain is a major cause of gastroenteritis outbreaks in Australia. J. Med. Virol. 2002, 68, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Guix, S.; Asanaka, M.; Katayama, K.; Crawford, S.E.; Neill, F.H.; Atmar, R.L.; Estes, M.K. Norwalk virus RNA is infectious in mammalian cells. J. Virol. 2007, 81, 12238–12248. [Google Scholar] [CrossRef] [PubMed]
- Mead, P.S.; Slutsker, L.; Dietz, V.; McCaig, L.F.; Bresee, J.S.; Shapiro, C.; Griffin, P.M.; Tauxe, R.V. Food-related illness and death in the United States. Emerg. Infect. Dis. 1999, 5, 607–625. [Google Scholar] [CrossRef] [PubMed]
- Duizer, E.; Schwab, K.J.; Neill, F.H.; Atmar, R.L.; Koopmans, M.P.; Estes, M.K. Laboratory efforts to cultivate noroviruses. J. Gen. Virol. 2004, 85, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Le Pendu, J.; Ruvoen-Clouet, N.; Kindberg, E.; Svensson, L. Mendelian resistance to human norovirus infections. Semin. Immunol. 2006, 18, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Jiang, X. Norovirus-host interaction: implications for disease control and prevention. Exp. Rev. Mol. Med. 2007, 9, 1–22. [Google Scholar] [CrossRef]
- Tamura, M.; Natori, K.; Kobayashi, M.; Miyamura, T.; Takeda, N. Genogroup II noroviruses efficiently bind to heparan sulfate proteoglycan associated with the cellular membrane. J. Virol. 2004, 78, 3817–3826. [Google Scholar] [CrossRef] [PubMed]
- Rydell, G.E.; Nilsson, J.; Rodriguez-Diaz, J.; Ruvoen-Clouet, N.; Svensson, L.; Le Pendu, J.; Larson, G. Human noroviruses recognize sialyl Lewis x neoglycoprotein. Glycobiology 2009, 19, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Clausen, H.; Hakomori, S. ABH and related histo-blood group antigens; immunochemical differences in carrier isotypes and their distribution. Vox Sang. 1989, 56, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Henry, S.M.; Samuelsson, B.E.; Oriol, R. Immunochemical and immunohistological expression of Lewis histo-blood group antigens in small intestine including individuals of the Le(a+b+) and Le(a-b-) nonsecretor phenotypes. Glycoconj. J. 1994, 11, 600–607. [Google Scholar] [CrossRef] [PubMed]
- Marionneau, S.; Cailleau-Thomas, A.; Rocher, J.; Le Moullac-Vaidye, B.; Ruvoen, N.; Clement, M.; Le Pendu, J. ABH and Lewis histo-blood group antigens, a model for the meaning of oligosaccharide diversity in the face of a changing world. Biochimie 2001, 83, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Le Pendu, J. Histo-blood group antigen and human milk oligosaccharides: genetic polymorphism and risk of infectious diseases. Adv. Exp. Med. Biol. 2004, 554, 135–143. [Google Scholar] [PubMed]
- Hakomori, S. Antigen structure and genetic basis of histo-blood groups A, B and O: their changes associated with human cancer. Biochim. Biophys. Acta 1999, 1473, 247–266. [Google Scholar] [PubMed]
- Taube, S.; Perry, J.W.; Yetming, K.; Patel, S.P.; Auble, H.; Shu, L.; Nawar, H.F.; Lee, C.H.; Connell, T.D.; Shayman, J.A.; Wobus, C.E. Ganglioside-linked terminal sialic acid moieties on murine macrophages function as attachment receptors for murine noroviruses. J. Virol. 2009, 83, 4092–4101. [Google Scholar] [CrossRef] [PubMed]
- Zakhour, M.; Ruvoen-Clouet, N.; Charpilienne, A.; Langpap, B.; Poncet, D.; Peters, T.; Bovin, N.; Le Pendu, J. The alphaGal epitope of the histo-blood group antigen family is a ligand for bovine norovirus Newbury2 expected to prevent cross-species transmission . PLoS Path. 2009, 5, e1000504. [Google Scholar] [CrossRef]
- Ruvoen-Clouet, N.; Ganiere, J.P.; Andre-Fontaine, G.; Blanchard, D.; Le Pendu, J. Binding of rabbit hemorrhagic disease virus to antigens of the ABH histo-blood group family. J. Virol. 2000, 74, 11950–11954. [Google Scholar] [CrossRef] [PubMed]
- Isa, P.; Gutierrez, M.; Arias, C.F.; Lopez, S. Rotavirus cell entry. Future Medicine 2008, 3, 135–146. [Google Scholar]
- Rolsma, M.D.; Gelberg, H.B.; Kuhlenschmidt, M.S. Assay for evaluation of rotavirus-cell interactions: identification of an enterocyte ganglioside fraction that mediates group A porcine rotavirus recognition. J. Virol. 1994, 68, 258–268. [Google Scholar] [PubMed]
- Rolsma, M.D.; Kuhlenschmidt, T.B.; Gelberg, H.B.; Kuhlenschmidt, M.S. Structure and function of a ganglioside receptor for porcine rotavirus. J. Virol. 1998, 72, 9079–9091. [Google Scholar] [PubMed]
- Haselhorst, T.; Fleming, F.E.; Dyason, J.C.; Hartnell, R.D.; Yu, X.; Holloway, G.; Santegoets, K.; Kiefel, M.J.; Blanchard, H.; Coulson, B.S.; von Itzstein, M. Sialic acid dependence in rotavirus host cell invasion. Nat. Chem. Biol. 2009, 5, 91–93. [Google Scholar] [CrossRef] [PubMed]
- Delorme, C.; Brussow, H.; Sidoti, J.; Roche, N.; Karlsson, K.A.; Neeser, J.R.; Teneberg, S. Glycosphingolipid binding specificities of rotavirus: identification of a sialic acid-binding epitope. J. Virol. 2001, 75, 2276–2287. [Google Scholar] [CrossRef] [PubMed]
- Dormitzer, P.R.; Sun, Z.Y.; Blixt, O.; Paulson, J.C.; Wagner, G.; Harrison, S.C. Specificity and affinity of sialic acid binding by the rhesus rotavirus VP8* core. J. Virol. 2002, 76, 10512–10517. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.T.; Nakagomi, O.; Mochizuki, M.; Ishida, H.; Kiso, M.; Ohta, Y.; Suzuki, T.; Miyamoto, D.; Hidari, K.I.; Suzuki, Y. Ganglioside GM(1a) on the cell surface is involved in the infection by human rotavirus KUN and MO strains. J. Biochem. (Tokyo). 1999, 126, 683–688. [Google Scholar] [CrossRef]
- Tsai, B.; Gilbert, J.M.; Stehle, T.; Lencer, W.; Benjamin, T.L.; Rapoport, T.A. Gangliosides are receptors for murine polyoma virus and SV40. EMBO J. 2003, 22, 4346–4355. [Google Scholar] [CrossRef] [PubMed]
- Stehle, T.; Harrison, S.C. Crystal structures of murine polyomavirus in complex with straight-chain and branched-chain sialyloligosaccharide receptor fragments. Structure 1996, 4, 183–194. [Google Scholar] [CrossRef]
- Caruso, M.; Belloni, L.; Sthandier, O.; Amati, P.; Garcia, M.I. Alpha4beta1 integrin acts as a cell receptor for murine polyomavirus at the postattachment level. J. Virol. 2003, 77, 3913–3921. [Google Scholar] [CrossRef] [PubMed]
- Caruso, M.; Busanello, A.; Sthandier, O.; Cavaldesi, M.; Gentile, M.; Garcia, M.I.; Amati, P. Mutation in the VP1-LDV motif of the murine polyomavirus affects viral infectivity and conditions virus tissue tropism in vivo. J. Mol. Biol. 2007, 367, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Breau, W.C.; Atwood, W.J.; Norkin, L.C. Class I major histocompatibility proteins are an essential component of the simian virus 40 receptor. J. Virol. 1992, 66, 2037–2045. [Google Scholar] [PubMed]
- Atwood, W.J.; Norkin, L.C. Class I major histocompatibility proteins as cell surface receptors for simian virus 40. J. Virol. 1989, 63, 4474–4477. [Google Scholar] [PubMed]
- Low, J.A.; Magnuson, B.; Tsai, B.; Imperiale, M.J. Identification of gangliosides GD1b and GT1b as receptors for BK virus. J. Virol. 2006, 80, 1361–1366. [Google Scholar] [CrossRef] [PubMed]
- Dugan, A.S.; Eash, S.; Atwood, W.J. An N-linked glycoprotein with alpha(2,3)-linked sialic acid is a receptor for BK virus. J. Virol. 2005, 79, 14442–14445. [Google Scholar] [CrossRef] [PubMed]
- Komagome, R.; Sawa, H.; Suzuki, T.; Suzuki, Y.; Tanaka, S.; Atwood, W.J.; Nagashima, K. Oligosaccharides as receptors for JC virus. J. Virol. 2002, 76, 12992–13000. [Google Scholar] [CrossRef] [PubMed]
- Elphick, G.F.; Querbes, W.; Jordan, J.A.; Gee, G.V.; Eash, S.; Manley, K.; Dugan, A.; Stanifer, M.; Bhatnagar, A.; Kroeze, W.K.; Roth, B.L.; Atwood, W.J. The human polyomavirus, JCV, uses serotonin receptors to infect cells. Science 2004, 306, 1380–1383. [Google Scholar] [CrossRef] [PubMed]
- Dugan, A.S.; Gasparovic, M.L.; Atwood, W.J. Direct correlation between sialic acid binding and infection of cells by two human polyomaviruses (JC virus and BK virus). J. Virol. 2008, 82, 2560–2564. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.K.; Wei, G.; Atwood, W.J. Infection of glial cells by the human polyomavirus JC is mediated by an N-linked glycoprotein containing terminal alpha(2-6)-linked sialic acids. J. Virol. 1998, 72, 4643–4649. [Google Scholar] [PubMed]
- Erickson, K.D.; Garcea, R.L.; Tsai, B. Ganglioside GT1b is a putative host cell receptor for the Merkel cell polyomavirus. J. Virol. 2009, 83, 10275–10279. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.E.; Anderson, S.M.; Young, N.S. Erythrocyte P antigen: cellular receptor for B19 parvovirus. Science 1993, 262, 114–117. [Google Scholar] [PubMed]
- Cooling, L.L.; Koerner, T.A.; Naides, S.J. Multiple glycosphingolipids determine the tissue tropism of parvovirus B19. J. Infect. Dis. 1995, 172, 1198–1205. [Google Scholar] [PubMed]
- Weigel-Kelley, K.A.; Yoder, M.C.; Srivastava, A. Alpha5beta1 integrin as a cellular coreceptor for human parvovirus B19: requirement of functional activation of beta1 integrin for viral entry. Blood 2003, 102, 3927–3933. [Google Scholar] [CrossRef] [PubMed]
- Munakata, Y.; Saito-Ito, T.; Kumura-Ishii, K.; Huang, J.; Kodera, T.; Ishii, T.; Hirabayashi, Y.; Koyanagi, Y.; Sasaki, T. Ku80 autoantigen as a cellular coreceptor for human parvovirus B19 infection. Blood 2005, 106, 3449–3456. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.E.; Liu, Z.; Gallinella, G.; Wong, S.; Mills, I.P.; O'Sullivan, M.G. Simian parvovirus infection: a potential zoonosis. J. Infect. Dis. 2004, 190, 1900–1907. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.; Chiorini, J.A. Gangliosides are essential for bovine adeno-associated virus entry. J. Virol. 2006, 80, 5516–5522. [Google Scholar] [CrossRef] [PubMed]
- Hakomori, S.I. Structure and function of glycosphingolipids and sphingolipids: recollections and future trends. Biochim. Biophys. Acta 2008, 1780, 325–346. [Google Scholar] [PubMed]
- Ravn, V.; Dabelsteen, E. Tissue distribution of histo-blood group antigens. APMIS 2000, 108, 1–28. [Google Scholar] [PubMed]
- Kelly, R.J.; Ernst, L.K.; Larsen, R.D.; Bryant, J.G.; Robinson, J.S.; Lowe, J.B. Molecular basis for H blood group deficiency in Bombay (Oh) and para-Bombay individuals. Proc. Natl. Acad. Sci. U. S. A. 1994, 91, 5843–5847. [Google Scholar] [CrossRef] [PubMed]
- Harrington, P.R.; Lindesmith, L.; Yount, B.; Moe, C.L.; Baric, R.S. Binding of Norwalk virus-like particles to ABH histo-blood group antigens is blocked by antisera from infected human volunteers or experimentally vaccinated mice. J. Virol. 2002, 76, 12335–12343. [Google Scholar] [CrossRef] [PubMed]
- Harrington, P.R.; Vinje, J.; Moe, C.L.; Baric, R.S. Norovirus capture with histo-blood group antigens reveals novel virus-ligand interactions. J. Virol. 2004, 78, 3035–3045. [Google Scholar] [CrossRef] [PubMed]
- Hutson, A.M.; Atmar, R.L.; Marcus, D.M.; Estes, M.K. Norwalk virus-like particle hemagglutination by binding to h histo-blood group antigens. J. Virol. 2003, 77, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Lindesmith, L.; Moe, C.; Marionneau, S.; Ruvoen, N.; Jiang, X.; Lindblad, L.; Stewart, P.; LePendu, J.; Baric, R. Human susceptibility and resistance to Norwalk virus infection. Nat. Med. 2003, 9, 548–553. [Google Scholar] [CrossRef] [PubMed]
- Marionneau, S.; Ruvoen, N.; Le Moullac-Vaidye, B.; Clement, M.; Cailleau-Thomas, A.; Ruiz-Palacois, G.; Huang, P.; Jiang, X.; Le Pendu, J. Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology 2002, 122, 1967–1977. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Farkas, T.; Marionneau, S.; Zhong, W.; Ruvoen-Clouet, N.; Morrow, A.L.; Altaye, M.; Pickering, L.K.; Newburg, D.S.; LePendu, J.; Jiang, X. Noroviruses bind to human ABO, Lewis, and secretor histo-blood group antigens: identification of 4 distinct strain-specific patterns. J. Infect. Dis. 2003, 188, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Hutson, A.M.; Atmar, R.L.; Graham, D.Y.; Estes, M.K. Norwalk virus infection and disease is associated with ABO histo-blood group type. J. Infect. Dis. 2002, 185, 1335–1337. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, J.; Rydell, G.E.; Le Pendu, J.; Larson, G. Norwalk virus-like particles bind specifically to A, H and difucosylated Lewis but not to B histo-blood group active glycosphingolipids. Glycoconj. J. 2009. [Google Scholar]
- Rydell, G.E.; Dahlin, A.B.; Hook, F.; Larson, G. QCM-D studies of human norovirus VLPs binding to glycosphingolipids in supported lipid bilayers reveal strain-specific characteristics. Glycobiology 2009, 19, 1176–1184. [Google Scholar] [CrossRef] [PubMed]
- Shirato, H.; Ogawa, S.; Ito, H.; Sato, T.; Kameyama, A.; Narimatsu, H.; Xiaofan, Z.; Miyamura, T.; Wakita, T.; Ishii, K.; Takeda, N. Noroviruses distinguish between type 1 and type 2 histo-blood group antigens for binding. J. Virol. 2008. [Google Scholar]
- Rockx, B.H.; Vennema, H.; Hoebe, C.J.; Duizer, E.; Koopmans, M.P. Association of histo-blood group antigens and susceptibility to norovirus infections. J. Infect. Dis. 2005, 191, 749–754. [Google Scholar] [CrossRef] [PubMed]
- Hennessy, E.P.; Green, A.D.; Connor, M.P.; Darby, R.; MacDonald, P. Norwalk virus infection and disease is associated with ABO histo-blood group type. J. Infect. Dis. 2003, 188, 176–177. [Google Scholar] [CrossRef] [PubMed]
- Marionneau, S.; Airaud, F.; Bovin, N.V.; Le Pendu, J.; Ruvoen-Clouet, N. Influence of the combined ABO, FUT2, and FUT3 polymorphism on susceptibility to Norwalk virus attachment. J. Infect. Dis. 2005, 192, 1071–1077. [Google Scholar] [CrossRef] [PubMed]
- Larsson, M.M.; Rydell, G.E.; Grahn, A.; Rodriguez-Diaz, J.; Akerlind, B.; Hutson, A.M.; Estes, M.K.; Larson, G.; Svensson, L. Antibody prevalence and titer to norovirus (genogroup II) correlate with secretor (FUT2) but not with ABO phenotype or Lewis (FUT3) genotype. J. Infect. Dis. 2006, 194, 1422–1427. [Google Scholar] [CrossRef] [PubMed]
- Lindesmith, L.; Moe, C.; Lependu, J.; Frelinger, J.A.; Treanor, J.; Baric, R.S. Cellular and humoral immunity following Snow Mountain virus challenge. J. Virol. 2005, 79, 2900–2909. [Google Scholar] [CrossRef] [PubMed]
- Lindesmith, L.C.; Donaldson, E.F.; Lobue, A.D.; Cannon, J.L.; Zheng, D.P.; Vinje, J.; Baric, R.S. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 2008, 5, e31. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, E.F.; Lindesmith, L.C.; Lobue, A.D.; Baric, R.S. Norovirus pathogenesis: mechanisms of persistence and immune evasion in human populations. Immunol. Rev. 2008, 225, 190–211. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.C.; Wobus, C.E.; Steffen, E.K.; Riley, L.K.; Livingston, R.S. Development of a microsphere-based serologic multiplexed fluorescent immunoassay and a reverse transcriptase PCR assay to detect murine norovirus 1 infection in mice. Clin. Diagn. Lab. Immunol. 2005, 12, 1145–1151. [Google Scholar] [PubMed]
- Muller, B.; Klemm, U.; Mas Marques, A.; Schreier, E. Genetic diversity and recombination of murine noroviruses in immunocompromised mice. Arch. Virol. 2007, 152, 1709–1719. [Google Scholar] [CrossRef] [PubMed]
- Henderson, K.S. Murine norovirus, a recently discovered and highly prevalent viral agent of mice. Lab Anim (NY) 2008, 37, 314–320. [Google Scholar] [CrossRef] [PubMed]
- Mahler, M.; Kohl, W. A serological survey to evaluate contemporary prevalence of viral agents and Mycoplasma pulmonis in laboratory mice and rats in western Europe. Lab Anim (NY) 2009, 38, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Kitajima, M.; Oka, T.; Tohya, Y.; Katayama, H.; Takeda, N.; Katayama, K. Development of a broadly reactive nested reverse transcription-PCR assay to detect murine noroviruses, and investigation of the prevalence of murine noroviruses in laboratory mice in Japan. Microbiol. Immunol. 2009, 53, 531–534. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Lee, H.; Chang, K.O.; Ko, G. Molecular characterization of murine norovirus isolates from South Korea. Virus Res. 2010, 147, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Wobus, C.E.; Thackray, L.B.; Virgin IV, H.W. Murine norovirus: a model system to study norovirus biology and pathogenesis. J. Virol. 2006, 80, 5104–5112. [Google Scholar] [CrossRef] [PubMed]
- Woode, G.N.; Bridger, J.C. Isolation of small viruses resembling astroviruses and caliciviruses from acute enteritis of calves. J. Med. Microbiol. 1978, 11, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Guillon, P.; Ruvoen-Clouet, N.; Le Moullac-Vaidye, B.; Marchandeau, S.; Le Pendu, J. Association between expression of the H histo-blood group antigen, alpha1,2fucosyltransferases polymorphism of wild rabbits, and sensitivity to rabbit hemorrhagic disease virus. Glycobiology 2009, 19, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Guittre, C.; Ruvoen-Clouet, N.; Barraud, L.; Cherel, Y.; Baginski, I.; Prave, M.; Ganiere, J.P.; Trepo, C.; Cova, L. Early stages of rabbit haemorrhagic disease virus infection monitored by polymerase chain reaction. Zentralbl. Veterinarmed. [B]. 1996, 43, 109–118. [Google Scholar] [PubMed]
- Park, J.H.; Ochiai, K.; Itakura, C. Detection of rabbit haemorrhagic disease virus particles in the rabbit liver tissues. J. Comp. Pathol. 1992, 107, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Laurent, S.; Vautherot, J.F.; Madelaine, M.F.; Le Gall, G.; Rasschaert, D. Recombinant rabbit hemorrhagic disease virus capsid protein expressed in baculovirus self-assembles into viruslike particles and induces protection. J. Virol. 1994, 68, 6794–6798. [Google Scholar] [PubMed]
- Rademacher, C.; Krishna, N.R.; Palcic, M.; Parra, F.; Peters, T. NMR experiments reveal the molecular basis of receptor recognition by a calicivirus. J. Am. Chem. Soc. 2008, 130, 3669–3675. [Google Scholar] [CrossRef] [PubMed]
- Morisse, J.P.; Le Gall, G.; Boilletot, E. Hepatitis of viral origin in Leporidae: introduction and aetiological hypotheses. Rev. Sci. Tech. 1991, 10, 269–310. [Google Scholar] [PubMed]
- Rodak, L.; Smid, B.; Valicek, L. Application of control measures against viral haemorrhagic disease of rabbits in the Czech and Slovak Federal Republic. Rev. Sci. Tech. 1991, 10, 513–524. [Google Scholar] [PubMed]
- Lopez, S.; Arias, C.F. Early steps in rotavirus cell entry. Curr. Top. Microbiol. Immunol. 2006, 309, 39–66. [Google Scholar] [PubMed]
- Lorrot, M.; Vasseur, M. How do the rotavirus NSP4 and bacterial enterotoxins lead differently to diarrhea? Virol. J. 2007, 4, 31. [Google Scholar] [CrossRef]
- Isa, P.; Arias, C.F.; Lopez, S. Role of sialic acids in rotavirus infection. Glycoconj. J. 2006, 23, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Ciarlet, M.; Crawford, S.E.; Cheng, E.; Blutt, S.E.; Rice, D.A.; Bergelson, J.M.; Estes, M.K. VLA-2 (alpha2beta1) integrin promotes rotavirus entry into cells but is not necessary for rotavirus attachment. J. Virol. 2002, 76, 1109–1123. [Google Scholar] [PubMed]
- Trask, S.D.; Kim, I.S.; Harrison, S.C.; Dormitzer, P.R. A rotavirus spike protein conformational intermediate binds lipid bilayers. J. Virol. 2010, 84, 1764–1770. [Google Scholar] [CrossRef] [PubMed]
- Dowling, W.; Denisova, E.; LaMonica, R.; Mackow, E.R. Selective membrane permeabilization by the rotavirus VP5* protein is abrogated by mutations in an internal hydrophobic domain. J. Virol. 2000, 74, 6368–6376. [Google Scholar] [CrossRef] [PubMed]
- Keljo, D.J.; Smith, A.K. Characterization of binding of simian rotavirus SA-11 to cultured epithelial cells. J. Pediatr. Gastroenterol. Nutr. 1988, 7, 249–256. [Google Scholar] [PubMed]
- Bastardo, J.W.; Holmes, I.H. Attachment of SA-11 rotavirus to erythrocyte receptors. Infect. Immun. 1980, 29, 1134–1140. [Google Scholar] [PubMed]
- Fauvel, M.; Spence, L.; Babiuk, L.A.; Petro, R.; Bloch, S. Hemagglutination and hemagglutination-inhibition studies with a strain of Nebraska calf diarrhea virus (bovine rotavirus). Intervirology 1978, 9, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Spence, L.; Fauvel, M.; Petro, R.; Babiuk, L.A. Comparison of rotavirus strains by hemagglutination inhibition. Can. J. Microbiol. 1978, 24, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Spence, L.; Fauvel, M.; Petro, R.; Bloch, S. Haemagglutinin from Rotavirus. Lancet 1976, 2, 1023. [Google Scholar] [CrossRef]
- Ciarlet, M.; Estes, M.K. Human and most animal rotavirus strains do not require the presence of sialic acid on the cell surface for efficient infectivity. J. Gen. Virol. 1999, 80, 943–948. [Google Scholar] [PubMed]
- Banda, K.; Kang, G.; Varki, A. 'Sialidase sensitivity' of rotaviruses revisited. Nat. Chem. Biol. 2009, 5, 71–72. [Google Scholar] [CrossRef] [PubMed]
- Realpe, M.; Espinosa, R.; Lopez, S.; Arias, C.F. Rotaviruses require basolateral molecules for efficient infection of polarized MDCKII cells. Virus Res. 2010, 147, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Blutt, S.E.; Conner, M.E. Rotavirus: to the gut and beyond! Curr. Opin. Gastro. 2007, 23, 39–43. [Google Scholar] [CrossRef]
- Blutt, S.E.; Matson, D.O.; Crawford, S.E.; Staat, M.A.; Azimi, P.; Bennett, B.L.; Piedra, P.A.; Conner, M.E. Rotavirus antigenemia in children is associated with viremia. PLoS Med. 2007, 4, e121. [Google Scholar] [CrossRef] [PubMed]
- Kuhlenschmidt, T.B.; Hanafin, W.P.; Gelberg, H.B.; Kuhlenschmidt, M.S. Sialic acid dependence and independence of group A rotaviruses. Adv. Exp. Med. Biol. 1999, 473, 309–317. [Google Scholar] [PubMed]
- Lopez, J.A.; Maldonado, A.J.; Gerder, M.; Abanero, J.; Murgich, J.; Pujol, F.H.; Liprandi, F.; Ludert, J.E. Characterization of neuraminidase-resistant mutants derived from rotavirus porcine strain OSU. J. Virol. 2005, 79, 10369–10375. [Google Scholar] [CrossRef] [PubMed]
- Svensson, L. Group C rotavirus requires sialic acid for erythrocyte and cell receptor binding. J. Virol. 1992, 66, 5582–5585. [Google Scholar] [PubMed]
- Kuhlenschmidt, M.S.; Rolsma, M.D.; Kuhlenschmidt, T.B.; Gelberg, H.B. Characterization of a porcine enterocyte receptor for group A rotavirus. Adv. Exp. Med. Biol. 1997, 412, 135–143. [Google Scholar] [PubMed]
- Willoughby, R.E.; Yolken, R.H.; Schnaar, R.L. Rotaviruses specifically bind to the neutral glycosphingolipid asialo-GM1. J. Virol. 1990, 64, 4830–4835. [Google Scholar] [PubMed]
- Superti, F.; Donelli, G. Gangliosides as binding sites in SA-11 rotavirus infection of LLC-MK2 cells. J. Gen. Virol. 1991, 72, 2467–2474. [Google Scholar] [CrossRef] [PubMed]
- Srnka, C.A.; Tiemeyer, M.; Gilbert, J.H.; Moreland, M.; Schweingruber, H.; de Lappe, B.W.; James, P.G.; Gant, T.; Willoughby, R.E.; Yolken, R.H.; et al Cell surface ligands for rotavirus: mouse intestinal glycolipids and synthetic carbohydrate analogs. Virology 1992, 190, 794–805. [Google Scholar] [CrossRef] [PubMed]
- Fukudome, K.; Yoshie, O.; Konno, T. Comparison of human, simian, and bovine rotaviruses for requirement of sialic acid in hemagglutination and cell adsorption. Virology 1989, 172, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Ludert, J.E.; Feng, N.; Yu, J.H.; Broome, R.L.; Hoshino, Y.; Greenberg, H.B. Genetic mapping indicates that VP4 is the rotavirus cell attachment protein in vitro and in vivo. J. Virol. 1996, 70, 487–493. [Google Scholar] [PubMed]
- Ciarlet, M.; Crawford, S.E.; Estes, M.K. Differential infection of polarized epithelial cell lines by sialic acid-dependent and sialic acid-independent rotavirus strains. J. Virol. 2001, 75, 11834–11850. [Google Scholar] [CrossRef] [PubMed]
- Kraschnefski, M.J.; Bugarcic, A.; Fleming, F.E.; Yu, X.; von Itzstein, M.; Coulson, B.S.; Blanchard, H. Effects on sialic acid recognition of amino acid mutations in the carbohydrate-binding cleft of the rotavirus spike protein. Glycobiology 2009, 19, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Dormitzer, P.R.; Sun, Z.Y.; Wagner, G.; Harrison, S.C. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J. 2002, 21, 885–897. [Google Scholar] [CrossRef] [PubMed]
- Monnier, N.; Higo-Moriguchi, K.; Sun, Z.Y.; Prasad, B.V.; Taniguchi, K.; Dormitzer, P.R. High-resolution molecular and antigen structure of the VP8* core of a sialic acid-independent human rotavirus strain. J. Virol. 2006, 80, 1513–1523. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, H.; Yu, X.; Coulson, B.S.; von Itzstein, M. Insight into host cell carbohydrate-recognition by human and porcine rotavirus from crystal structures of the virion spike associated carbohydrate-binding domain (VP8*). J. Mol. Biol. 2007, 367, 1215–1226. [Google Scholar] [CrossRef] [PubMed]
- Imperiale, M.J.; Major, E.O. Polyomaviruses. In Fields Virology, 5th; Knipe, D. M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, 2007. [Google Scholar]
- Liddington, R.C.; Yan, Y.; Moulai, J.; Sahli, R.; Benjamin, T.L.; Harrison, S.C. Structure of simian virus 40 at 3.8-A resolution. Nature 1991, 354, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Meneguzzi, G.; Pignatti, P.F.; Barbanti-Brodano, G.; Milanesi, G. Minichromosome from BK virus as a template for transcription in vitro. Proc. Natl. Acad. Sci. U. S. A. 1978, 75, 1126–1130. [Google Scholar] [CrossRef] [PubMed]
- Muller, U.; Zentgraf, H.; Eicken, I.; Keller, W. Higher order structure of simian virus 40 chromatin. Science 1978, 201, 406–415. [Google Scholar] [PubMed]
- Stehle, T.; Gamblin, S.J.; Yan, Y.; Harrison, S.C. The structure of simian virus 40 refined at 3.1 A resolution. Structure 1996, 4, 165–182. [Google Scholar] [CrossRef]
- Salunke, D.M.; Caspar, D.L.; Garcea, R.L. Self-assembly of purified polyomavirus capsid protein VP1. Cell 1986, 46, 895–904. [Google Scholar] [CrossRef] [PubMed]
- Stehle, T.; Harrison, S.C. High-resolution structure of a polyomavirus VP1-oligosaccharide complex: implications for assembly and receptor binding. EMBO J. 1997, 16, 5139–5148. [Google Scholar] [CrossRef] [PubMed]
- Stehle, T.; Yan, Y.; Benjamin, T.L.; Harrison, S.C. Structure of murine polyomavirus complexed with an oligosaccharide receptor fragment. Nature 1994, 369, 160–163. [Google Scholar] [CrossRef] [PubMed]
- Neu, U.; Woellner, K.; Gauglitz, G.; Stehle, T. Structural basis of GM1 ganglioside recognition by simian virus 40. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 5219–5224. [Google Scholar] [CrossRef] [PubMed]
- Dubensky, T.W.; Freund, R.; Dawe, C.J.; Benjamin, T.L. Polyomavirus replication in mice: influences of VP1 type and route of inoculation. J. Virol. 1991, 65, 342–349. [Google Scholar] [PubMed]
- Bauer, P.H.; Bronson, R.T.; Fung, S.C.; Freund, R.; Stehle, T.; Harrison, S.C.; Benjamin, T.L. Genetic and structural analysis of a virulence determinant in polyomavirus VP1. J. Virol. 1995, 69, 7925–7931. [Google Scholar] [PubMed]
- Chen, B.J.; Atwood, W.J. Construction of a novel JCV/SV40 hybrid virus (JCSV) reveals a role for the JCV capsid in viral tropism. Virology 2002, 300, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Neu, U.; Stehle, T.; Atwood, W.J. The Polyomaviridae: Contributions of virus structure to our understanding of virus receptors and infectious entry. Virology 2009, 384, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, T.; Marquez, J.P.; Wakatsuki, T.; Sorokin, A. Caveolar endocytosis is critical for BK virus infection of human renal proximal tubular epithelial cells. J. Virol. 2007, 81, 8552–8562. [Google Scholar] [CrossRef] [PubMed]
- Eash, S.; Querbes, W.; Atwood, W.J. Infection of vero cells by BK virus is dependent on caveolae. J. Virol. 2004, 78, 11583–11590. [Google Scholar] [CrossRef] [PubMed]
- Pho, M.T.; Ashok, A.; Atwood, W.J. JC virus enters human glial cells by clathrin-dependent receptor-mediated endocytosis. J. Virol. 2000, 74, 2288–2292. [Google Scholar] [CrossRef] [PubMed]
- Norkin, L.C.; Anderson, H.A.; Wolfrom, S.A.; Oppenheim, A. Caveolar endocytosis of simian virus 40 is followed by brefeldin A-sensitive transport to the endoplasmic reticulum, where the virus disassembles. J. Virol. 2002, 76, 5156–5166. [Google Scholar] [CrossRef] [PubMed]
- Pelkmans, L.; Kartenbeck, J.; Helenius, A. Caveolar endocytosis of simian virus 40 reveals a new two-step vesicular-transport pathway to the ER. Nat. Cell Biol. 2001, 3, 473–483. [Google Scholar] [CrossRef]
- Richterova, Z.; Liebl, D.; Horak, M.; Palkova, Z.; Stokrova, J.; Hozak, P.; Korb, J.; Forstova, J. Caveolae are involved in the trafficking of mouse polyomavirus virions and artificial VP1 pseudocapsids toward cell nuclei. J. Virol. 2001, 75, 10880–10891. [Google Scholar] [CrossRef] [PubMed]
- Magnuson, B.; Rainey, E.K.; Benjamin, T.; Baryshev, M.; Mkrtchian, S.; Tsai, B. ERp29 triggers a conformational change in polyomavirus to stimulate membrane binding. Mol. Cell 2005, 20, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Schelhaas, M.; Malmstrom, J.; Pelkmans, L.; Haugstetter, J.; Ellgaard, L.; Grunewald, K.; Helenius, A. Simian Virus 40 depends on ER protein folding and quality control factors for entry into host cells. Cell 2007, 131, 516–529. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Abend, J.R.; Tsai, B.; Imperiale, M.J. Early events during BK virus entry and disassembly. J. Virol. 2009, 83, 1350–1358. [Google Scholar] [CrossRef] [PubMed]
- Lilley, B.N.; Gilbert, J.M.; Ploegh, H.L.; Benjamin, T.L. Murine polyomavirus requires the endoplasmic reticulum protein Derlin-2 to initiate infection. J. Virol. 2006, 80, 8739–8744. [Google Scholar] [CrossRef] [PubMed]
- Qian, M.; Cai, D.; Verhey, K.J.; Tsai, B. A lipid receptor sorts polyomavirus from the endolysosome to the endoplasmic reticulum to cause infection. PLoS Path. 2009, 5, e1000465. [Google Scholar] [CrossRef]
- Gross, L. A filterable agent, recovered from Ak leukemic extracts, causing salivary gland carcinomas in C3H mice. Proc. Soc. Exp. Biol. Med. 1953, 83, 414–421. [Google Scholar] [PubMed]
- Cahan, L.D.; Singh, R.; Paulson, J.C. Sialyloligosaccharide receptors of binding variants of polyoma virus. Virology 1983, 130, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Fried, H.; Cahan, L.D.; Paulson, J.C. Polyoma virus recognizes specific sialyligosaccharide receptors on host cells. Virology 1981, 109, 188–192. [Google Scholar] [CrossRef] [PubMed]
- Cahan, L.D.; Paulson, J.C. Polyoma virus adsorbs to specific sialyloligosaccharide receptors on erythrocytes. Virology 1980, 103, 505–509. [Google Scholar] [CrossRef] [PubMed]
- Freund, R.; Calderone, A.; Dawe, C.J.; Benjamin, T.L. Polyomavirus tumor induction in mice: effects of polymorphisms of VP1 and large T antigen. J. Virol. 1991, 65, 335–341. [Google Scholar] [PubMed]
- Freund, R.; Garcea, R.L.; Sahli, R.; Benjamin, T.L. A single-amino-acid substitution in polyomavirus VP1 correlates with plaque size and hemagglutination behavior. J. Virol. 1991, 65, 350–355. [Google Scholar] [PubMed]
- Gilbert, J.; Dahl, J.; Riney, C.; You, J.; Cui, C.; Holmes, R.; Lencer, W.; Benjamin, T. Ganglioside GD1a restores infectibility to mouse cells lacking functional receptors for polyomavirus. J. Virol. 2005, 79, 615–618. [Google Scholar] [CrossRef] [PubMed]
- Sweet, B.H.; Hilleman, M.R. The vacuolating virus, S.V. 40. Proc. Soc. Exp. Biol. Med. 1960, 105, 420–427. [Google Scholar] [PubMed]
- Garcea, R.L.; Imperiale, M.J. Simian virus 40 infection of humans. J. Virol. 2003, 77, 5039–5045. [Google Scholar] [CrossRef] [PubMed]
- Clayson, E.T.; Compans, R.W. Characterization of simian virus 40 receptor moieties on the surfaces of Vero C1008 cells. J. Virol. 1989, 63, 1095–1100. [Google Scholar] [PubMed]
- Campanero-Rhodes, M.A.; Smith, A.; Chai, W.; Sonnino, S.; Mauri, L.; Childs, R.A.; Zhang, Y.; Ewers, H.; Helenius, A.; Imberty, A.; Feizi, T. N-glycolyl GM1 ganglioside as a receptor for simian virus 40. J. Virol. 2007, 81, 12846–12858. [Google Scholar] [CrossRef] [PubMed]
- Norkin, L.C. Simian virus 40 infection via MHC class I molecules and caveolae. Immunol. Rev. 1999, 168, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Damm, E.M.; Pelkmans, L.; Kartenbeck, J.; Mezzacasa, A.; Kurzchalia, T.; Helenius, A. Clathrin- and caveolin-1-independent endocytosis: entry of simian virus 40 into cells devoid of caveolae. J. Cell Biol. 2005, 168, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Kukura, P.; Ewers, H.; Muller, C.; Renn, A.; Helenius, A.; Sandoghdar, V. High-speed nanoscopic tracking of the position and orientation of a single virus. Nat. Methods. 2009, 6, 923–927. [Google Scholar] [CrossRef]
- Ewers, H.; Romer, W.; Smith, A.E.; Bacia, K.; Dmitrieff, S.; Chai, W.; Mancini, R.; Kartenbeck, J.; Chambon, V.; Berland, L.; Oppenheim, A.; Schwarzmann, G.; Feizi, T.; Schwille, P.; Sens, P.; Helenius, A.; Johannes, L. GM1 structure determines SV40-induced membrane invagination and infection. Nat. Cell Biol. 2010, 12 sup pp 11-12, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Gardner, S.D.; Field, A.M.; Coleman, D.V.; Hulme, B. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet 1971, 1, 1253–1257. [Google Scholar] [CrossRef] [PubMed]
- Padgett, B.L.; Walker, D.L.; ZuRhein, G.M.; Eckroade, R.J.; Dessel, B.H. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet 1971, 1, 1257–1260. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Abend, J.R.; Johnson, S.F.; Imperiale, M.J. The role of polyomaviruses in human disease. Virology 2009, 384, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Sawa, H.; Komagome, R. The JC virus-like particle overlay assay. Methods Mol. Biol. 2005, 292, 175–186. [Google Scholar] [PubMed]
- Feng, H.; Shuda, M.; Chang, Y.; Moore, P.S. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 2008, 319, 1096–1100. [Google Scholar] [CrossRef] [PubMed]
- Berns, K.; Parish, C.R. Parvoviridae. In Fields Virology, 5th; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, 2007; pp. 2437–2477. [Google Scholar]
- Brown, K.E.; Young, N.S. Parvovirus B19 infection and hematopoiesis. Blood Rev. 1995, 9, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Murai, C.; Shibata, S.; Munakata, Y.; Ishii, T.; Ishii, K.; Saitoh, T.; Sawai, T.; Sugamura, K.; Sasaki, T. Human parvovirus B19 as a causative agent for rheumatoid arthritis. Proc. Natl. Acad. Sci. U. S. A. 1998, 95, 8227–8232. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.E.; Hibbs, J.R.; Gallinella, G.; Anderson, S.M.; Lehman, E.D.; McCarthy, P.; Young, N.S. Resistance to parvovirus B19 infection due to lack of virus receptor (erythrocyte P antigen). N. Engl. J. Med. 1994, 330, 1192–1196. [Google Scholar] [CrossRef] [PubMed]
- Chipman, P.R.; Agbandje-McKenna, M.; Kajigaya, S.; Brown, K.E.; Young, N.S.; Baker, T.S.; Rossmann, M.G. Cryo-electron microscopy studies of empty capsids of human parvovirus B19 complexed with its cellular receptor. Proc. Natl. Acad. Sci. U. S. A. 1996, 93, 7502–7506. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.A. Simian parvoviruses: biology and implications for research. Comp. Med. 2008, 58, 47–50. [Google Scholar] [PubMed]
- Grimm, D.; Kay, M.A. From virus evolution to vector revolution: use of naturally occurring serotypes of adeno-associated virus (AAV) as novel vectors for human gene therapy. Curr. Gene Ther. 2003, 3, 281–304. [Google Scholar] [CrossRef] [PubMed]
- Romer, W.; Berland, L.; Chambon, V.; Gaus, K.; Windschiegl, B.; Tenza, D.; Aly, M.R.; Fraisier, V.; Florent, J.C.; Perrais, D.; Lamaze, C.; Raposo, G.; Steinem, C.; Sens, P.; Bassereau, P.; Johannes, L. Shiga toxin induces tubular membrane invaginations for its uptake into cells. Nature 2007, 450, 670–675. [Google Scholar] [CrossRef] [PubMed]
© 2010 by the authors; licensee MDPI, Basel, Switzerland This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Share and Cite
Taube, S.; Jiang, M.; Wobus, C.E. Glycosphingolipids as Receptors for Non-Enveloped Viruses. Viruses 2010, 2, 1011-1049. https://doi.org/10.3390/v2041011
Taube S, Jiang M, Wobus CE. Glycosphingolipids as Receptors for Non-Enveloped Viruses. Viruses. 2010; 2(4):1011-1049. https://doi.org/10.3390/v2041011
Chicago/Turabian StyleTaube, Stefan, Mengxi Jiang, and Christiane E. Wobus. 2010. "Glycosphingolipids as Receptors for Non-Enveloped Viruses" Viruses 2, no. 4: 1011-1049. https://doi.org/10.3390/v2041011
APA StyleTaube, S., Jiang, M., & Wobus, C. E. (2010). Glycosphingolipids as Receptors for Non-Enveloped Viruses. Viruses, 2(4), 1011-1049. https://doi.org/10.3390/v2041011





