SaDA: From Sampling to Data Analysis—An Extensible Open Source Infrastructure for Rapid, Robust and Automated Management and Analysis of Modern Ecological High-Throughput Microarray Data
Abstract
:1. Introduction
State-of-Art Technology
2. Material and Methods
2.1. Implementation
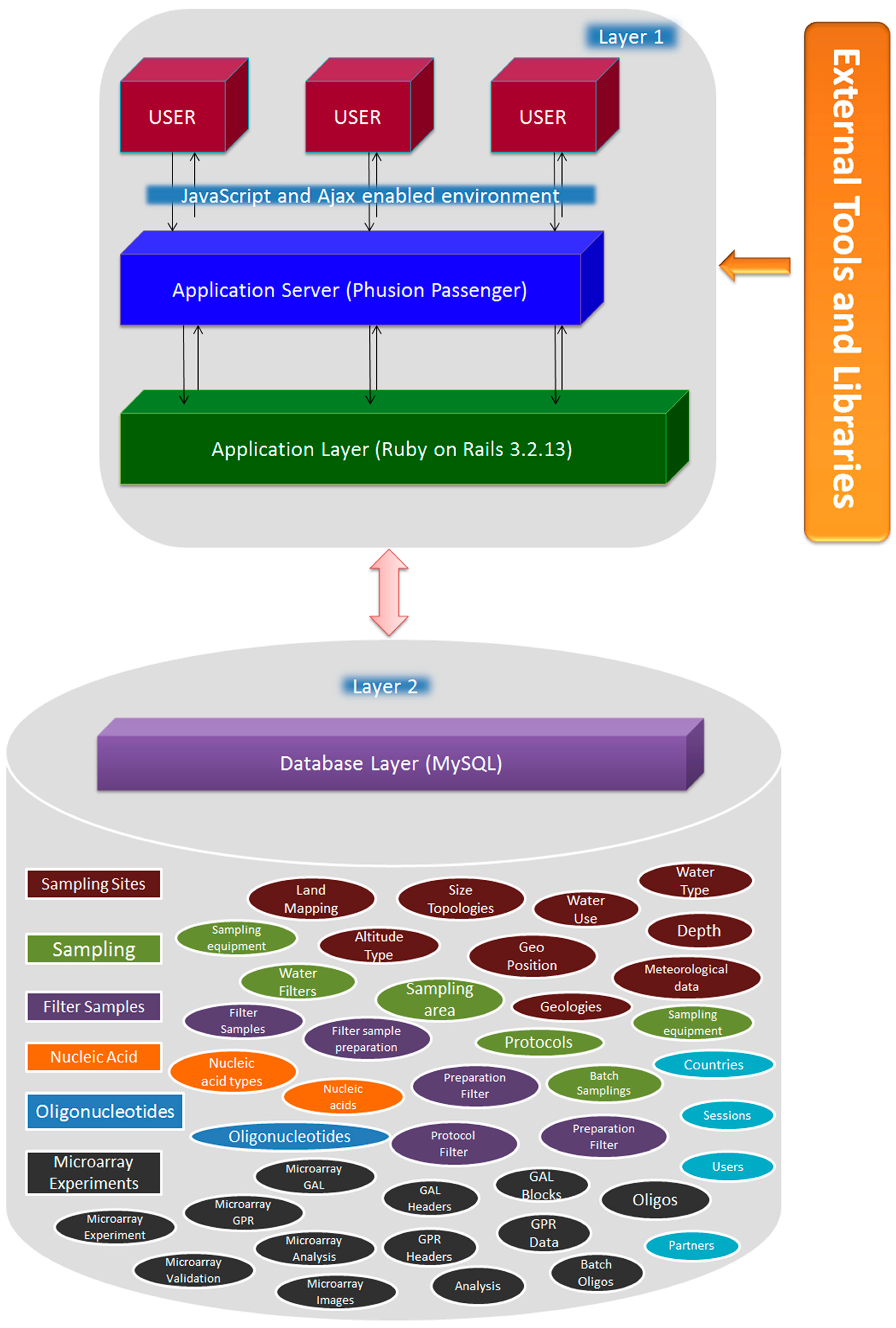
2.2. Experimental Design and Data Acquisition

2.3. The Features of SaDA
2.3.1. Samples and Filter Tracking
2.3.2. Management of project specific experimental data
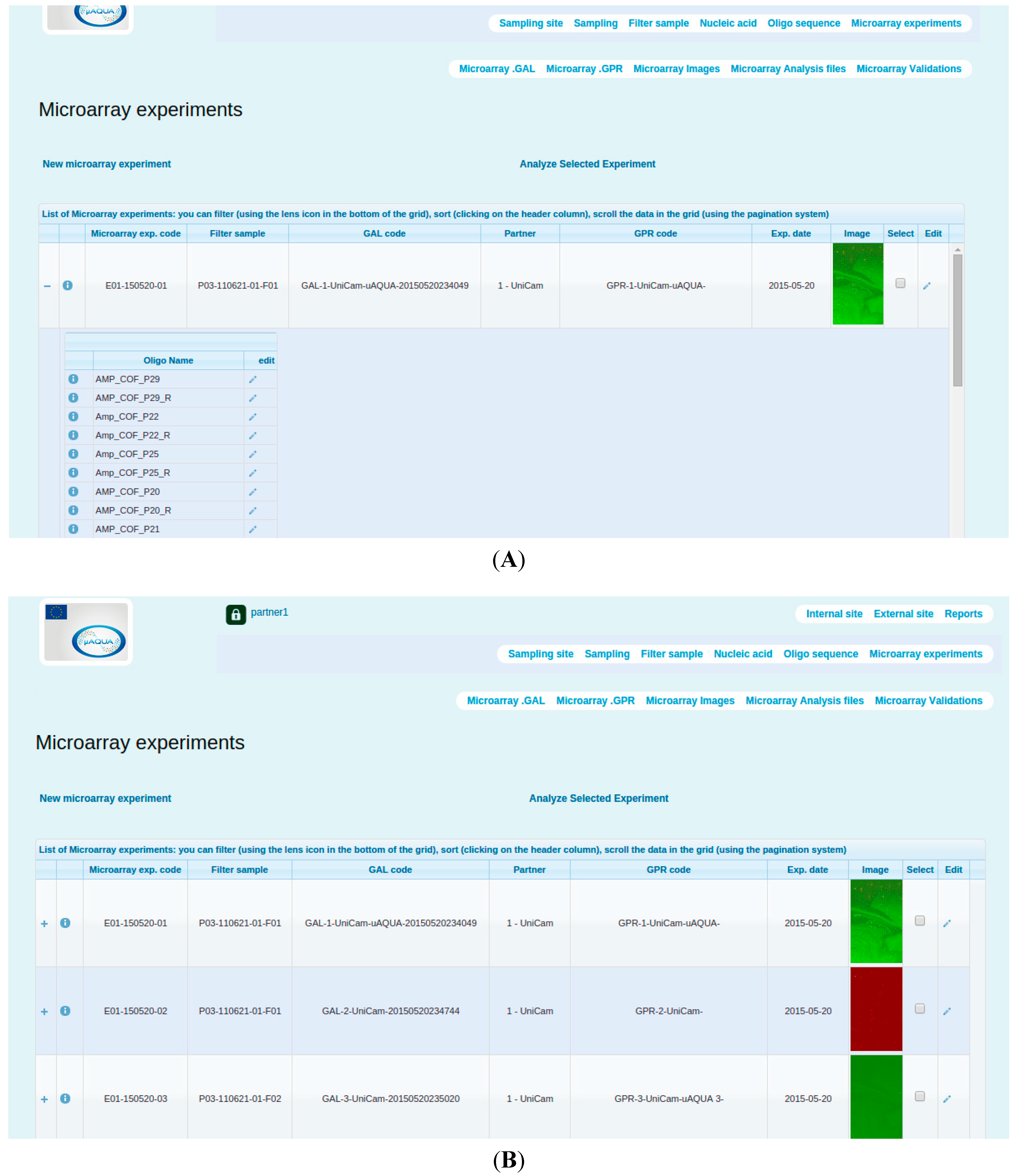
2.3.3. Microarray data management and analysis
- The scanned images (raw data) available in images sub-module.
- The quantitative outputs from the image analysis procedure (microarray quantitation matrices), available as GPR files that can be uploaded via Microarray .GPR section.
- The derived measurements (data matrices) that could be viewed from Microarray Analysis section.
2.4. Database Schema
2.5. Batch Uploads and Downloads of Data
3. Results and Discussion
3.1. Utility of the SaDA Infrastructure
3.3. Limitations of SaDA
3.4. Availability and Future Directions
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Rüegg, J.; Gries, C.; Bond-Lamberty, B.; Bowen, G.J.; Felzer, B.S.; McIntyre, N.E.; Soranno, P.; Vanderbilt, K.L.; Weathers, K.C. Completing the data life cycle: Using information management in macrosystems ecology research. Front. Ecol. Environ. 2014, 12, 24–30. [Google Scholar] [CrossRef]
- Heffernan, J.B.; Soranno, P.; Angilletta, M.J.; Buckley, L.B.; Gruner, D.S.; Keitt, T.H.; Kellner, J.R.; Kominoski, J.S.; Rocha, A.V.; Xiao, J.; et al. Macrosystems ecology: Understanding ecological patterns and processes at continental scales. Front. Ecol. Environ. 2014, 12, 5–14. [Google Scholar] [CrossRef]
- Goring, S.J.; Weathers, K.C.; Dodds, W.K.; Soranno, P.; Sweet, L.C.; Cheruvelil, K.S.; Kominoski, J.S.; Rüegg, J.; Thorn, A.M.; Utz, R.M. Improving the culture of interdisciplinary collaboration in ecology by expanding measures of success. Front. Ecol. Environ. 2014, 12, 39–47. [Google Scholar] [CrossRef]
- Bushmann, F.; Meunier, R.; Rohnert, H. Pattern-Oriented Software Architecture: A System of Patterns; John Wiley & Sons: West Sussex, England, 1996; Volume 1, p. 476. [Google Scholar]
- Maurer, M.; Molidor, R.; Sturn, A.; Hartler, J.; Hackl, H.; Stocker, G.; Prokesch, A.; Scheideler, M.; Trajanoski, Z. MARS: Microarray analysis, retrieval, and storage system. BMC Bioinform. 2005, 6. [Google Scholar] [CrossRef]
- Stocker, G.; Fischer, M.; Rieder, D.; Bindea, G.; Kainz, S.; Oberstolz, M.; McNally, J.G.; Trajanoski, Z. iLAP: A workflow-driven software for experimental protocol development, data acquisition and analysis. BMC Bioinform. 2009, 10. [Google Scholar] [CrossRef] [PubMed]
- Vallon-Christersson, J.; Nordborg, N.; Svensson, M.; Häkkinen, J. BASE—2nd generation software for microarray data management and analysis. BMC Bioinform. 2009, 10. [Google Scholar] [CrossRef] [PubMed]
- Nelson, E.K.; Piehler, B.; Eckels, J.; Rauch, A.; Bellew, M.; Hussey, P.; Ramsay, S.; Nathe, C.; Lum, K.; Krouse, K.; et al. LabKey Server: An open source platform for scientific data integration, analysis and collaboration. BMC bioinform. 2011, 12. [Google Scholar] [CrossRef] [PubMed]
- Nix, D.; di Sera, T.L.; Dalley, B.K.; Milash, B.; Cundick, R.M.; Quinn, K.S.; Courdy, S.J. Next generation tools for genomic data generation, distribution, and visualization. BMC Bioinform. 2010, 11. [Google Scholar] [CrossRef]
- Colmsee, C.; Flemming, S.; Klapperstück, M.; Lange, M.; Scholz, U. A case study for efficient management of high throughput primary lab data. BMC Res. Notes 2011, 4. [Google Scholar] [CrossRef] [PubMed]
- Bauch, A.; Adamczyk, I.; Buczek, P.; Elmer, F.-J.; Enimanev, K.; Glyzewski, P.; Kohler, M.; Pylak, T.; Quandt, A.; Ramakrishnan, C.; et al. OpenBIS: A flexible framework for managing and analyzing complex data in biology research. BMC Bioinform. 2011, 12. [Google Scholar] [CrossRef] [PubMed]
- Venco, F.; Ceol, A.; Muller, H. SLIMS: A LIMS for handling next-generation sequencing workflows. EMBnet. J. 2013, 15, 85–87. [Google Scholar] [CrossRef]
- McLellan, A.S.; Dubin, R.; Jing, Q.; Broin, P.Ó.; Moskowitz, D.; Suzuki, M.; Calder, R.B.; Hargitai, J.; Golden, A.; Greally, J.M. The Wasp System: An open source environment for managing and analyzing genomic data. Genomics 2012, 100, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Giardine, B.; Riemer, C.; Hardison, R.C.; Burhans, R.; Elnitski, L.; Shah, P.; Zhang, Y.; Blankenberg, D.; Albert, I.; Taylor, J.; et al. Galaxy: A platform for interactive large-scale genome analysis. Genome Res. 2005, 15, 1451–1455. [Google Scholar] [CrossRef] [PubMed]
- Scholtalbers, J.; Rößler, J.; Sorn, P.; de Graaf, J.; Boisguérin, V.; Castle, J.; Sahin, U. Galaxy LIMS for next-generation sequencing. Bioinformatics (Oxf., Engl.) 2013, 29, 1233–1234. [Google Scholar] [CrossRef] [PubMed]
- CASAVA. Available online: http://support.illumina.com/sequencing/documentation.html (accessed on 2 June 2015).
- Van Rossum, T.; Tripp, B.; Daley, D. SLIMS—A user-friendly sample operations and inventory management system for genotyping labs. Bioinformatics 2010, 26, 1808–1810. [Google Scholar] [CrossRef] [PubMed]
- Protopsaltou, A. Model Driven Development with Ruby on Rails Ruby on Rails and Reverse Engineering. Master Thesis, IT University of Göteborg, Göteborg, Sweden, 27 June 2007; pp. 1–34. [Google Scholar]
- Rosen, L.S.R. Web Application Architecture: Principles, Protocol and Practices; John Wiley & Sons Ltd.: West Sussex, England, 2009. [Google Scholar]
- MicroAQUA Project Consortium. Available online: www.microaqua.eu (accessed on 2 June 2015).
- YM4R/GM Plugin, for Utilizing Google Map in Rails Applications. Available online: https://github.com/queso/ym4r-gm (accessed on 2 June 2015).
- Viksna, J.; Celms, E.; Opmanis, M.; Podnieks, K.; Rucevskis, P.; Zarins, A.; Barrett, A.; Neogi, S.G.; Krestyaninova, M.; McCarthy, M.I.; et al. PASSIM—An open source software system for managing information in biomedical studies. BMC Bioinform. 2007, 8. [Google Scholar] [CrossRef]
- Saeed, A.I.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thia-garajan, M.; et al. TM4: A free, open-source system for microarray data management and analysis. BioTechniques 2003, 34, 374–378. [Google Scholar] [PubMed]
- Pieler, R.; Sanchez-Cabo, F.; Hackl, H.; Thallinger, G.G.; Trajanoski, Z. ArrayNorm: Comprehensive normalization and analysis of microarray data. Bioinformatics 2004, 20, 1971–1973. [Google Scholar] [CrossRef] [PubMed]
- Hokamp, K.; Roche, F.M.; Acab, M.; Rousseau, M.E.; Kuo, B.; Goode, D.; Aeschliman, D.; Bryan, J.; Babiuk, L.; Hancock, R.E.W.; et al. ArrayPipe: A flexible processing pipeline for microarray data. Nucleic Acids Res. 2004, 32, 457–459. [Google Scholar] [CrossRef] [PubMed]
- Dittami, S.M.; Edvardsen, B. GPR-Analyzer: A simple tool for quantitative analysis of hierarchical multispecies microarrays. Environ. Sci. Pollut. Res. 2013, 20, 6808–6815. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, P.; Kottmann, R.; Field, D.; Knight, R.; Cole, J.R.; Amaral-Zettler, L.; Gilbert, J.; Karsch-Mizrachi, I.; Johnston, A.; Cochrane, G.; et al. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat. Biotechnol. 2011, 29, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Rayner, T.F.; Rocca-Serra, P.; Spellman, P.T.; Causton, H.C.; Farne, A.; Holloway, E.; Irizarry, R.; Liu, J.; Maier, D.S.; Miller, M.; et al. A simple spreadsheet-based, MIAME-supportive format for microarray data: MAGE-TAB. BMC Bioinform. 2006, 7. [Google Scholar] [CrossRef]
- MAGE-TAB Specification. 2011. Available online: http://fged.org/site_media/pdf/MAGE-TABv1.1_2011_07_28.pdf (accessed on 2 June 2015).
- jQWidgets. Available online: http://www.jqwidgets.com/ (accessed on 2 June 2015).
- Crawford, S. RinRuby: Accessing the R interpreter from pure ruby. J. Stat. Softw. 2009, 29, 4. [Google Scholar]
- jqGrid. Available online: http://www.trirand.com/blog/ (accessed on 2 June 2015).
- S2C Source Code. Available online: https://github.com/kumarsaurabh20/PlotApp (accessed on 2 June 2015).
- Mitsuteru, N.G.; Mitsuteru, N.G.; Goto, N.; Goto, N.; Nakao, M.C.; Nakao, M.C.; Kawashima, S.; Kawashima, S.; Katayama, T.; Katayama, T.; et al. BioRuby: Open-source bioinformatics library. Genome Inform. 2003, 14, 629–630. [Google Scholar]
- Apache 2.0 License. Available online: http://www.apache.org/licenses/LICENSE-2.0 (accessed on 2 June 2015).
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, K.S.; Thual, D.; Spurio, R.; Cannata, N. SaDA: From Sampling to Data Analysis—An Extensible Open Source Infrastructure for Rapid, Robust and Automated Management and Analysis of Modern Ecological High-Throughput Microarray Data. Int. J. Environ. Res. Public Health 2015, 12, 6352-6366. https://doi.org/10.3390/ijerph120606352
Singh KS, Thual D, Spurio R, Cannata N. SaDA: From Sampling to Data Analysis—An Extensible Open Source Infrastructure for Rapid, Robust and Automated Management and Analysis of Modern Ecological High-Throughput Microarray Data. International Journal of Environmental Research and Public Health. 2015; 12(6):6352-6366. https://doi.org/10.3390/ijerph120606352
Chicago/Turabian StyleSingh, Kumar Saurabh, Dominique Thual, Roberto Spurio, and Nicola Cannata. 2015. "SaDA: From Sampling to Data Analysis—An Extensible Open Source Infrastructure for Rapid, Robust and Automated Management and Analysis of Modern Ecological High-Throughput Microarray Data" International Journal of Environmental Research and Public Health 12, no. 6: 6352-6366. https://doi.org/10.3390/ijerph120606352
APA StyleSingh, K. S., Thual, D., Spurio, R., & Cannata, N. (2015). SaDA: From Sampling to Data Analysis—An Extensible Open Source Infrastructure for Rapid, Robust and Automated Management and Analysis of Modern Ecological High-Throughput Microarray Data. International Journal of Environmental Research and Public Health, 12(6), 6352-6366. https://doi.org/10.3390/ijerph120606352




