Nucleic Acid-based Detection of Bacterial Pathogens Using Integrated Microfluidic Platform Systems
Abstract
:1. Introduction
2. Nucleic Acid-Based Detection
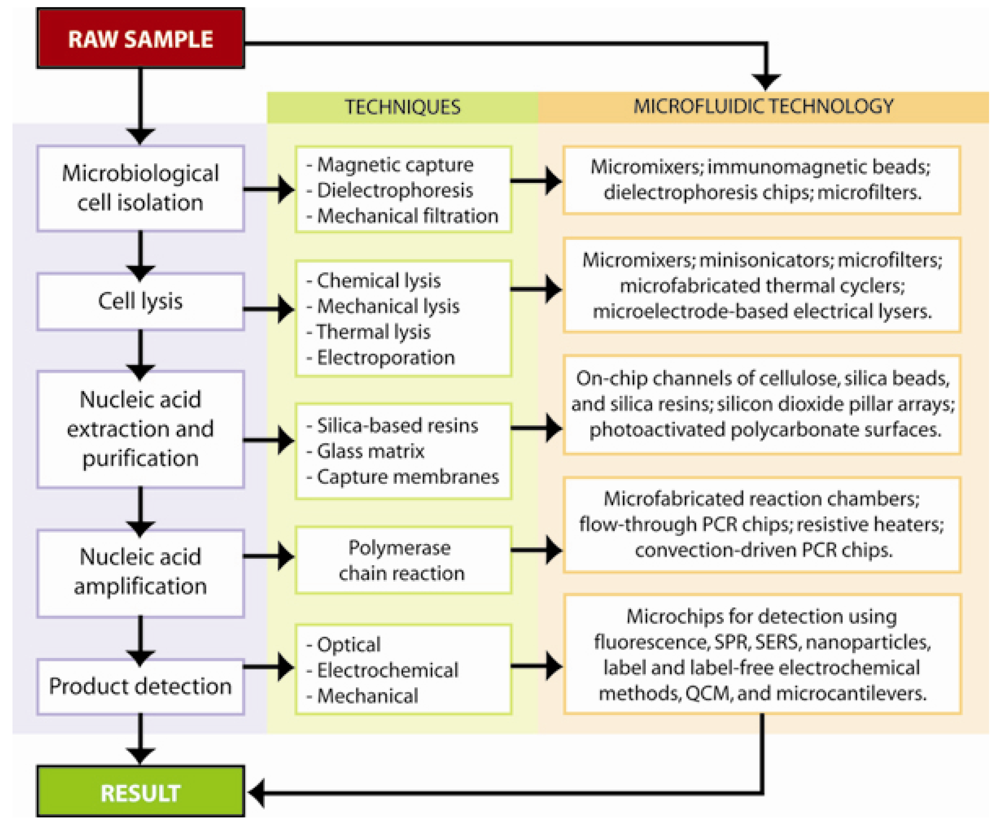
3. Microfluidic Nucleic-Acid Based Pathogen Detection Systems
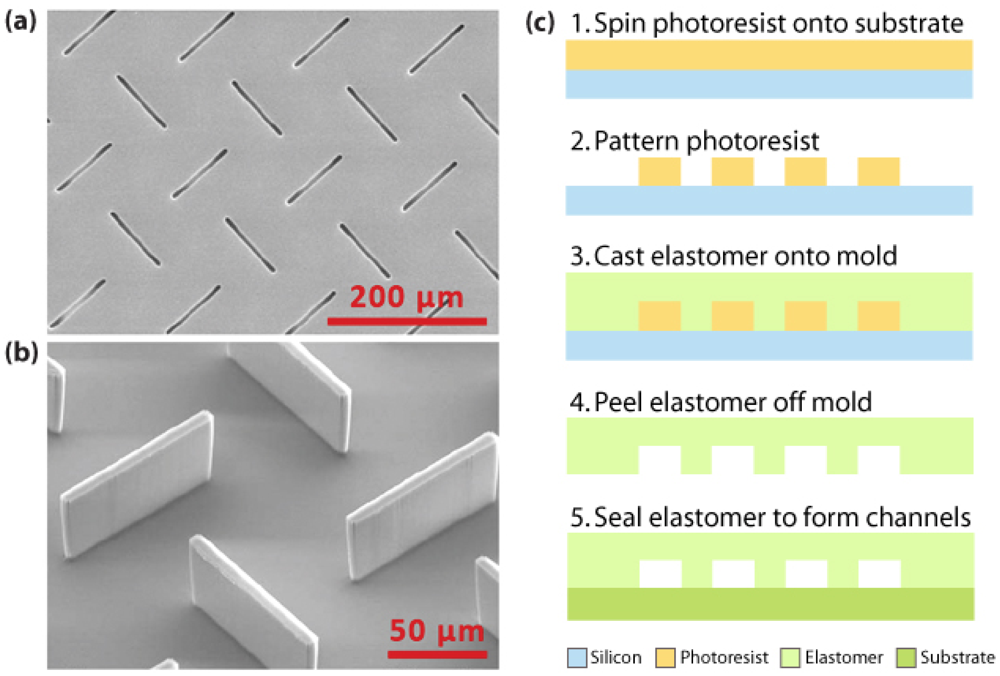
3.1. Materials and fabrication for microfluidic sensors
3.2. On-chip fluid and reagent handling
3.2.1. Microfluidic Pumping
3.2.2. Microfluidic Valving
3.2.3. Microfluidic Mixing
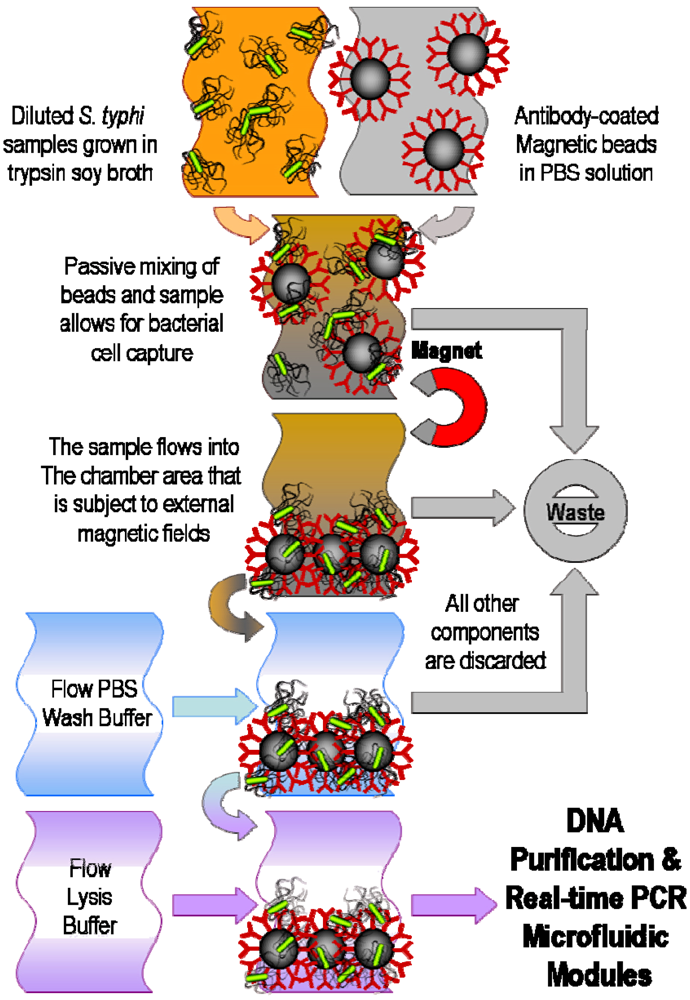
4. Filtration and Separation of Bacterial Cells
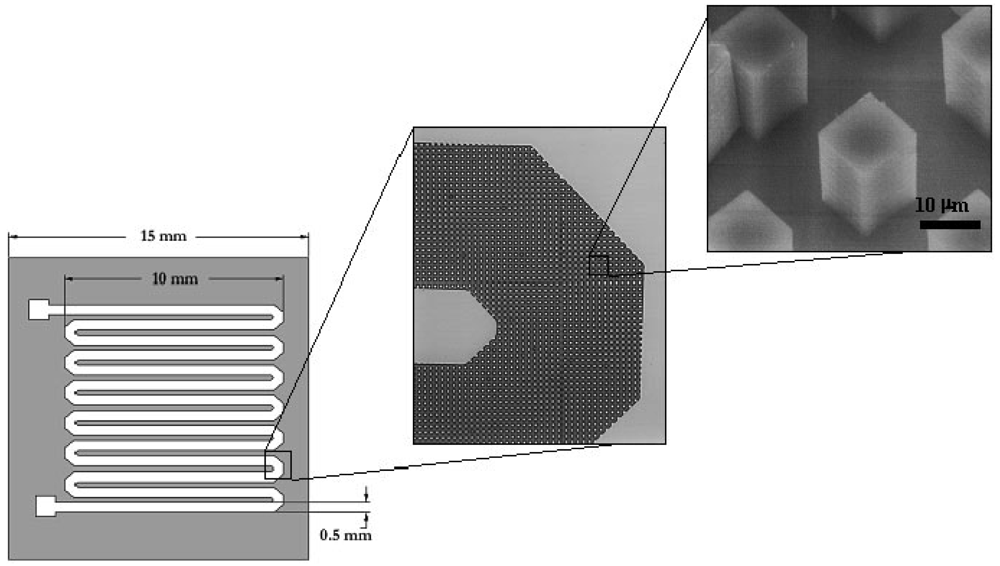
5. Pathogenic DNA Extraction and Purification
6. Pathogenic DNA Detection
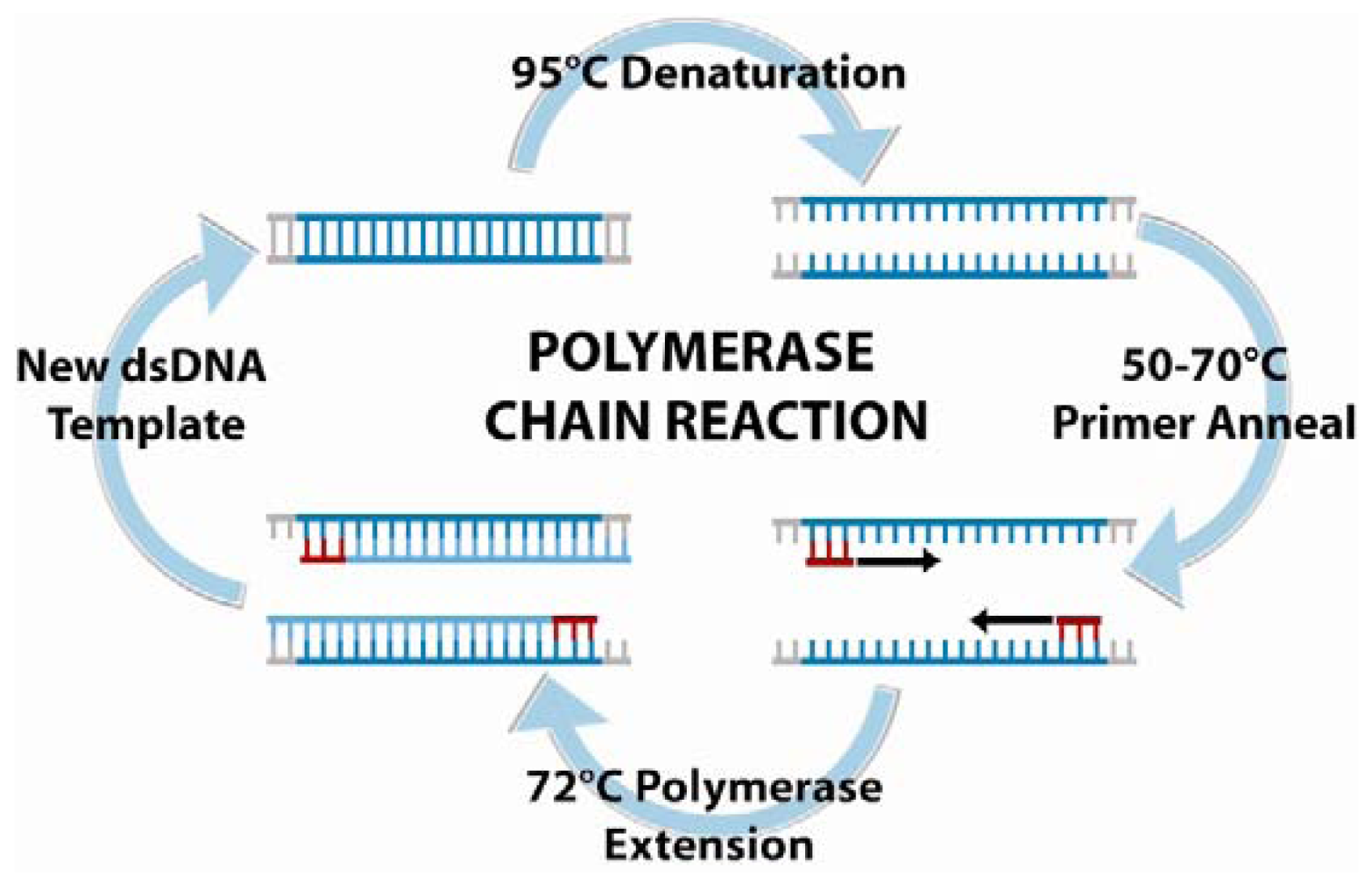
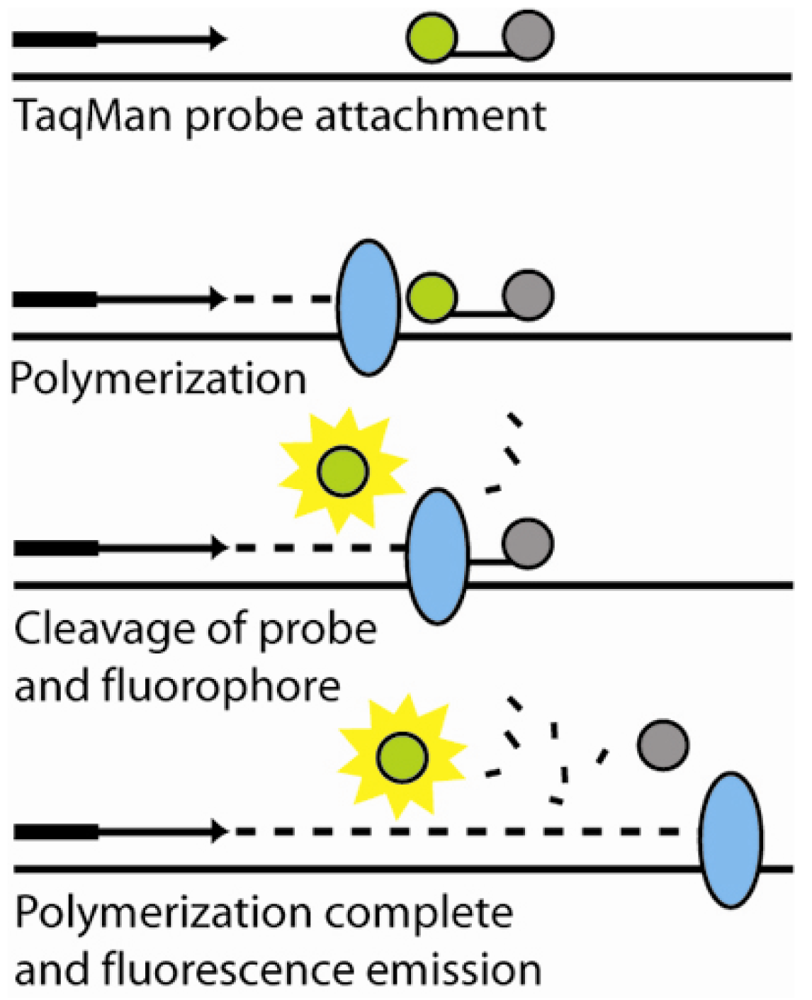
6.1. Polymerase chain reaction amplification and detection
6.2. Optical methods in nucleic acid-based detection
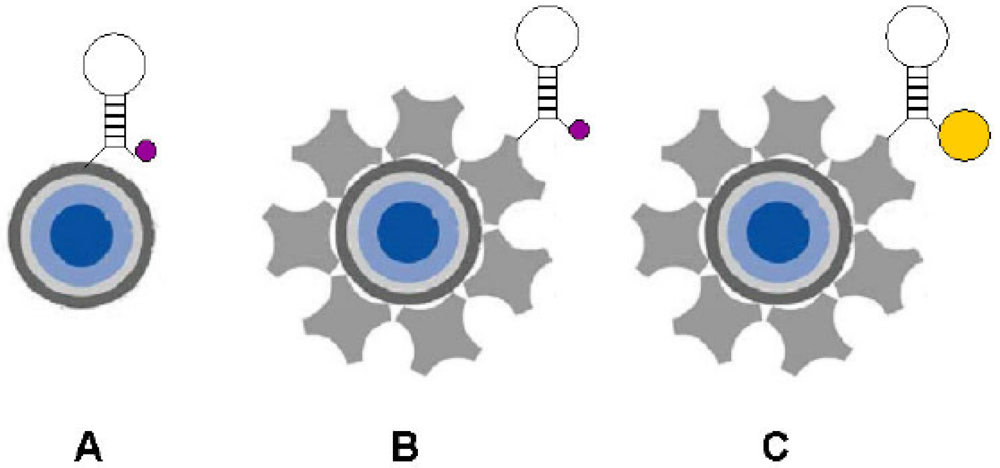
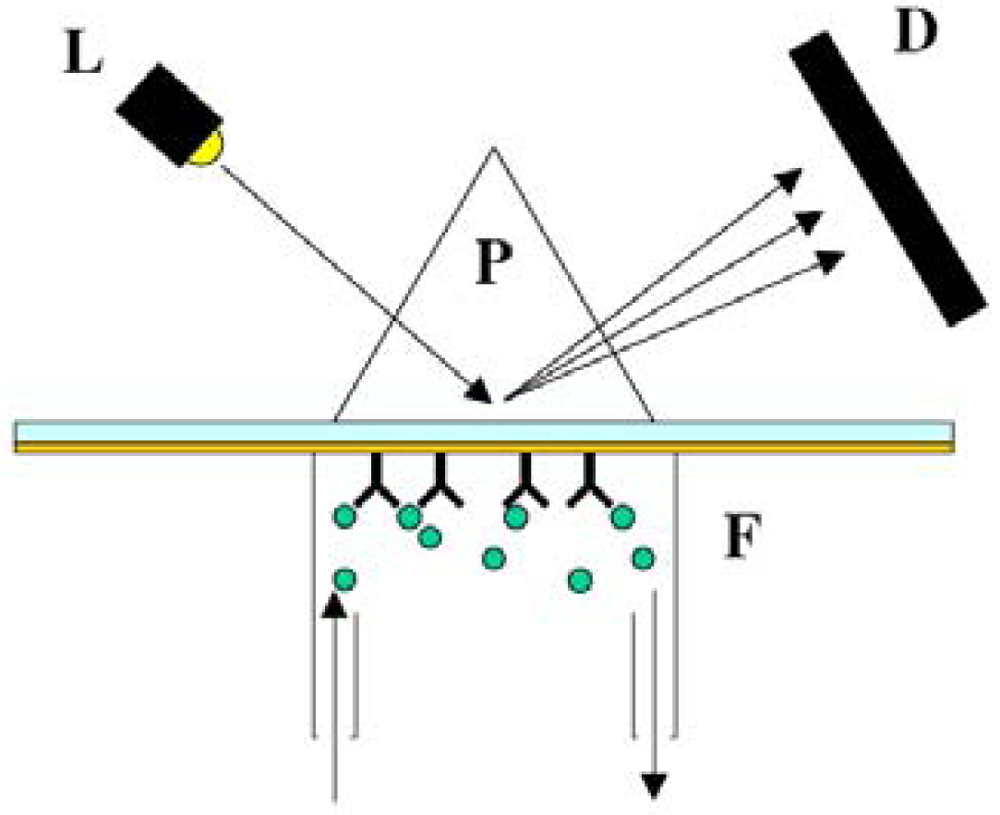
6.2.1. Fluorescence-based detection
6.2.2. Surface plasmon resonance
6.2.3. Raman detection
6.3. Electrochemical methods in nucleic acid-based detection
6.3.1. Labeling techniques
6.3.2. Amperometric detection
6.3.3. Potentiometric detection
6.3.4. Conductimetric and Impedimetric Detection
6.4. Mechanical methods in nucleic acid-based detection
6.4.1. Quartz crystal microbalance
6.4.2. Cantilever-based detection
7. Integrated Pathogen Detection Systems
8. Conclusion and Future Directions
References and Notes
- Lazcka, O.; Del Campo, F.J.; Muñoz, F.X. Pathogen detection: A perspective of traditional methods and biosensors. Biosens. Bioelectron. 2006, 22, 1205–1217. [Google Scholar]
- Leonard, P.; Hearty, S.; Brennan, J.; Dunne, L.; Quinn, J.; Chakraborty, T.; O'Kennedy, R. Advances in biosensors for detection of pathogens in food and water. Enzyme Microb. Technol. 2003, 32, 3–13. [Google Scholar]
- Belgrader, P.; Benett, W.; Hadley, D.; Long, G.; Raymond Mariella, J.; Milanovich, F.; Nasarabadi, S.; Nelson, W.; Richards, J.; Stratton, P. Rapid pathogen detection using a microchip PCR array instrument. Clin. Chem. 1998, 44, 2191–2194. [Google Scholar]
- Chroni, C.; Kyriacou, A.; Georgaki, I.; Manios, T.; Kotsou, M.; Lasaridi, K. Microbial characterization during composting of biowaste. Waste Manag. 2009, 29, 1520–1525. [Google Scholar]
- Arora, K.; Chand, S.; Malhotra, B.D. Recent developments in bio-molecular electronics techniques for food pathogens. Anal. Chim. Acta 2006, 568, 259–274. [Google Scholar]
- Auroux, P.A.; Koc, Y.; deMello, A.; Manz, A.; Day, P.J.R. Miniaturized nucleic acid analysis. Lab Chip 2004, 4, 534–546. [Google Scholar]
- Becker, H.; Gartner, C. Polymer microfabrication methods for microfluidic analytical applications. Electrophoresis 2000, 21, 12–26. [Google Scholar]
- Campàs, M.; Katakis, I. DNA biochip arraying, detection, and amplification strategies. Trends. Analyt. Chem. 2004, 23, 49–62. [Google Scholar]
- Haeberle, S.; Zengerle, R. Microfluidic platforms for lab-on-a-chip applications. Lab Chip 2007, 7, 1094–1110. [Google Scholar]
- Mothershed, E.A.; Whitney, A.M. Nucleic acid-based methods for the detection of bacterial pathogens: Present and future considerations for the clinical laboratory. Clin. Chim. Acta 2006, 363, 206–220. [Google Scholar]
- Palchetti, I.; Mascini, M. Electroanalytical biosensors and their potential for food pathogen and toxin detection. Anal. Bioanal. Chem. 2008, 391, 455–471. [Google Scholar]
- Ricci, F.; Volpe, G.; Micheli, L.; Palleschi, G. A review on novel developments and applications of immunosensors in food analysis. Anal. Chim. Acta 2007, 605, 111–129. [Google Scholar]
- Sapsford, K.E.; Bradburne, C.; Delehanty, J.B.; Medintz, I.L. Sensors for detecting biological agents. Materials Today 2008, 11, 38–49. [Google Scholar]
- Sassolas, A.; Leco-Bouvier, B.D.; Blum, L.J. DNA Biosensors and Microarrays. Chem. Rev. 2008, 108, 109–139. [Google Scholar]
- Sun, Y.; Kwok, Y.C. Polymeric microfluidic system for DNA analysis. Anal. Chim. Acta 2006, 556, 80–96. [Google Scholar]
- Viskari, P.J.; Landers, J.P. Unconventional detection methods for microfluidic devices. Electrophoresis 2006, 27, 1797–1810. [Google Scholar]
- Weigl, B.; Domingo, G.; LaBarre, P.; Gerlach, J. Towards non- and minimally instrumented, microfluidics-based diagnostic devices. Lab Chip 2008, 8, 1999–2014. [Google Scholar]
- Yi, C.; Li, C.W.; Ji, S.; Yang, M. Microfluidics technology for manipulation and analysis of biological cells. Anal. Chim. Acta 2006, 560, 1–23. [Google Scholar]
- Zhang, C.; Xu, J.; Ma, W.; Zheng, W. PCR microfluidic devices for DNA amplification. Biotechnol. Adv. 2006, 24, 243–284. [Google Scholar]
- Jarvis, B.; Hedges, A.J.; Corry, J.E.L. Assessment of measurement uncertainty for quantitative methods of analysis: Comparative assessment of the precision (uncertainty) of bacterial colony counts. Int. J. Food Microbiol 2007, 116, 44–51. [Google Scholar]
- Zhang, C.; Chen, W.B.; Liu, W.L.; Chen, C.B. An automated bacterial colony counting system. IEEE International Conference on Sensor Networks, Ubiquitous, and Trustworthy Computing, Taichung, Taiwan; 2008; pp. 233–240. [Google Scholar]
- Lamprecht, M.R.; Sabatini, D.M.; Carpenter, A.E. Cell Profiler™: free, versatile software for automated biological image analysis. BioTechniques 2007, 42, 71–75. [Google Scholar]
- Wang, X.; Yamaguchi, N.; Someya, T.; Nasu, M. Rapid and automated enumeration of viable bacteria in compost using a micro-colony auto counting system. J. Microb. Methods 2007, 71, 1–6. [Google Scholar]
- Yamazaki, W.; Taguchi, M.; Kawai, T.; Kawatsu, K.; Sakata, J.; Inoue, K.; Misawa, N. Comparison of loop-mediated isothermal amplification assay and conventional culture methods for detection of Campylobacter jejuni and Campylobacter coli in naturally contaminated chicken meat samples. Appl. Environ. Microbiol 2009, 75, 1597–1603. [Google Scholar]
- Maglott, D.; Ostell, J.; Priott, K.D.; Tatusova, T. Entrez gene: gene-centered information at NCBI. Nucleic Acids Res. 2005, 33, D54–D58. [Google Scholar]
- Winfield, M.D.; Groisman, E.A. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl. Environ. Microbiol. 2003, 69, 3689–3694. [Google Scholar]
- Rahman, I.; Shahamat, M.; Chowdhury, M.A.; Colwell, R.R. Potential virulence of viable but nonculturable Shigella dysenteriae type 1. Appl. Environ. Microbiol 1996, 62, 115–120. [Google Scholar]
- Effendi, I.; Austin, B. Dormant/unculturable cells of the fish pathogen Aeromonas salmonicida. Microb. Ecol. 1995, 30, 183–192. [Google Scholar]
- Rovery, C.; Greub, G.; Lepidi, H.; Casalta, J.P. PCR Detection of Bacteria on Cardiac Valves of Patients with Treated Bacterial Endocarditis. J. Clin. Microb. 2005, 43, 163–167. [Google Scholar]
- Bartosch, S.; Fite, A.; Macfarlane, G.T.; McMurdo, M.E.T. Characterization of Bacterial Communities in Feces from Healthy Elderly Volunteers and Hospitalized Elderly Patients by Using Real-Time PCR and Effects of Antibiotic Tratement on the Fecal Microbiota. Appl. Environ. Microbiol 2004, 70, 3575–3581. [Google Scholar]
- Rosamond, J.; Allsop, A. Harnessing the power of the genome in the search for new antibiotics. Science 2000, 287, 1973–1976. [Google Scholar]
- Cady, N.C.; Stelick, S.; Kunnavakkam, M.V.; Batt, C.A. Real-time PCR detection of Listeria monocytogenes using an integrated microfluidics platform. Sens. Actuat. B Chem. 2005, 107, 332–341. [Google Scholar]
- Dineva, M.A.; Mahilum-Tapay, L.; Lee, H. Sample preparation: a challenge in the development of point-of-care nucleic acid-based assays for resource-limited settings. Analyst 2007, 132, 1193–1199. [Google Scholar]
- Chin, C.D.; Linder, V.; Sia, S.K. Lab-on-a-chip devices for global health: Past studies and future opportunities. Lab Chip 2007, 7, 41–57. [Google Scholar]
- Linder, V. Microfluidics at the crossroad with point-of-care diagnostics. Analyst 2007, 132, 1186–1192. [Google Scholar]
- Manz, A.; Graber, N.; Wildmer, H.M. Miniaturized total chemical analysis systems: a novel concept for chemical sensing. Sens. Actuat. B Chem. 1990, 1, 240–248. [Google Scholar]
- Liu, Y.; Cady, N.C.; Batt, C.A. A plastic microchip for nucleic acid purification. Biomed. Microdevices 2007, 9, 769–776. [Google Scholar]
- Qi, S.; Liu, X.; Ford, S.; Barrows, J.; Thomas, G.; Kelly, K.; McCandless, A.; Lian, K.; Goettert, J.; Soper, S.A. Microfluidic devices fabricated in poly(methyl methacrylate) using hot-embossing with integrated sampling capillary and fiber optics for fluorescence detection. Lab Chip 2002, 2, 88–95. [Google Scholar]
- Narasimhan, J.; Paputsky, I. Polymer embossing tools for rapid prototyping of plastic microfluidic devices. J. Micromech. Microeng. 2004, 14, 96–103. [Google Scholar]
- Mair, D.A.; Geiger, E.; Pisano, A.P.; Frechet, J.M.J.; Svec, F. Injection molded microfluidic chips featuring integrated interconnects. Lab Chip 2006, 6, 1346–1354. [Google Scholar]
- Xia, Y.; Whitesides, G.M. Soft lithography. Angew. Chem. 1998, 37, 550–575. [Google Scholar]
- Duncan, A.C.; Weisbuch, F.; Rouais, F.; Lazare, S.; Baquey, C. Laser microfabricated model surfaces for controlled cell growth. Biosens. Bioelectron. 2002, 17, 413–426. [Google Scholar]
- Mappes, T.; Achenbach, S.; Mohr, J. X-ray lithography for devices with high aspect ratio polymer submicron structures. Microelec. Eng. 2007, 84, 1235–1239. [Google Scholar]
- Garstecki, P.; Fuerstman, M.J.; Fischbach, M.A.; Sia, S.K.; Whitesides, G.M. Mixing with bubbles: a practical technology for use with portable microfluidic devices. Lab Chip 2006, 6, 207–212. [Google Scholar]
- Laser, D.J.; Santiago, J.G. A review of micropumps. J. Micromech. Microeng. 2004, 14, R35–R64. [Google Scholar]
- Kruger, J.; Singh, K.; O'Neill, A.; Jackson, C.; Morrison, A.; O'Brien, P. Development of a microfluidic device for fluorescence activated cell sorting. J. Micromech. Microeng. 2002, 12, 486–494. [Google Scholar]
- Cabrera, C.R.; Yager, P. Continuous concentration of bacteria in a microfluidic flow cell using electrokinetic techniques. Electrophoresis 2000, 22, 355–362. [Google Scholar]
- Cady, N.C.; Stelick, S.; Batt, C.A. Nucleic acid purification using microfabricated silicon structures. Biosens. Bioelectron. 2003, 19, 59–66. [Google Scholar]
- Li, S.; Fozdar, D.; Ali, M.F.; Li, H.; Shao, D.; Vykoukal, D.M.; Vykoukal, J.; Floriano, P.N.; Olsen, M.; McDevitt, J.T.; Gascoyne, P.R.C.; Chen, S. A continuous-flow polymerase chain reaction microchip with regional velocity control. J. Microelectromech. Syst. 2006, 15, 223–236. [Google Scholar]
- Roxhed, N.; Rydholm, S.; Samuel, B.; Van der Wijngaart, W.; Griss, P.; Stemme, G. A compact, low-cost microliter-range liquid dispenser based on expandable microspheres. J. Micromech. Microeng. 2006, 16, 2740–2746. [Google Scholar]
- Cooney, C.G.; Towe, B.C. A thermopneumatic dispensing micropump. Sens. Actuat. A Phys. 2004, 116, 519–524. [Google Scholar]
- Zeng, S.; chen, C.H.; James, C.; Mikkelsen, J.; Santiago, J.G. Fabrication and characterization of electroosmotic micropumps. Sens. Actuat. B Chem. 2001, 79, 107–114. [Google Scholar]
- Machauf, A.; Nemirovsky, Y.; Dinnar, U. A membrane micropump electrostatically actuated across the working fluid. J. Micromech. Microeng. 2005, 15, 2309–2316. [Google Scholar]
- Sounart, T.L.; Michalske, T.A.; Zavadil, K.R. Frequency-dependent electrostatic acutation in microfluidic MEMS. J. Microelectromech. Syst. 2005, 14, 125–133. [Google Scholar]
- Kock, M.; Harris, N.; Evans, A.G.R.; White, N.M.; Brunnschweiler, A. A novel micromachined pump based on thick-film piezoelectric actuation. Sens. Actuat. A Phys. 1998, 70, 98–103. [Google Scholar]
- Yang, Z.; Matsumoto, S.; Goto, H.; Matsumoto, M.; Maeda, R. Ultrasonic micromixer for microfluidic systems. Sens. Actuat. A Phys. 2001, 93, 266–272. [Google Scholar]
- Graf, N.J.; Bowser, M.T. A soft-polymer piezoelectric bimorph cantilever-actuated persitaltic micropump. Lab Chip 2008, 8, 1664–1670. [Google Scholar]
- Lemoff, A.V.; Lee, A.P. An AC magnetohydrodynamic micropump. Sens. Actuat. B Chem. 2000, 63, 178–185. [Google Scholar]
- Rinderknecht, D.; Hickerson, A.I. A valveless micro impedance pump driven by electromagnetic actuation. J. Micromech. Microeng. 2005, 15, 861–866. [Google Scholar]
- Yamamata, C.; Lotto, C.; Al-Assaf, E.; Gijs, M.A.M. A PMMA valveless micropump using electromagnetic actuation. Microfluid. Nanofluid. 2005, 1, 197–207. [Google Scholar]
- Harmon, M.E.; Tang, M.; Frank, C.W. A microfluidic actuator based on thermoresponsive hydrogels. Polymer 2003, 44, 4547–4556. [Google Scholar]
- Bassetti, M.J.; Chatterjee, A.N.; Aluru, N.R.; Beebe, D.J. Development and modeling of electrically triggered hydrogels for microfluidic applications. J. Microelectromech. Syst. 2005, 14, 1198–1207. [Google Scholar]
- Bohm, S.; Timmer, B.; Olthuis, W.; Bergveld, P. A closed-loop controlled electrochemically actuated micro-dosing system. J. Micromech. Microeng. 2000, 10, 498–504. [Google Scholar]
- Metref, L.; Herrera, F.; Berdat, D.; Gijs, M.A.M. Contactless Electrochemical Actuator for Microfluidic Dosing. J. Microelectromech. Syst. 2007, 16, 885–892. [Google Scholar]
- Squires, T.M.; Quake, S.R. Microfluidics: Fluid physics at the nanoliter scale. Rev. Mod. Phys. 2005, 77, 977–1026. [Google Scholar]
- Yu, Q.; Bauer, J.M.; Moore, J.S.; Beebe, D.J. Responsive biomimetic hydrogel valve for microfluidics. Appl. Phys. Lett. 2001, 78, 2589–2591. [Google Scholar]
- Studer, V.; Hang, G.; Pandolfi, A.; Ortiz, M.; Anderson, W.F.; Quake, S.R. Scaling properties of a low-actuation pressure microfluidic valve. J. Appl. Phys. 2004, 95, 393–398. [Google Scholar]
- Selvaganapathy, P.; Carlen, E.T.; Mastangelo, C.H. Electrothermally actuated inline microfluidic valve. Sens. Actuat. A Phys. 2003, 104, 275–282. [Google Scholar]
- Grover, W.H.; Skelley, A.M.; Liu, C.N.; Lagally, E.T.; Mathies, R.A. Monolithic membrane valves and diaphram pumps for practical large-scale integration into glass microfluidic devices. Sens. Actuat. B Chem. 2003, 89, 315–323. [Google Scholar]
- Liu, R.H.; Bonanno, J.; Yang, J.; Lenigk, R.; Grodzinski, P. Single-use, thermally actuated paraffin valves for microfluidic applications. Sens. Actuat. B Chem. 2004, 98, 328–336. [Google Scholar]
- Feng, Y.; Zhou, Z.; Ye, X.; Xiong, J. Passive valves based on hydrophobic microfluidics. Sens. Actuat. A Phys. 2003, 108, 138–143. [Google Scholar]
- Hua, S.Z.; Sachs, F.; Yang, D.X.; Chopra, H.D. Microfluidic Actuation Using Electrochemically Generated Bubbles. Anal. Chem. 2002, 74, 6392–6396. [Google Scholar]
- Stoeber, B.; Yang, Z.; Liepmann, D.; Muller, S.J. Flow control in microdevices using thermally responsive triblock copolymers. J. Microelectromech. Syst. 2005, 14, 207–213. [Google Scholar]
- Lu, L.H.; Ryu, K.S.; Liu, C. A magnetic microstirrer and array for microfluidic mixing. J. Microelectromech. Syst. 2002, 11, 462–469. [Google Scholar]
- Ryu, K.S.; Shaikh, K.; Goluch, E.; Fan, Z.; Liu, C. Micro magnetic stir-bar mixer integrated with parylene microfluidic channels. Lab Chip 2004, 4, 604–613. [Google Scholar]
- Tsai, J.H.; Lin, L. Active microfluidic mixer and gas bubble filter driven by thermal bubble micropump. Sens. Actuat. A Phys. 2002, 97-98, 665–671. [Google Scholar]
- Suzuki, H.; Ho, C.M.; Kasagi, N. A Chaotic Mixer for Magnetic Bead-Based Micro Cell Sorter. J. Microelectromech. Syst. 2004, 13, 779–790. [Google Scholar]
- Stroock, A.D.; Dertinger, S.K.W.; Ajdari, A.; Mezic, I.; Stone, H.A.; Whitesides, G.M. Chaotic Mixer for Microchannels. Science 2002, 295, 647–651. [Google Scholar]
- Hong, C.C.; Choi, J.W.; Ahn, C.H. A novel in-plane passive microfluidic mixer with modified Tesla structures. Lab Chip 2004, 4, 109–113. [Google Scholar]
- Lin, Y.C.; Chung, Y.C.; Wu, C.Y. Mixing enhancement of the passive microfluidic mixer with J-shaped baffles in the tee channel. Biomed. Microdevices 2007, 9, 215–221. [Google Scholar]
- Jacobson, S.C.; McKnight, T.E.; Ramsey, J.M. Microfluidic devices for electrokinetically driven parallel and serial mixing. Anal. Chem. 1999, 71, 4455–4459. [Google Scholar]
- Song, S.; Singh, A.K. On-chip sample preconcentration for integrated microfluidic analysis. Anal. Bioanal. Chem. 2005, 38, 41–43. [Google Scholar]
- Han, K.H.; Frazier, A.B. Continuous magnetophoretic separation of blood cells in microdevice format. J. Appl. Phys. 2004, 96, 5797. [Google Scholar]
- Inglis, D.W.; Riehn, R.; Austin, R.H.; Sturm, J.C. Continuous microfluidic immunomagnetic cell separation. Appl. Phys. Lett. 2004, 85, 5093–5095. [Google Scholar]
- Lee, H.; Purdon, A.M.; Westervelt, R.M. Manipulation of biological cells using a microelectromagnet matrix. Appl. Phys. Lett. 2004, 85, 1063–1065. [Google Scholar]
- Grodzinski, P.; Yang, J.; Liu, R.H.; Ward, M.D. A Modular Microfluidic System for Cell Pre-concentration and Genetic Sample Preparation. Biomed. Microdev. 2003, 5, 303–310. [Google Scholar]
- Cui, L.; Zhang, T.; Morgan, H. Optical particle detection integrated in a dielectrophoretic lab-on-a-chip. J. Micromech. Microeng. 2002, 12, 7–12. [Google Scholar]
- Huang, Y.; Rubinsky, B. Flow-through micro-electroporation chip for high efficiency single-cell genetic manipulation. Sens. Actuat. A Phys. 2003, 104, 205–212. [Google Scholar]
- Li, H.; Bashir, R. On the Design and Optimization of Micro-Fluidic Dielectrophoretic Devices: A Dynamic Simulation Study. Biomed. Microdev. 2004, 6, 289–295. [Google Scholar]
- Cui, L.; Holmes, D.; Morgan, H. The dielectrophoretic levitation and separation of latex beads in microchips. Electrophoresis 2001, 22, 3893–3901. [Google Scholar]
- Zhu, L.; Zhang, Q.; Feng, H.; Ang, S.; Chau, F.S.; Liu, W.-T. Filter-based microfluidic device as a platform for immunofluorescent assay of microbial cells. Lab Chip 2004, 4, 337–341. [Google Scholar]
- Khademhosseini, A.; Yeh, J.; Eng, S.J.G.; Suh, K.Y.; Burdick, J.A.; Langer, R. Molded polyethylene glycol microstructures for capturing cells within microfluidic channels. Lab Chip 2004, 4, 425–430. [Google Scholar]
- Tani, H.; Maehana, K.; Kamidate, T. Chip-based bioassay using bacterial sensor strians immobilized in three-dimensional microfluidic network. Anal. Chem. 2004, 76, 6693–6697. [Google Scholar]
- Chang, W.C.; Lee, L.P.; Liepmann, D. Biomimetic technique for adhesion-based collection and separation of cels in a microfluidic channel. Lab Chip 2005, 5, 64–73. [Google Scholar]
- Chen, T.; Small, D.A.; McDermott, M.K.; Bentley, W.E.; Payne, G.F. Enzymatic Methods for in situ Cell Entrapment and Cell Release. Biomacromolecules 2003, 4, 1558–1563. [Google Scholar]
- Irimia, D.; Tompkins, R.G.; Toner, M. Single-cell chemical lysis in picoliter-scale closed volumes using a microfabricated device. Anal. Chem. 2004, 76, 6137–6143. [Google Scholar]
- Carlo, D.D.; Ionescu-Zanetta, C.; Zhang, Y.; Hung, P.; Lee, L.P. On-chip cell lysis by local hydroxide generation. Lab Chip 2005, 5, 171–178. [Google Scholar]
- Huang, Y.; Maher, E.L.; Bell, J.L.; Madou, M. MEMS-based sample preparation for molecular diagnostics. Anal. Bioanal. Chem. 2002, 372, 49–65. [Google Scholar]
- Griffiths, L.J.; Anyim, M.; Doffman, S.R.; Wilks, M.; Millar, M.R.; Agrawal, S.G. Comparison of DNA extraction methods for Aspergillus fumigatus using real-time PCR. J. Med. Microb. 2006, 55, 1187–1191. [Google Scholar]
- Lee, J.G.; Cheong, K.H.; Huh, N.; Kim, S.; Choi, J.W.; Ko, C. Microchip-based one step DNA extraction and real-time PCR in one chamber for rapid pathogen identification. Lab Chip 2006, 6, 886–895. [Google Scholar]
- Lu, H.; Schmitdt, M.A.; Jensen, K.F. A microfluidic electroporation device for cell lysis. Lab Chip 2005, 5, 23–29. [Google Scholar]
- Wang, H.-Y.; Bhunia, A.K.; Lu, C. A microfluidic flow-through device for high throughput electrical lysis of bacterial cells based on continuous dc voltage. Biosens. Bioelectron. 2006, 22, 582–588. [Google Scholar]
- Fox, M.B.; Esveld, D.C.; Valero, A.; Luttge, R.; Mastwijk, H.C.; Bartels, P.V.; Van den Berg, A.; Boom, R.M. Electroporation of cells in microfluidic devices: a review. Anal. Bioanal. Chem. 2006, 385, 474–485. [Google Scholar]
- Higgins, J.A.; Nasarabadi, S.; Karns, J.S.; Shelton, D.R.; Cooper, M.; Gbakima, A.; Koopman, R.P. A handheld real time thermal cycler for bacterial pathogen detection. Biosens. Bioelectron. 2003, 18, 1115–1123. [Google Scholar]
- Koh, C.G.; Tan, W.; Zhao, M.Q.; Ricco, A.J.; Fan, Z.H. Integrating polymerase chain reaction, valving, and electrophoresis in a plastic device for bacterial detection. Anal. Chem. 2003, 75, 4591–4598. [Google Scholar]
- Liu, J.; Enzelberger, M.; Quake, S. A nanoliter rotary device for polymerase chain reaction. Electrophoresis 2002, 23, 1531–1536. [Google Scholar]
- Saiki, R.K.; Scharf, S.; Faloona, F.; Mullis, K.B.; Horn, G.T.; Erlich, H.A.; Arnheim, N. Enzymatic amplification of betaglobin genomic sequences and restruction site analysis for diagnosis of sickle cell anemia. Science 1985, 230, 1350–1354. [Google Scholar]
- Northrup, M.A.; Ching, M.T.; White, R.M.; Watson, R.T. DNA amplification in a microfabricated reaction chamber. Proceedings of the 7th International Conference on Solid-State Sensors and Actuators (Transducers '93), Yokohama, Japan; 1993; pp. 924–926. [Google Scholar]
- Giordano, B.C.; Ferrance, J.; Swedberg, S.; Huhmer, A.F.R.; Landers, J.P. Polymerase chain reaction in polymeric microchips: DNA amplification in less than 240 seconds. Anal. Biochem. 2001, 291, 124–132. [Google Scholar]
- Lin, Y.C.; Huang, M.Y.; Young, K.C.; Chang, T.T.; Wu, C.Y. A rapid micro-polymerase chain reaction system for hepatitis C virus amplification. Sens. Actuat. B Chem. 2000, 71, 2–8. [Google Scholar]
- Schneegass, I.; Kohler, J.M. Flow-through polymerase chain reactions in chip thermocyclers. Mol. Biotechnol. 2001, 82, 101–121. [Google Scholar]
- Sun, K.; Yamaguchi, A.; Ishida, Y.; Matsuo, S.; Misawa, H. A heater-integrated transparent microchannel chip for continuous-flow PCR. Sens. Actuat. B Chem. 2002, 84, 283–289. [Google Scholar]
- Ottesen, E.A.; Hong, J.W.; Quake, S.R.; Leadbetter, J.R. Microfluidic Digital PCR Enables Multigene Analysis of Inidividual Environmental Bacteria. Science 2006, 314, 1464–1467. [Google Scholar]
- Marcus, J.S.; Anderson, W.F.; Quake, S.R. Parallel Picoliter RT-PCR Assays Using Microfluidics. Anal. Chem. 2006, 78, 956–958. [Google Scholar]
- Lagally, E.T.; Emrich, C.A.; Mathies, R.A. Fully integrated PCR-capillary electrophoresis microsystem for DNA analysis. Lab Chip 2001, 1, 102–107. [Google Scholar]
- Waggoner, A. Fluorescent labels for proteomics and genomics. Curr. Opin. Chem. Biol. 2006, 10, 62–66. [Google Scholar]
- LePecq, J.B.; Paoletti, C. A fluorescent complex between ethidium bromide and nucleic acids. Physical-chemical characterization. J. Mol. Biol. 1967, 27, 87–106. [Google Scholar]
- Rye, H.S.; Dabora, J.M.; Quesada, M.A.; Mathies, R.A.; Glazer, A.N. Fluorometrix assay using dimeric dyes for double and single stranded DNA and RNA with picogram sensitivity. Anal. Biochem. 1993, 208, 144–150. [Google Scholar]
- Rengarajan, K.; Cristol, S.M.; Mehta, M.; Nickerson, J.M. Quantifying DNA concentrations using fluorometry: a comparison of fluorophores. Mol. Vis. 2002, 8, 416–421. [Google Scholar]
- Mackay, J.; Landt, O. Real-time PCR fluorescent chemistries. In Protocols for Nucleic Acid Analysis by Nonradioactive Probes, 2nd Ed.; Hilario, E., Mackay, J., Eds.; Humana Press: Clifton, NJ, USA, 2007; Volume 353, pp. 237–261. [Google Scholar]
- Taylor, J.R.; Fang, M.M.; Nie, S. Probing specific sequences on single DNA moleucles with bioconjugated fluorescent nanoparticles. Anal. Chem. 2000, 72, 1979–1986. [Google Scholar]
- Cady, N.C.; Strickland, A.D.; Batt, C.A. Optimized linkage and quenching strategies for quantum dot molecular beacons. Mol. Cell. Probes 2006, 21, 116–124. [Google Scholar]
- Hong, J.W.; Fujui, T.; Seki, M.; Yamamoto, T.; Endo, I. Integration of gene amplification and capillary gel electrophoresis on a polydimethylsiloxane-glass hybrid microchip. Electrophoresis 2001, 22, 328–333. [Google Scholar]
- Dasgupta, P.K.; Eom, I.; Morris, K.J.; Li, J. Light emitting diode-based detectors absorbance, fluorescence, and spectroelectrochemical measurements in a planar flow-through cell. Anal. Chim. Acta 2003, 500, 337–364. [Google Scholar]
- Belgrader, P.; Young, S.; Yuan, B.; Primeau, M.; Pourahmadi, L.A.C.F.; Northrup, M.A. A Battery-Powered Notebook Thermal Cycler for Rapid Multiplex Real-Time PCR Analysis. Anal. Chem. 2001, 73, 286–289. [Google Scholar]
- Paegel, B.M.; Blazej, R.G.; Mathies, R.A. Microfluidic devices for DNA sequencing: sample preparation and electrophoretic analysis. Curr. Opin. Chem. Biol. 2003, 14, 42–50. [Google Scholar]
- Brockman, J.M.; Frutos, A.G.; Corn, R.M. A multistep chemical modification procedure to create DNA arrays on gold surfaces for the study of protein-DNA interactions with surface plasmon resonance imaging. J. Am. Chem. Soc. 1999, 121, 8044–8051. [Google Scholar]
- Geudon, P.; Livache, T.; Martin, F.; Lesbre, F.; Roget, A.; Bidan, G.; Levy, Y. Characterization and optimization of a real-time, parallel, label-free, polypyrrole-based DNA sensor by surface plasmon resonance imaging. Appl. Chem. 2000, 72, 6003–6009. [Google Scholar]
- Lehr, H.P.; Reimann, M.; Brandenburg, A.; Sulz, G.; Klapproth, H. Real-time detection of nucleic acid interactions by total internal reflection fluorescence. Anal. Chem. 2003, 75, 2414–2420. [Google Scholar]
- McDonnell, J.M. Surface plasmon resonance: towards an understanding of the mechanisms of biological molecular recognition. Curr. Opin. Chem. Biol. 2001, 5, 572–577. [Google Scholar]
- Lesuffleur, A.; Im, H.; Lindquist, N.C.; Lim, K.S.; Oh, S.-H. Laser-illuminated nanohole arrays for multiplex plasmonic microarray sensing. Opt. Express 2008, 16, 219–224. [Google Scholar]
- Usui-Aoki, K.; Shimada, K.; Nagano, M.; Kawai, M.; Koga, H. A novel approach to protein expression profiling using antibody microarrays combined with surface plasmon resonance technology. Proteomics 2005, 5, 2396–2401. [Google Scholar]
- Cady, N.C. Nucleic acid-based sensing; Department of Microbiology, Cornell University: Ithaca, NY, USA, 2006. [Google Scholar]
- Cao, Y.C.; Jin, R.; Mirkin, C.A. Nanoparticles with Raman Spectroscopic Fingerprints for DNA and RNA Detection. Science 2002, 297, 1536–1540. [Google Scholar]
- Fabris, L.; Dante, M.; Braun, G.; lee, S.J.; Reich, N.O.; Moskovits, M.; Nguyen, T.Q.; Bazan, G.C. A heterogeneous PNA-based SERS method for DNA detection. J. Am. Chem. Soc. 2007, 129, 6086–6087. [Google Scholar]
- Docherty, F.T.; Monaghan, P.B.; Keir, R.; Graham, D. The first SERRS multiplexing from labelled oligonucleotides in a microfluidics lab-on-a-chip. Chem. Commun. 2004, 118–119. [Google Scholar]
- Erdemm, A.; Pividori, M.I.; Lermo, A.; Bonanni, A.; Del Valle, M.; Alegret, S. Genomagnetic assay based on label-free electrochemical detection using magneto-composite electrodes. Sens. Actuat. B Chem. 2005, 114, 591–598. [Google Scholar]
- Berney, H.; West, J.; Haefele, E.; Alderman, J.; Lane, W.; Collins, J.K. A DNA diagnostic biosensor: development, characterisation and performance. Sens. Actuat. B Chem. 2000, 68, 100–108. [Google Scholar]
- Wang, J.; Rivas, G.; Cai, X. Screen-printed electrochemcial hybridization biosensor for the detection of DNA sequences from the Escherichia coli pathogen. Electroanalysis 2005, 9, 395–398. [Google Scholar]
- Pingarron, J.M.; Yanez-Sedeno, P.; Gonzalez-Cortes, A. Gold nanoparticle-based electrochemical biosensors. Electrochim. Acta 2008, 53, 5848–5866. [Google Scholar]
- Guo, S.; Wang, E. Synthesis and electrochemical applications of gold nanoparticles. Anal. Chim. Acta 2007, 598, 181–192. [Google Scholar]
- Castaneda, M.T.; Merkoci, A.; Pumera, M.; Alegret, S. Electrochemical genosensors for biomedical applications based on gold nanoparticles. Biosens. Bioelectron. 2006, 22, 1961–1967. [Google Scholar]
- Cheng, G.; Zhao, J.; Tu, Y.; He, P.; Fang, Y. A sensitive DNA electrochemical biosensor based on magnetite with a glassy carbon electrode modified by multi-walled carbon nanotubes in polypyrrole. Anal. Chim. Acta 2005, 533, 11–16. [Google Scholar]
- Eggins, B.R. Chemical Sensors and Biosensors; John Wiley & Sons Ltd.: Hoboken, NJ, USA, 2002. [Google Scholar]
- Abdel-Hamid, I.; Ivnitski, D.; Atanasov, P.; Wilkins, E. Flow-through immunofiltration assay system for rapid detection of E. coli O157:H7. Biosens. Bioelectron. 1999, 14, 309–316. [Google Scholar]
- Baeumner, A.J.; Cohen, R.N.; Miksic, V.; Min, J. RNA biosensor for the rapid detection of viable Escherichia coli in drinking water. Biosens. Bioelectron. 2002, 18, 405–413. [Google Scholar]
- Bergveld, P. Thirty years of ISFETOLOGY: What happened in the past 30 years and what may happen in the next 30 years. Sens. Actuat. B Chem. 2003, 88, 1–20. [Google Scholar]
- Alocilja, E.C.; Radke, S.M. Market analysis of biosensors for food safety. Biosens. Bioelectron. 2003, 18, 841–846. [Google Scholar]
- Barssoukov, E.; Macdonald, J.R. Impedance Spectroscopy Theory, Experiment and Applications; John Wiley & Sons Ltd: Hoboken, NJ, USA, 2005. [Google Scholar]
- Radke, S.M.; Alocilja, E.C. A high density microelectrode array biosensor for detection of E. coli O157:H7. Biosens. Bioelectron. 2005, 20, 1662–1667. [Google Scholar]
- Muhammad-Tahir, Z.; Alocilja, E.C. A conductometric biosensor for biosecurity. Biosens. Bioelectron. 2003, 18, 813–819. [Google Scholar]
- Pal, S.; Alocilja, E.C.; Downes, F.P. Nanowire labeled direct-charge transfer biosensor for detection Bacillus species. Biosens. Bioelectron. 2007, 22, 2329–2336. [Google Scholar]
- Gheorghe, M.; Guiseppi-Elie, A. Electrical frequency dependent characterization of DNA hybridization. Biosens. Bioelectron. 2003, 19, 95–102. [Google Scholar]
- Hang, T.C.; Guiseppi-Elie, A. Frequency dependent and surface characterization of DNA immobilization and hybridization. Biosens. Bioelectron. 2004, 19, 1537–1548. [Google Scholar]
- Tombelli, S.; Mascini, M.; Sacco, M.; Turner, A.P. A DNA piezoelectric biosensor assay coupled with a PCR for bacterial toxicity determination in environmental samples. Anal. Chim. Acta 2000, 418, 1–9. [Google Scholar]
- Liu, T.; Tang, J.; Jiang, L. Sensitivity enhancement of DNA sensors by nanogold surface modification. Biochem. Biophys. Res. Commun. 2002, 295, 14–16. [Google Scholar]
- Fritz, J.; Baller, M.K.; Lang, H.P.; Rothuizen, H.; Vettiger, P.; Meyer, E.; Guntherodt, H.; Gerber, C.; Gimzewski, J.K. Translating biomolecular recognition into nanomechanics. Science 2000, 288, 316–318. [Google Scholar]
- Liu, F.; Zhang, Y.; Ou-Yang, Z. Flexoelectric origin of nanomechanic deflection in DNA-microcantilever system. Biosens. Bioelectron. 2003, 18, 655–660. [Google Scholar]
- Hansen, K.M.; Ji, F.; Wi, G.; Datar, R.; Cote, R.; Majumdar, A.; Thundat, T. Cantilever-based optical deflection assay for discrimination of DNA single-nucleotide mismatches. Anal. Chem. 2001, 73, 1567–1571. [Google Scholar]
- Koh, C.G.; Tan, W.; Zhao, M.-q.; Ricco, A.J.; Fan, Z.H. Integrating polymerase chain reaction, valving, and electrophoresis in a plastic device for bacterial detection. Anal. Chem. 2003, 75, 4591–4598. [Google Scholar]
- Hong, J.W.; Studer, V.; Hang, G.; Anderson, W.F.; Quake, S.R. A nanoliter-scale nucleic acid processor with parallel architecture. Nat. Biotechnol. 2004, 22, 435–439. [Google Scholar]
- Taylor, M.T.; Belgrader, P.; Joshi, R.; Kintz, G.A.; Northrup, M.A. Fully automated sample preparation for pathogen detection performed in a microfluidic cassette. In Proceedings of the Micro Total Analysis Systems.; Monterey, California, USA, 2001; Kluwer: Boston, MA, USA, 2001; p. 670. [Google Scholar]
- Liu, R.H.; Yang, J.; Lenigk, R.; Bonanno, J.; Grodzinski, P. Self-contained, fully integrated biochip for sample preparation, polymerase chain reaction amplification, and DNA microarray detection. Anal. Chem. 2004, 76, 1824–1831. [Google Scholar]
- Yeung, S.S.W.; Lee, T.M.H.; Hsing, I.M. Electrochemistry-Based Real-Time PCR on a Microchip. Anal. Chem. 2008, 80, 363–368. [Google Scholar]
- Yeung, S.W.; Lee, T.M.H.; Cai, H.; Hsing, I.M. A DNA biochip for on-the-spot multiplexed pathogen identification. Nucleic Acids Res. 2006, 34, e118. [Google Scholar]
- Eckersten, A.; Orlefors, A.E.; Ellstrom, C.; Erickson, K.; Lofman, E.; Eriksson, A.; Eriksson, S.; Jorsback, A. High-throughput SNP scoring in a disposable microfabricated CD device. In Micro Total Analysis Systems (muTAS 2000); Kluwer: Enschede, the Netherlands, 2000; p. 521. [Google Scholar]
- Lagally, E.T.; Scherer, J.R.; Blazej, R.G.; Toriello, N.M.; Diep, B.A.; Ramchandani, M.; Sensabaugh, G.F.; Riley, L.W.; Mathies, R.A. Integrated portable genetic analysis microsystem for pathogen/infectious disease detection. Anal. Chem. 2004, 76, 3162–3170. [Google Scholar]
- Blazej, R.G.; Kumaresan, P.; Mathies, R.A. Microfabricated bioprocessor for integrated nanoliter-scale Sanger DNA sequencing. Proc. Natl. Acad. Sci. USA 2006, 103, 7240–7245. [Google Scholar]
- Holland, C.A.; Kiechle, F.L. Point-of-care molecular diagnostic systems - past, present, and future. Curr. Opin. Microbiol 2005, 8, 504–509. [Google Scholar]
- Ulrich, M.P.; Christensen, D.R.; Coyne, S.R.; Craw, P.D.; Henchal, E.A.; Sakai, S.H.; Swenson, D.; Tholath, J.; Tsai, J.; Weir, A.F.; Norwood, D.A. Evaluation of the Cepheid GeneXpert® system for detecting Bacillus anthracis. J. Appl. Microbiol. 2006, 100, 1011–1016. [Google Scholar]
- Lee, T.M.-H.; Hsing, I.-M. DNA-based bioanalytical microsystems for handheld devices applications. Anal. Chim. Acta 2006, 556, 26–37. [Google Scholar]
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Lui, C.; Cady, N.C.; Batt, C.A. Nucleic Acid-based Detection of Bacterial Pathogens Using Integrated Microfluidic Platform Systems. Sensors 2009, 9, 3713-3744. https://doi.org/10.3390/s90503713
Lui C, Cady NC, Batt CA. Nucleic Acid-based Detection of Bacterial Pathogens Using Integrated Microfluidic Platform Systems. Sensors. 2009; 9(5):3713-3744. https://doi.org/10.3390/s90503713
Chicago/Turabian StyleLui, Clarissa, Nathaniel C. Cady, and Carl A. Batt. 2009. "Nucleic Acid-based Detection of Bacterial Pathogens Using Integrated Microfluidic Platform Systems" Sensors 9, no. 5: 3713-3744. https://doi.org/10.3390/s90503713



