Electrochemical Interrogation of Interactions between Surface-Confined DNA and Methylene Blue
Abstract
:1. Introduction
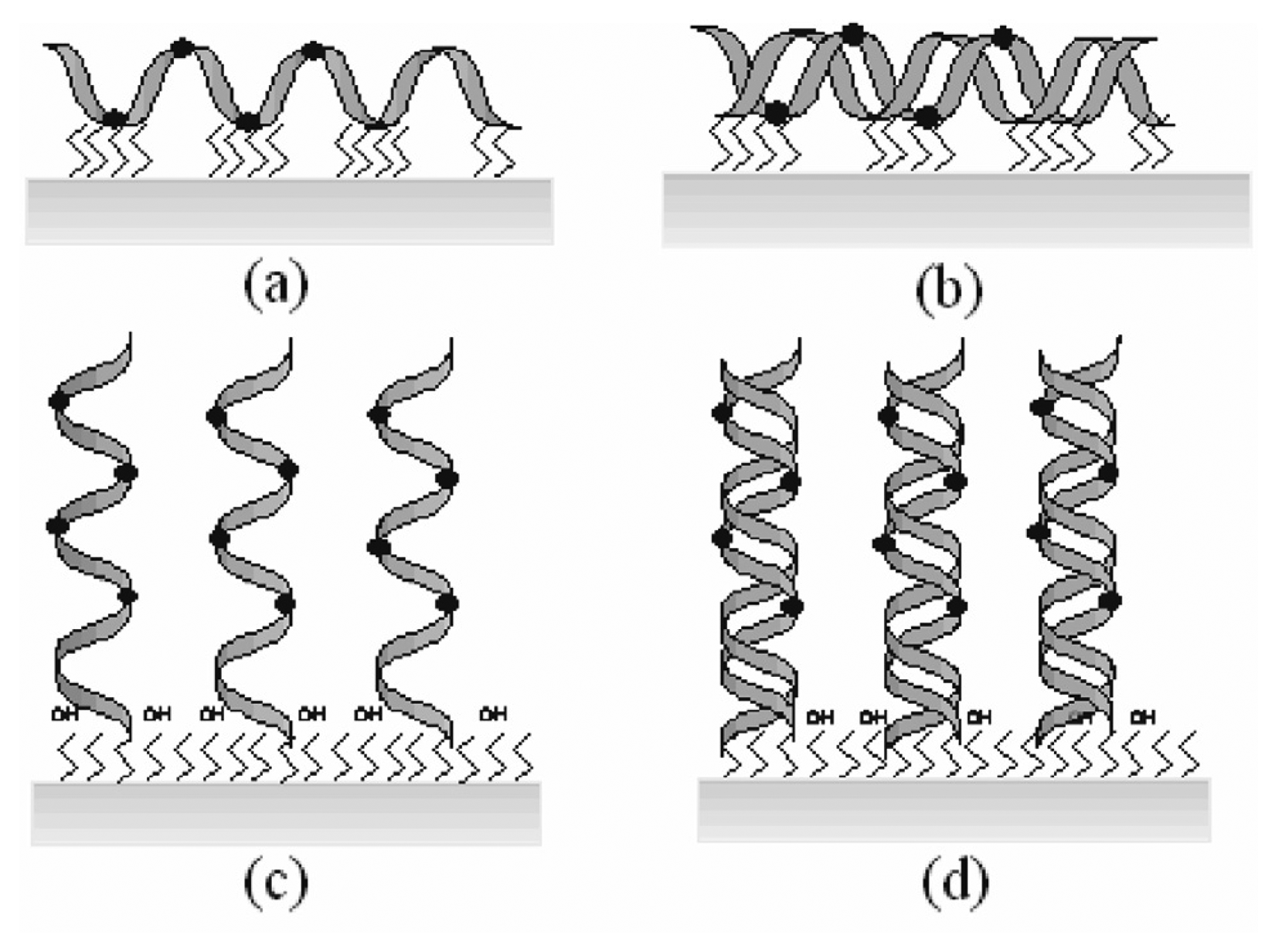
2. Results and Discussion
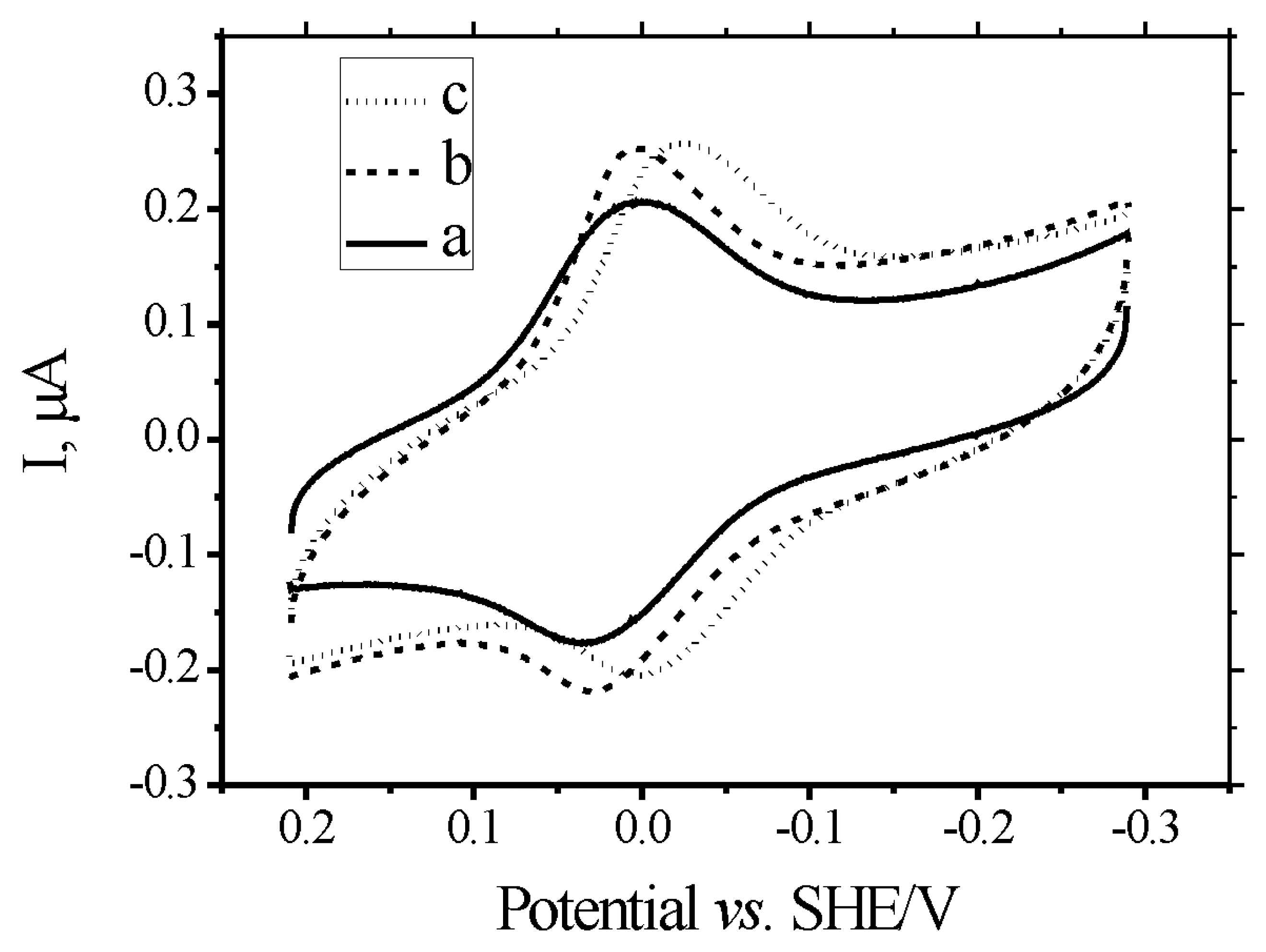
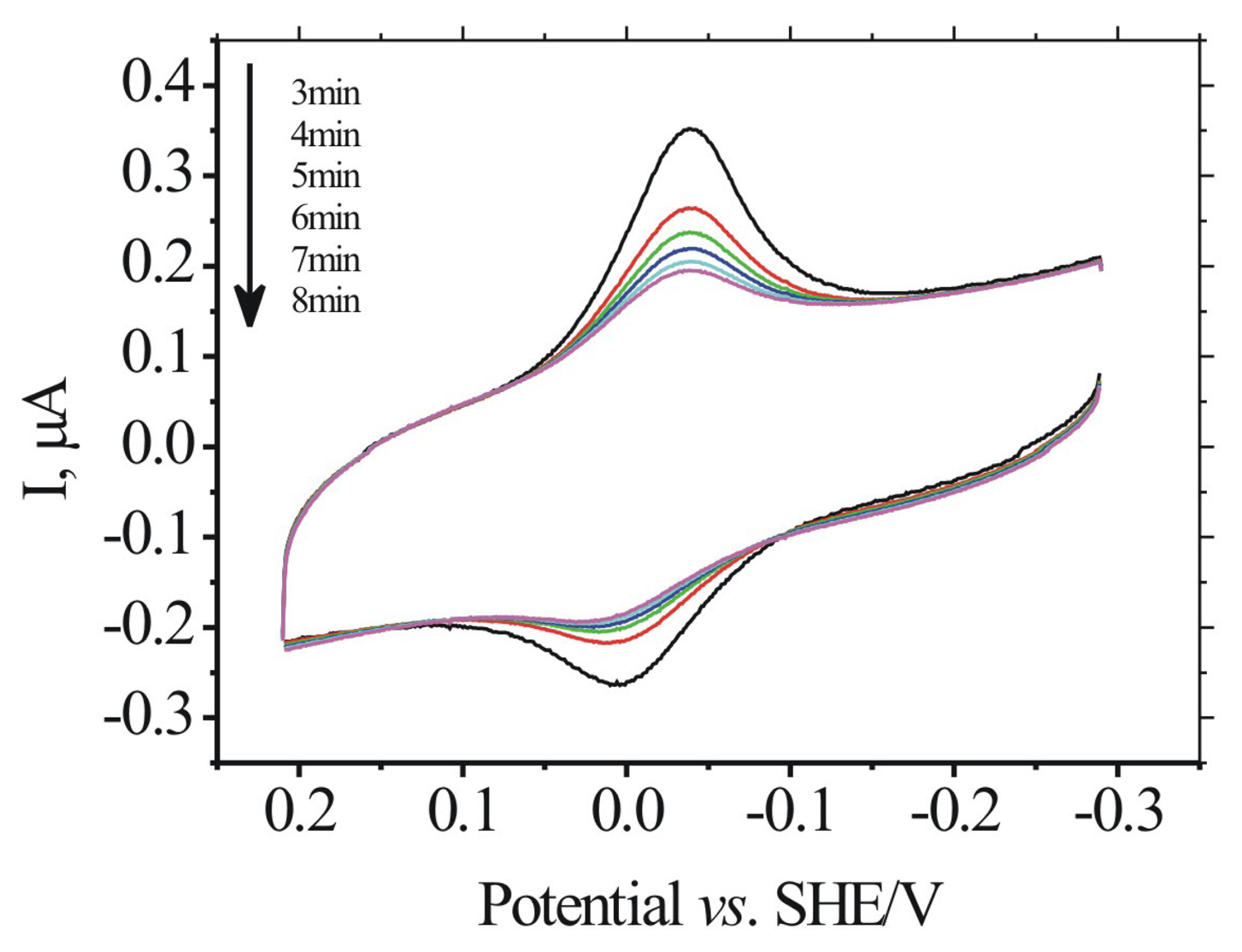
2.1. Electrochemistry of MB at DNA-modified gold electrodes
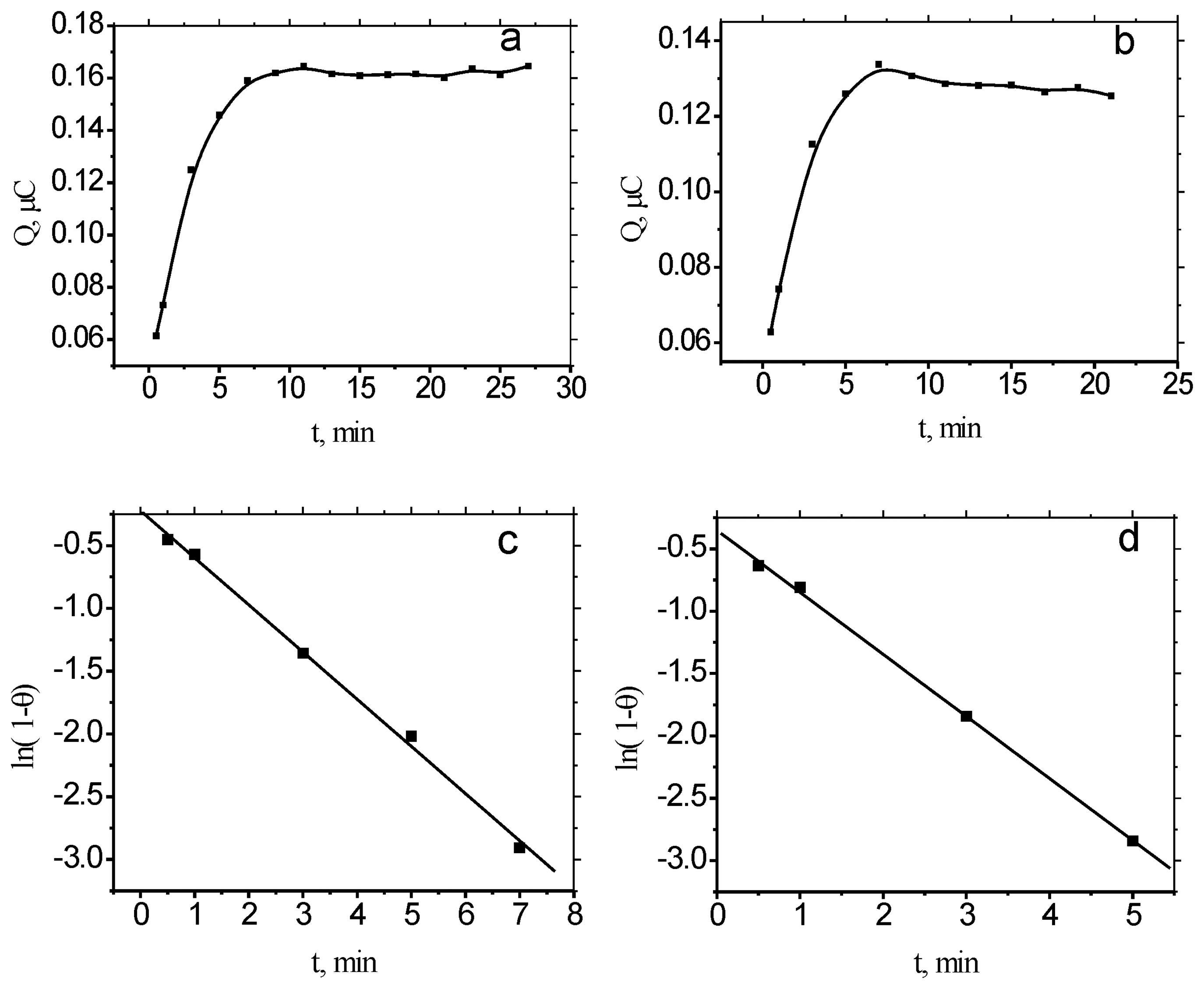
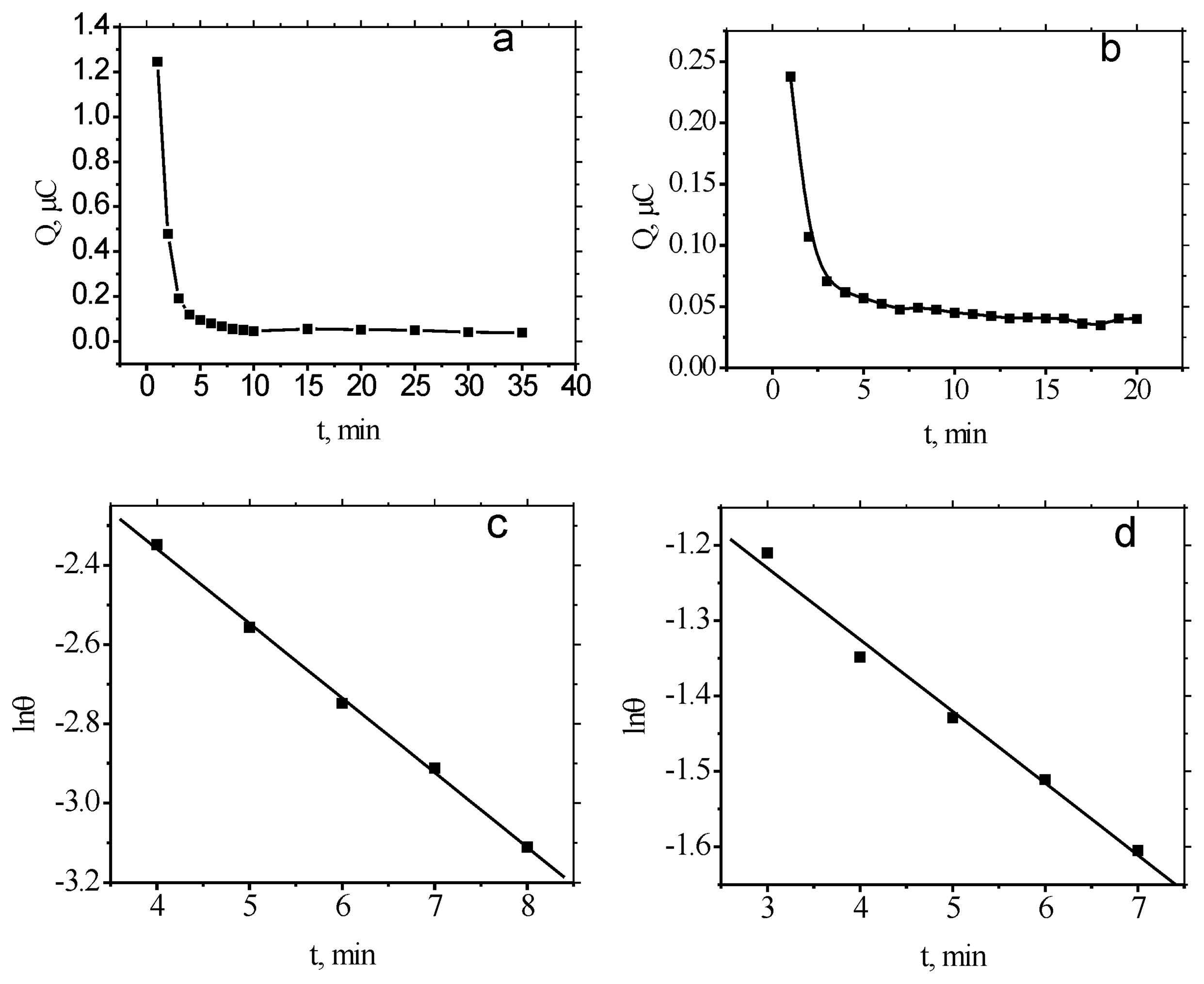
2.2. Binding equilibrium time of MB to ds-DNA/Au and ss-DNA/Au
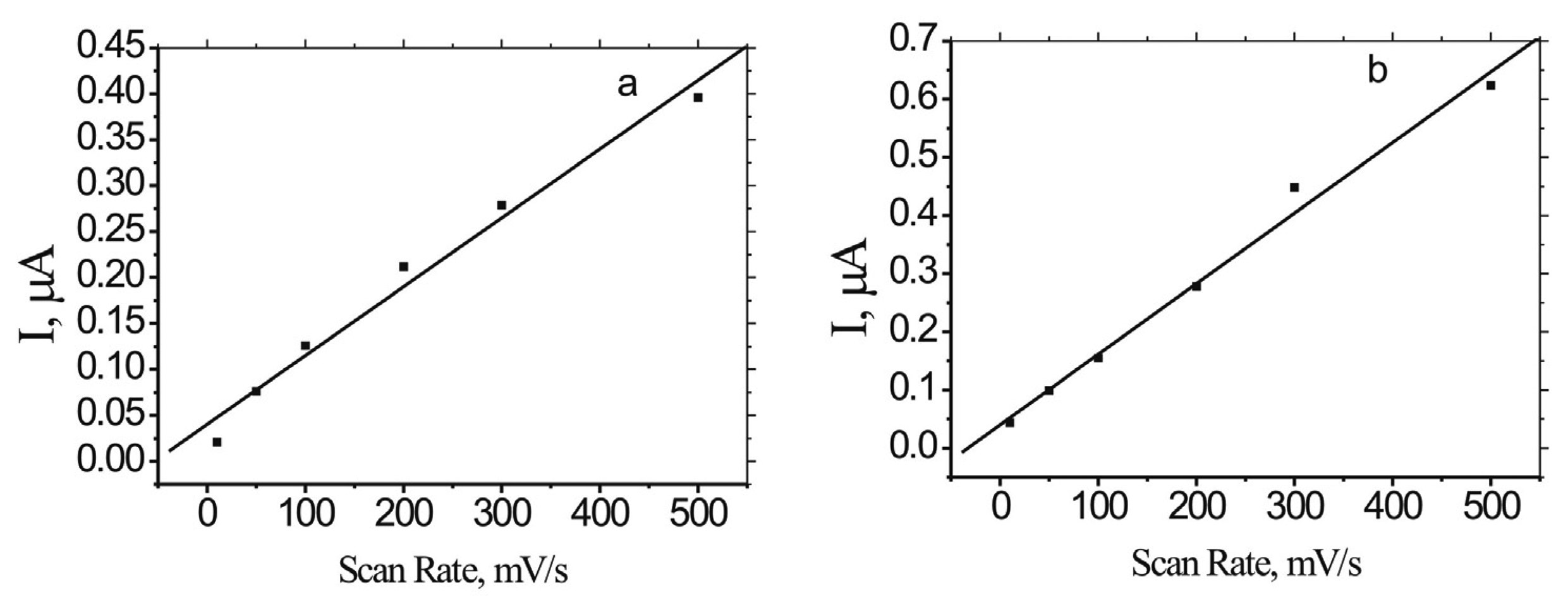
2.3. Binding kinetics of MB to ds-DNA/Au and ss-DNA/Au
2.4. Dissociation process of MB to ss-DNA/Au and ds-DNA/Au
3. Experimental Section
3.1. Materials
3.2. Pretreatment of Electrodes and DNA Immobilization
3.3. Electrochemical Measurements
3.4. Procedures of MB binding and dissociation
4. Conclusions
Acknowledgments
References
- Heller, M. J. DNA microarray technology: devices, systems, and applications. Annu. Rev. Biomed. Eng. 2002, 4, 129–153. [Google Scholar]
- Wang, J.; Liu, G.; Merkoci, A. Electrochemical coding technology for simultaneous detection of multiple DNA targets. J. Am. Chem. Soc. 2003, 125, 3214–3215. [Google Scholar]
- Fan, C.; Plaxco, K. W.; Heeger, A. J. Biosensors based on binding-modulated donor-acceptor distances. Trends Biotechnol. 2005, 23, 186–192. [Google Scholar]
- Zhang, J.; Song, S.; Zhang, L.; Wang, L.; Wu, H.; Pan, D.; Fan, C. Sequence-specific detection of femtomolar DNA via a chronocoulometric DNA sensor (CDS): effects of nanoparticle-mediated amplification and nanoscale control of DNA assembly at electrodes. J. Am. Chem. Soc. 2006, 128, 8575–8580. [Google Scholar]
- Whiteley, G. R. Proteomic patterns for cancer diagnosis - promise and challenges. Molecular Biosystems 2006, 2, 358–363. [Google Scholar]
- Okamoto, A.; Tainaka, K.; Ochi, Y.; Kanatani, K.; Saito, I. Simple SNP typing assay using a base-discriminating fluorescent probe. Molecular Biosystems 2006, 2, 122–126. [Google Scholar]
- Staiano, M.; Bazzicalupo, P.; Rossi, M.; D'Auria, S. Glucose biosensors as models for the development of advanced protein-based biosensors. Molecular Biosystems 2005, 1, 354–362. [Google Scholar]
- Usui, K.; Tomizaki, K.; Ohyama, T.; Nokihara, K.; Mihara, H. A novel peptide microarray for protein detection and analysis utilizing a dry peptide array system. Molecular Biosystems 2006, 2, 113–121. [Google Scholar]
- Workman, P. Genomics and the second golden era of cancer drug development. Molecular Biosystems 2005, 1, 17–26. [Google Scholar]
- Yang, J.; Wang, B. Q.; Rusling, J. F. Genotoxicity sensor response correlated with DNA nucleobase damage rates measured by LC-MS. Molecular Biosystems 2005, 1, 251–259. [Google Scholar]
- Pastor, J.; Siro, J. G.; Garcia-Navio, J. L.; Vaquero, J. J.; Alvarez-Builla, J.; Gago, F.; de Pascual-Teresa, B.; Pastor, M.; Rodrigo, M. M. Azino-Fused Benzimidazolium Salts as DNA Intercalating Agents. 2. J. Org. Chem. 1997, 62, 5476–5483. [Google Scholar]
- Cao, Y.; He, X. W. Studies of interaction between safranine T and double helix DNA by spectral methods. Spectrochim Acta A Mol. Biomol. Spectrosc. 1998, 54A, 883–892. [Google Scholar]
- Coates, C. G.; Jacquet, L.; McGarvey, J. J.; Bell, S. E. J.; Al-Obaidi, A. H. R.; Kelly, J. M. Resonance Raman Probing of the Interaction between Dipyridophenazine Complexes of Ru(II) and DNA. J. Am. Chem. Soc. 1997, 119, 7130–7136. [Google Scholar]
- Pasternack, R. F.; Bustamante, C.; Collings, P. J.; Giannetto, A.; Gibbs, E. J. Porphyrin assemblies on DNA as studied by a resonance light-scattering technique. J. Am. Chem. Soc. 1993, 115, 5393–5399. [Google Scholar]
- Pasternack, R. F.; Collings, P. J. Resonance light scattering: a new technique for studying chromophore aggregation. Science 1995, 269, 935–939. [Google Scholar]
- Huang, C. Z.; Li, Y. F.; Hu, X. L.; Li, N. B. Three-dimensional spectra of the long-range assembly of Nile Blue sulfate on the molecular surface of DNA and determination of DNA by light-scattering. Anal. Chim. Acta 1999, 395, 187–197. [Google Scholar]
- Yu, X.; Kim, S. N.; Papadimitrakopoulos, F.; Rusling, J. F. Protein immunosensor using single-wall carbon nanotube forests with electrochemical detection of enzyme labels. Molecular Biosystems 2005, 1, 70–78. [Google Scholar]
- Wang, J. Electroanalysis and Biosensors. Anal. Chem. 1999, 71, 328–332. [Google Scholar]
- Wang, J. From DNA biosensors to gene chips. Nucleic Acids Res. 2000, 28, 3011–3016. [Google Scholar]
- Ozkan, D.; Kara, P.; Kerman, K.; Meric, B.; Erdem, A.; Jelen, F.; Nielsen, P. E.; Ozsoz, M. DNA and PNA sensing on mercury and carbon electrodes by using methylene blue as an electrochemical label. Bioelectrochemistry 2002, 58, 119–126. [Google Scholar]
- Gu, J. Y.; Lu, X. J.; Ju, H. X. DNA sensor for recognition of native yeast DNA sequence with methylene blue as an electrochemical hybridization indicator. Electroanalysis 2002, 14, 949–954. [Google Scholar]
- Boon, E. M.; Barton, J. K. DNA electrochemistry as a probe of base pair stacking in A-, B-, and Z-form DNA. Bioconjug. Chem. 2003, 14, 1140–1147. [Google Scholar]
- Steel, A. B.; Herne, T. M.; Tarlov, M. J. Electrochemical quantitation of DNA immobilized on gold. Anal. Chem. 1998, 70, 4670–4677. [Google Scholar]
- Lao, R.; Song, S.; Wu, H.; Wang, L.; Zhang, Z.; He, L.; Fan, C. Electrochemical interrogation of DNA monolayers on gold surfaces. Anal. Chem. 2005, 77, 6475–6480. [Google Scholar]
- Tani, A.; Thomson, A. J.; Butt, J. N. Methylene blue as an electrochemical discriminator of single- and double-stranded oligonucleotides immobilised on gold substrates. Analyst 2001, 126, 1756–1759. [Google Scholar]
- Kelley, S. O.; Boon, E. M.; Barton, J. K.; Jackson, N. M.; Hill, M. G. Single-base mismatch detection based on charge transduction through DNA. Nucleic Acids Res. 1999, 27, 4830–4837. [Google Scholar]
- Liu, T.; Barton, J. K. DNA electrochemistry through the base pairs not the sugar-phosphate backbone. J. Am. Chem. Soc. 2005, 127, 10160–10161. [Google Scholar]
- Kelley, S. O.; Barton, J. K.; Jackson, N. M.; Hill, M. G. Electrochemistry of methylene blue bound to a DNA-modified electrode. Bioconjug. Chem. 1997, 8, 31–37. [Google Scholar]
- Pang, D. W.; Abruna, H. D. Interactions of Benzyl Viologen with Surface-Bound Single- and Double-Stranded DNA. Anal. Chem. 2000, 72, 4700–4706. [Google Scholar]
- Yu, H. Z.; Luo, C. Y.; Sankar, C. G.; Sen, D. Voltammetric procedure for examining DNA-modified surfaces: quantitation, cationic binding activity, and electron-transfer kinetics. Anal. Chem. 2003, 75, 3902–3907. [Google Scholar]
- Fan, C.; Zhong, J.; Guan, R.; Li, G. Direct electrochemical characterization of Vitreoscilla sp. hemoglobin entrapped in organic films. Biochim. Biophys. Acta 2003, 1649, 123–126. [Google Scholar]
- Hoogvliet, J. C.; Dijksma, M.; Kamp, B.; van Bennekom, W. P. Electrochemical pretreatment of polycrystalline gold electrodes to produce a reproducible surface roughness for self-assembly: a study in phosphate buffer pH 7.4. Anal. Chem. 2000, 72, 2016–2021. [Google Scholar]
- Janeak, R. P.; Fawcett, W. R.; Ulman, A. J. Impedance Spectroscopy of Self-Assembled Monolayers on Au(111): Evidence for Complex Double-Layer Structure in Aqueous NaClO4 at the Potential of Zero Charge. J. Phys. Chem. B 1997, 101, 8550–8558. [Google Scholar]
© 2007 by MDPI ( http://www.mdpi.org). Reproduction is permitted for noncommercial purposes.
Share and Cite
Pan, D.; Zuo, X.; Wan, Y.; Wang, L.; Zhang, J.; Song, S.; Fan, C. Electrochemical Interrogation of Interactions between Surface-Confined DNA and Methylene Blue. Sensors 2007, 7, 2671-2680. https://doi.org/10.3390/s7112671
Pan D, Zuo X, Wan Y, Wang L, Zhang J, Song S, Fan C. Electrochemical Interrogation of Interactions between Surface-Confined DNA and Methylene Blue. Sensors. 2007; 7(11):2671-2680. https://doi.org/10.3390/s7112671
Chicago/Turabian StylePan, Dun, Xiaolei Zuo, Ying Wan, Lihua Wang, Jiong Zhang, Shiping Song, and Chunhai Fan. 2007. "Electrochemical Interrogation of Interactions between Surface-Confined DNA and Methylene Blue" Sensors 7, no. 11: 2671-2680. https://doi.org/10.3390/s7112671
APA StylePan, D., Zuo, X., Wan, Y., Wang, L., Zhang, J., Song, S., & Fan, C. (2007). Electrochemical Interrogation of Interactions between Surface-Confined DNA and Methylene Blue. Sensors, 7(11), 2671-2680. https://doi.org/10.3390/s7112671



