Estimating 3D Leaf and Stem Shape of Nursery Paprika Plants by a Novel Multi-Camera Photography System
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
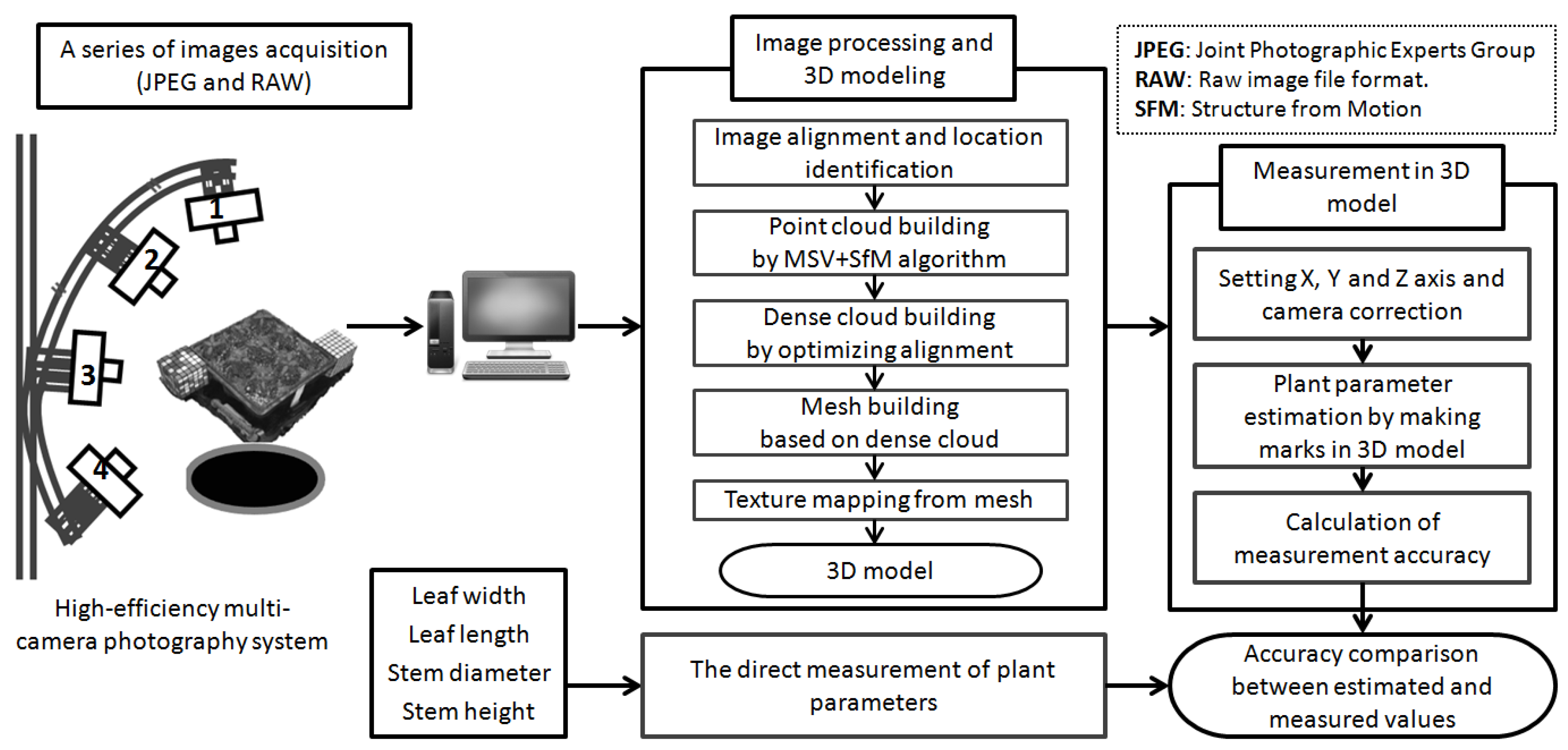
2.2. Multi-Camera Photography-Structure from Motion System
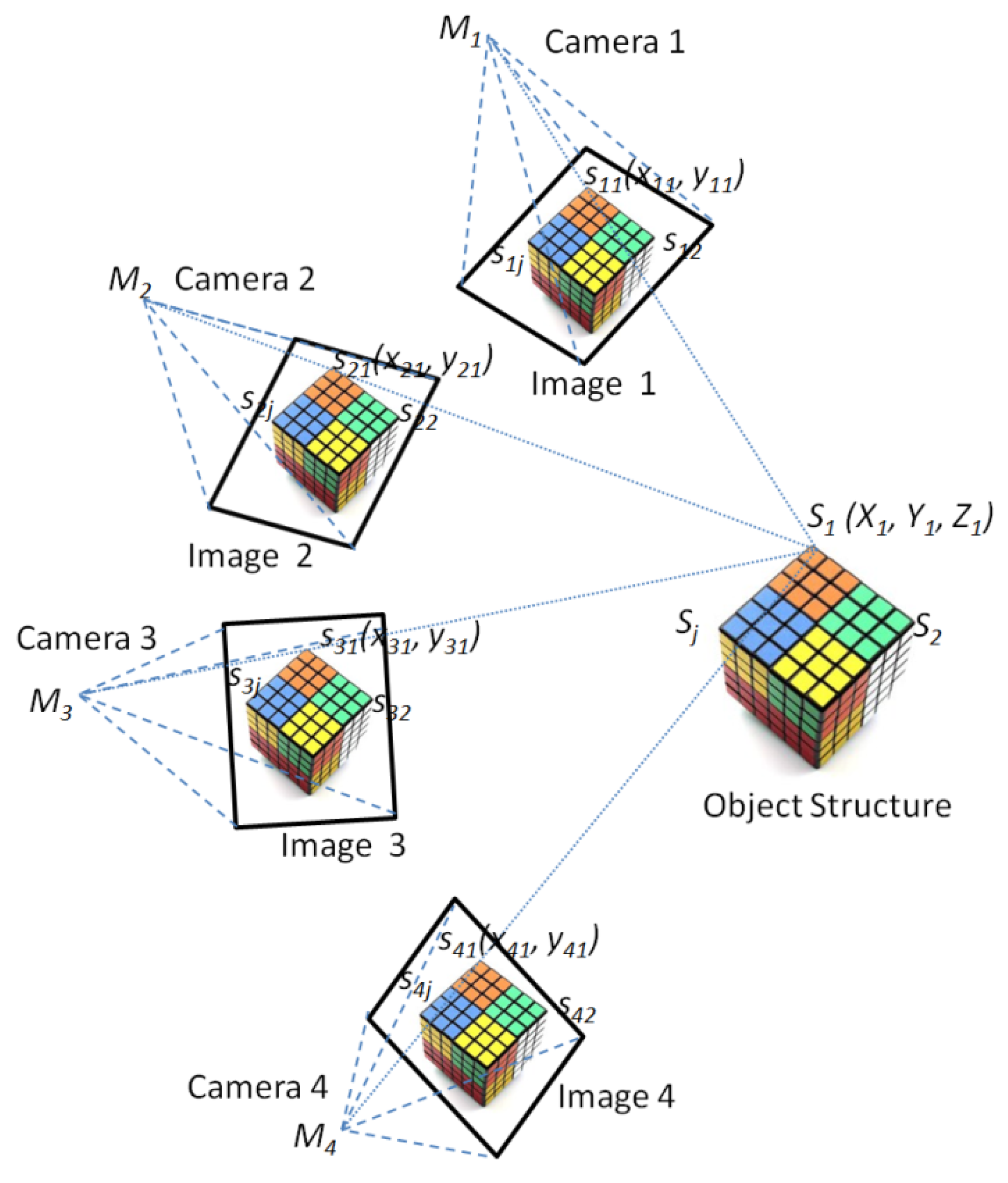
2.3. The MCP-SfM Approach
2.4. 3D Model Processing of Nursery Plants
2.5. 3D Model Measurement of Nursery Plants
3. Results
3.1. 3D Modeling Reconstruction of Paprika Plant
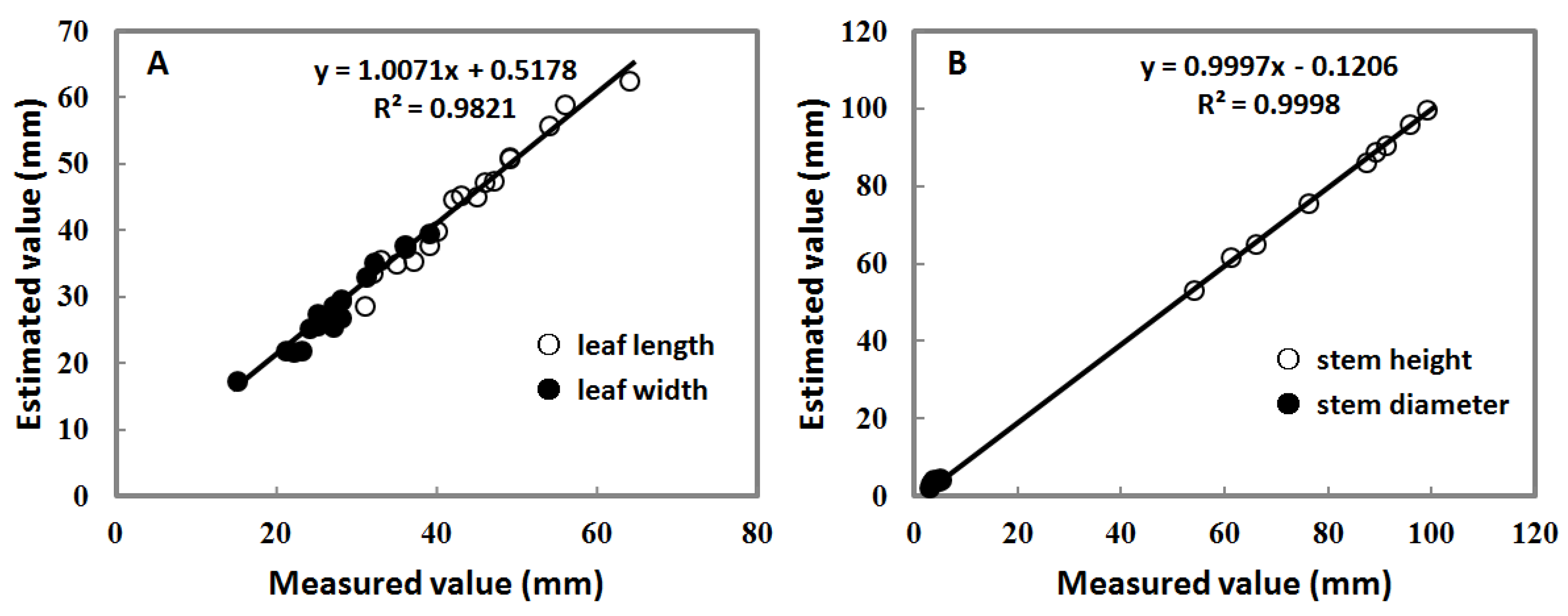
3.2. Parameters’ Estimation of the 3D Paprika Plant Model
3.3. Error Measurement of the 3D Paprika Plant Model
4. Discussions and Conclusions
4.1. Advantages of the MCP System Based on the MVS and SfM Methods
4.2. Error Assessment of the 3D Paprika Plant Model
4.3. Limitations and Issues of the Experiment
4.4. Future Improvements and Applications
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| MCP | Multi-Camera Photography |
| MVS | Multiple-View Stereovision |
| SfM | Structure from Motion |
| RMSE | Root-Mean-Square Error |
| SIFT | Scale-Invariant Feature Transform |
| PPF | Photosynthesis Photon Flux |
| PnP | Perspective N-Point |
| SSD | Sum of Squared Differences |
| DEM | Digital Elevation Model |
References
- Muller-Linow, M.; Pinto-Espinosa, F.; Scharr, H.; Rascher, U. The leaf angle distribution of natural plant populations: Assessing the canopy with a novel software tool. Plant Methods 2015, 11, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Omasa, K.; Hosoi, F.; Konishi, A. 3D lidar imaging for detecting and understanding plant responses and canopy structure. J. Exp. Bot. 2007, 58, 881–898. [Google Scholar] [CrossRef] [PubMed]
- Spalding, E.P.; Miller, N.D. Image analysis is driving a renaissance in growth measurement. Plant Biol. 2013, 16, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Omasa, K.; Takayama, K. Simultaneous measurement of stomatal conductance, non-photochemical quenching, and photochemical yield of photosystem II in intact leaves by yhermal and chlorophyll fluorescence imaging. Plant Cell Physiol. 2003, 44, 1290–1300. [Google Scholar] [CrossRef] [PubMed]
- Omasa, K. Image Analysis of Chlorophyll Fluorescence Transients of Cultured Carrot Tissues. Environ. Control Biol. 1992, 30, 127–131. [Google Scholar] [CrossRef]
- Omasa, K. Image Instrumentation Methods of Plant Analysis. In Modern Methods of Plant Analysis; Linskens, H.F., Jackson, J.F., Eds.; New Series, Physical Methods in Plant Sciences; Springer-Verlag: Berlin, Germany, 1990; Volume 11, pp. 203–243. [Google Scholar]
- Omasa, K.; Takayama, K.; Goto, E. Image Diagnosis of Photosynthetic Injuries Induced by Herbicide in Plants—Comparison of the Induction Method with the Saturation Pulse Method for Chlorophyll a Fluorescence Analysis. J. Soc. High Technol. Agric. 2001, 13, 29–37. [Google Scholar] [CrossRef]
- Barbagallo, R.P.; Oxborough, K.; Pallett, K.E.; Baker, N.R. Rapid, noninvasive screening for perturbations of metabolism and plant growth using chlorophyll fluorescence imaging. Plant Physiol. 2003, 132, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Garcia, R.; Francisco, J.; Sankaran, S.; Maja, J. Comparison of two aerial imaging platforms for identification of Huanglongbing-infected citrus trees. Comput. Electron. Agric. 2013, 91, 106–115. [Google Scholar] [CrossRef]
- Nuske, S.; Achar, S.; Bates, T.; Narasimhan, S.; Singh, S. Yield estimation in vineyards by visual grape detection. In Proceedings of the IEEE/RSJ International Conference on Intelligent Robots and Systems, San Francisco, CA, USA, 25–30 September 2011; pp. 2352–2358.
- Astrand, B.; Baerveldt, A. Plant recognition and localization using context information. In Proceedings of the IEEE Conference Mechatronics and Robotics, Aachen, Germany, 13–15 September 2004.
- Li, L.; Zhang, Q.; Huang, D. A Review of Imaging Techniques for Plant Phenotyping. Sensors 2014, 14, 20078–20111. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.S. Sensing technologies for precision specialty crop production. Comput. Electr. Agric. 2010, 74, 2–33. [Google Scholar] [CrossRef]
- Preuksakarn, C.; Boudon, F.; Ferrao, F.; Durand, J.-B.; Nikinmaa, E.; Godin, C. Reconstructing plant architecture from 3D laser scanner data. In Proceedings of the 6th International Workshop on Functional-Structural Plant Models, Davis, CA, USA, 12–17 September 2010; pp. 1999–2001.
- Takizawa, H.; Ezaki, N.; Mizuno, S.; Yamamoto, S. Plant Recognition by Integrating Color and Range Data Obtained Through Stereo Vision. J. Adv. Comput. Intell. Intelli. Inform. 2005, 9, 630–636. [Google Scholar]
- Hosoi, F.; Omasa, K. Estimating vertical plant area density profile and growth parameters of a wheat canopy at different growth stages using three-dimensional portable lidar imaging. ISPRS J. Photogramm. Remote Sens. 2009, 64, 151–158. [Google Scholar] [CrossRef]
- Hosoi, F.; Nakabayashi, K.; Omasa, K. 3-D Modeling of tomato canopies using a high-resolution portable scanning lidar for extracting structural information. Sensors 2011, 11, 2166–2174. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, M.; Andersen, H.; Granum, E. Comparative Study of Disparity Estimations with Multi-Camera Configurations in Relation to Descriptive Parameters of Complex Biological Objects. In Proceedings of the ISPRS Workshop BenCOS 2005: Towards Benchmarking Automated Calibration, Orientation and Surface Reconstruction from Images, Beijing, China, 15 October 2005.
- Hosoi, F.; Omasa, K. Estimating leaf inclination angle distribution of broad-leaved trees in each part of the canopies by a high-resolution portable scanning lidar. J. Agric. Meteorol. 2015, 71, 136–141. [Google Scholar] [CrossRef]
- Endres, F.; Hess, J.; Sturm, J.; Cremers, D.; Burgard, W. 3D Mapping with an RGB-D Camera. IEEE Trans. Robot. 2014, 30, 177–187. [Google Scholar] [CrossRef]
- Khoshelham, K.; Elberink, S.O. Accuracy and resolution of kinect depth data for indoor mapping applications. Sensors 2012, 12, 1437–1454. [Google Scholar] [CrossRef] [PubMed]
- Omasa, K.; Kouda, M.; Ohtani, Y. 3D microscopic measurement of seedlings using a shape-from-focus method. Trans. Soc. Instrum. Control Eng. 1997, 33, 752–758. [Google Scholar] [CrossRef]
- Mizuno, S.; Noda, K.; Ezaki, N.; Takizawa, H.; Yamamoto, S. Detection of Wilt by Analyzing Color and Stereo Vision Data of Plant. In Computer Vision/Computer Graphics Collaboration Techniques of Lecture Notes in Computer Science; Gagalowicz, A., Philips, W., Eds.; Springer Berlin Heidelberg: Berlin, Germany, 2007; Volume 4418, pp. 400–411. [Google Scholar]
- Scharstein, D.; Szeliski, R. A Taxonomy and Evaluation of Dense Two-Frame Stereo Correspondence Algorithms. Int. J. Comput. Vis. 2002, 47, 7–42. [Google Scholar] [CrossRef]
- Kyto, M.; Nuutinen, M.; Oittinen, P. Method for measuring stereo camera depth accuracy based on stereoscopic vision. Proc. SPIE 2011, 7864. [Google Scholar] [CrossRef]
- Van der Heijden, G.; Song, Y.; Horgan, G.; Polder, G.; Dieleman, A.; Bink, M.; Palloix, A.; van Eeuwijk, F.; Glasbey, C. SPICY: Towards automated phenotyping of large pepper plants in the greenhouse. Funct. Plant Biol. 2012, 39, 870–877. [Google Scholar] [CrossRef]
- Biskup, B.; Scharr, H.; Schurr, U.; Rascher, U. A stereo imaging system for measuring structural parameters of plant canopies. Plant Cell Environ. 2007, 30, 1299–1308. [Google Scholar] [CrossRef] [PubMed]
- Kise, M.; Zhang, Q. Development of a stereovision sensing system for 3D crop row structure mapping and tractor guidance. Biosyst. Eng. 2008, 101, 191–198. [Google Scholar] [CrossRef]
- Ivanov, N.; Boissard, P.; Chapron, M.; Valery, P. Estimation of the height and angles of orientation of the upper leaves in the maize canopy using stereovision. Agronomie 1994, 14, 183–194. [Google Scholar] [CrossRef]
- Kazmi, W.; Foix, S.; Alenya, G.; Andersen, H.J. Indoor and outdoor depth imaging of leaves with time of flight and stereo vision sensors: Analysis and comparison. ISPRS J. Photogramm. Remote Sens. 2014, 88, 128–146. [Google Scholar] [CrossRef] [Green Version]
- Okutomi, M.; Kanade, T. A multiple-baseline stereo. IEEE Trans. Pattern Anal. Mach. Intell. 1993, 15, 353–363. [Google Scholar] [CrossRef]
- Vogiatzis, G.; Hernández, C.; Torr, P.H.S.; Cipolla, R. Multiview stereo via volumetric graph-cuts and occlusion robust photoconsistency. IEEE Trans. Pattern Anal. Mach. Intell. 2007, 29, 2241–2246. [Google Scholar] [CrossRef] [PubMed]
- Vu, H.-H.; Labatut, P.; Keriven, R.; Pons, J.P. High accuracy and visibility-consistent dense multi-view stereo. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 34, 889–901. [Google Scholar]
- Harwin, S.; Lucieer, A. Assessing the Accuracy of Georeferenced Point Clouds Produced via Multi-View Stereopsis from Unmanned Aerial Vehicle (UAV) Imagery. Remote Sens. 2012, 4, 1573–1599. [Google Scholar] [CrossRef]
- Fonstad, M.A.; Dietrich, J.T.; Courville, B.C.; Jensen, J.L.; Carbonneau, P.E. Topographic structure from motion: A new development in photogrammetric measurement. Earth Surf. Proc. Landf. 2013, 38, 421–430. [Google Scholar] [CrossRef]
- Lowe, D. Distinctive image features from scale-invariant key points. Int. J. Comput. Vis. 2004, 60, 91–110. [Google Scholar] [CrossRef]
- Snavely, N.; Seitz, S.M.; Szeliski, R. Modeling the world from internet photo collections. Int. J. Comput. Vis. 2008, 80, 189–210. [Google Scholar] [CrossRef]
- Mathews, A.J.; Jensen, J.L.R. Three-dimensional building modeling using structure from motion: improving model results with telescopic pole aerial photography. In Proceedings of the 35th Applied Geography Conference, Minneapolis, MN, USA, 10–12 October 2012; pp. 98–107.
- Pollefeys, M.; Gool, L.V.; Vergauwen, M.; Verbiest, F.; Cornelis, K.; Tops, J. Visual modeling with a hand-held camera. Int. J. Comput. Vis. 2004, 59, 207–232. [Google Scholar] [CrossRef]
- Leberl, F.; Irschara, A.; Pock, T.; Meixner, P.; Gruber, M.; Scholz, S.; Weichert, A. Point clouds: Lidar versus 3D vision. Photogramm. Eng. Remote Sens. 2010, 76, 1123–1134. [Google Scholar] [CrossRef]
- Turner, D.; Lucieer, A.; Watson, C. Development of an unmanned aerial vehicle (UAV) for hyper resolution mapping based visible, multispectral, and thermal imagery. In Proceedings of the 34th International Symposium of Remote Sensing Environment, Sydney, Australia, 10–15 April 2011.
- Dey, A.; Mummet, L.; Sukthankar, R. Classification of plant structures from uncalibrated image sequences. In Proceedings of the IEEE Workshop on Applications of Computer Vision, Breckenridge, CO, USA, 9–11 January 2012; pp. 9–11.
- Dandois, J.P.; Ellis, E.C. Remote sensing of vegetation structure using computer vision. Remote Sens. 2010, 2, 1157–1176. [Google Scholar] [CrossRef]
- Wilburn, B.; Joshi, N.; Vaish, V.; Talvala, E.; Antunez, E.; Barth, A.; Adams, A.; Levoy, M.; Horowitz, M. High Performance Imaging Using Large Camera Arrays. Trans. Graph. ACM 2005, 24, 765–776. [Google Scholar] [CrossRef]
- Popovic, V.; Afshari, H.; Schmid, A.; Leblebici, Y. Real-time implementation of gaussian image blending in a spherical light field camera. In Proceedings of the IEEE International Conference on Industrial Technology, Cape Town, South Africa, 25–28 February 2013.
- Nguyen, T.T.; Slaughter, D.C.; Max, N.; Maloof, J.N.; Sinha, N. Structured Light-Based 3D Reconstruction System for Plants. Sensors 2015, 15, 18587–18612. [Google Scholar] [CrossRef] [PubMed]
- Barone, S.; Paoli, A.; Razionale, A.V. A Coded Structured Light System Based on Primary Color Stripe Projection and Monochrome Imaging. Sensors 2013, 13, 13802–13819. [Google Scholar] [CrossRef] [PubMed]
- Mathews, A.J.; Jensen, J.L.R. Visualizing and quantifying vineyard canopy lai using an unmanned aerial vehicle (UAV) collected high density structure from motion point cloud. Remote Sens. 2013, 5, 2164–2183. [Google Scholar] [CrossRef]
- Turner, D.; Lucieer, A.; Watson, C. An automated technique for generating georectified mosaics from ultra-high resolution unmanned aerial vehicle (UAV) imagery, based on structure from motion (SfM) point clouds. Remote Sens. 2012, 4, 1392–1410. [Google Scholar] [CrossRef]
- Snavely, N. Scene Reconstruction and Visualization from Internet Photo Collections. Ph.D. Thesis, University of Washington, Seattle, WA, USA, 2008. [Google Scholar]
- Tomasi, C.; Kanade, T. Shape and motion from image streams under orthography: A factorization method. Int. J. Comput. Vis. 1992, 9, 137–154. [Google Scholar] [CrossRef]
- Moreno-Noguer, F.; Lepetit, V.; Fua, P. Accurate Non-Iterative O(n) Solution to the PnP Problem. In Proceedings of the IEEE International Conference on Computer Vision, Rio de Janeiro, Brazil, 14–21 October 2007.
- Triggs, B.; Mclauchlan, P.; Hartley, R.; Fitzgibbon, A. Bundle adjustment—A modern synthesis. In Vision Algorithms: Theory and Practice; Springer Berlin Heidelberg: Berlin, Germany, 2000; Volume 1883, pp. 298–375. [Google Scholar]
- Koniaris, C.; Cosker, D.; Yang, X.S.; Mitchell, K. Survey of Texture Mapping Techniques for Representing and Rendering Volumetric Mesostructure. J. Comput. Graph. Tech. 2014, 3, 18–60. [Google Scholar]
- Paulus, S.; Behmann, J.; Mahlein, A.-K.; Plümer, L.; Kuhlmann, H. Low-Cost 3D Systems: Suitable Tools for Plant Phenotyping. Sensors 2014, 14, 3001–3018. [Google Scholar] [CrossRef] [PubMed]
- Paproki, A.; Sirault, X.; Berry, S.; Furbank, R.; Fripp, J. A novel mesh processing based technique for 3D plant analysis. BMC Plant Biol. 2012, 12. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, M.; Andersen, H.J.; Slaughter, D.C.; Granum, E. Ground truth evaluation of computer vision based 3D reconstruction of synthesized and real plant images. Precis. Agric. 2007, 8, 49–62. [Google Scholar] [CrossRef]
- Roberts, R.; Sinha, S.N.; Szeliski, R.; Steedly, D. Structure from motion for scenes with large duplicate structures. In Proceedings of the Computer Vision and Patter Recognition, Providence, RI, USA, 20–25 June 2011; pp. 3137–3144.



| Camera Positions | Focal Length of Lens | |||
|---|---|---|---|---|
| 24 mm | 28 mm | 35 mm | 50 mm | |
| 1 | 44 | 44 | 44 | 40 |
| 2 | 42 | 41 | 44 | 42 |
| 3 | 42 | 43 | 42 | 29 |
| 4 | 42 | 45 | 43 | 23 |
| Total | 170 | 173 | 173 | 134 |
| Camera Positions (see Figure 1) | Percentage of the Number of Category A or B to That of All Leaves (%) *1 | Percentage of the Number of Category A or B to That of All Stems (%) *2 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A *3 | B *4 | A *3 | B *4 | |||||||||||||
| >95 | >75 | >95 | >75 | |||||||||||||
| Focal Length of Lens (mm) | ||||||||||||||||
| 24 | 28 | 35 | 50 | 24 | 28 | 35 | 50 | 24 | 28 | 35 | 50 | 24 | 28 | 35 | 50 | |
| 1 | 34 | 47 | 31 | 32 | 57 | 76 | 74 | 62 | 0 | 0 | 0 | 0 | 11 | 11 | 11 | 0 |
| 2 | 20 | 27 | 27 | 22 | 54 | 59 | 64 | 68 | 44 | 57 | 44 | 33 | 67 | 78 | 78 | 67 |
| 1 + 2 | 39 | 45 | 28 | 22 | 59 | 68 | 66 | 64 | 56 | 78 | 44 | 33 | 67 | 78 | 56 | 56 |
| 3 | 3 | 5 | 1 | 0 | 11 | 24 | 9 | 5 | 78 | 100 | 100 | 33 | 89 | 100 | 100 | 44 |
| 1 + 2 + 3 | 9 | 26 | 27 | 0 | 49 | 50 | 69 | 0 | 78 | 89 | 33 | 0 | 89 | 89 | 56 | 0 |
| 4 | 4 | 5 | 4 | 3 | 14 | 19 | 22 | 5 | 44 | 56 | 44 | 22 | 56 | 56 | 78 | 44 |
| 1 + 2 + 3 + 4 | 7 | 14 | 9 | 16 | 42 | 46 | 41 | 68 | 89 | 89 | 22 | 33 | 89 | 100 | 44 | 44 |
| Camera Positions | Leaf | Stem | ||
|---|---|---|---|---|
| R2 | RMSE (mm) | R2 | RMSE (mm) | |
| 1 | 0.9821 | 1.65 | ||
| 3 | 0.9998 | 0.57 | ||
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Teng, P.; Shimizu, Y.; Hosoi, F.; Omasa, K. Estimating 3D Leaf and Stem Shape of Nursery Paprika Plants by a Novel Multi-Camera Photography System. Sensors 2016, 16, 874. https://doi.org/10.3390/s16060874
Zhang Y, Teng P, Shimizu Y, Hosoi F, Omasa K. Estimating 3D Leaf and Stem Shape of Nursery Paprika Plants by a Novel Multi-Camera Photography System. Sensors. 2016; 16(6):874. https://doi.org/10.3390/s16060874
Chicago/Turabian StyleZhang, Yu, Poching Teng, Yo Shimizu, Fumiki Hosoi, and Kenji Omasa. 2016. "Estimating 3D Leaf and Stem Shape of Nursery Paprika Plants by a Novel Multi-Camera Photography System" Sensors 16, no. 6: 874. https://doi.org/10.3390/s16060874
APA StyleZhang, Y., Teng, P., Shimizu, Y., Hosoi, F., & Omasa, K. (2016). Estimating 3D Leaf and Stem Shape of Nursery Paprika Plants by a Novel Multi-Camera Photography System. Sensors, 16(6), 874. https://doi.org/10.3390/s16060874






