Assessing and Correcting Topographic Effects on Forest Canopy Height Retrieval Using Airborne LiDAR Data
Abstract
:1. Introduction
2. Study Area and Data
2.1. Study Area
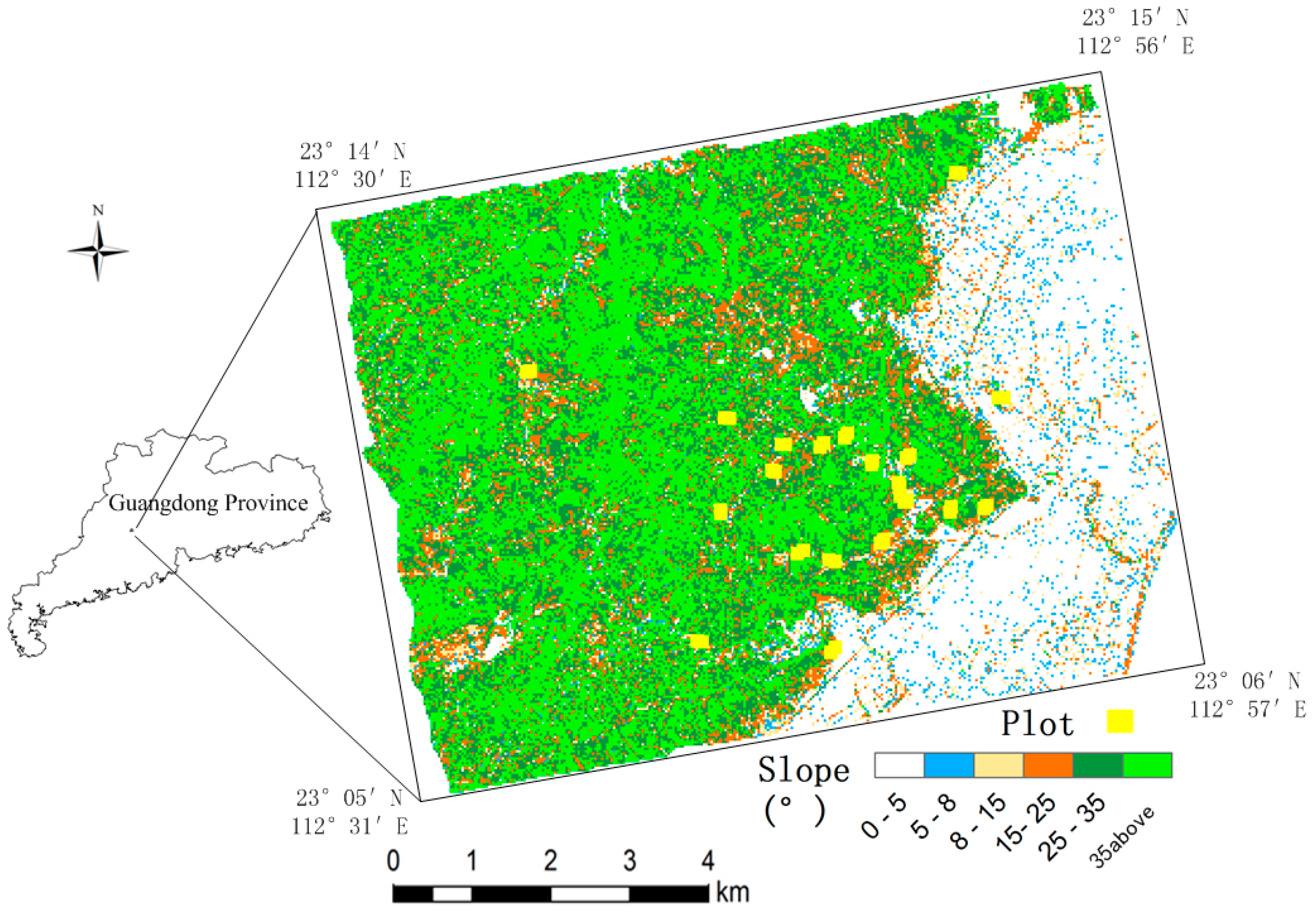
2.2. LiDAR Data
| Device Type | LiteMapper 6800 |
|---|---|
| Pulse repetition frequency | Up to 400 KHz |
| Laser wavelength | 1550 ns |
| Pulse length | 3.5 ns |
| Laser beam divergence | ≤0.5 mrad |
| Multiple target separation within single shot | 0.6 m |
| Return pulse width resolution | 0.15 m |
| Scan pattern | Parallel scan |
| Scan angle range | ±30° |
| Angle readout resolution | 0.001° |
| Ground sample spot diameter | 0.24 m (@800 m) |
| Horizontal accuracy | 0.08 m (@800 m) |
| Vertical accuracy | 0.04 m (@800 m) |
2.3. Field Inventory Data
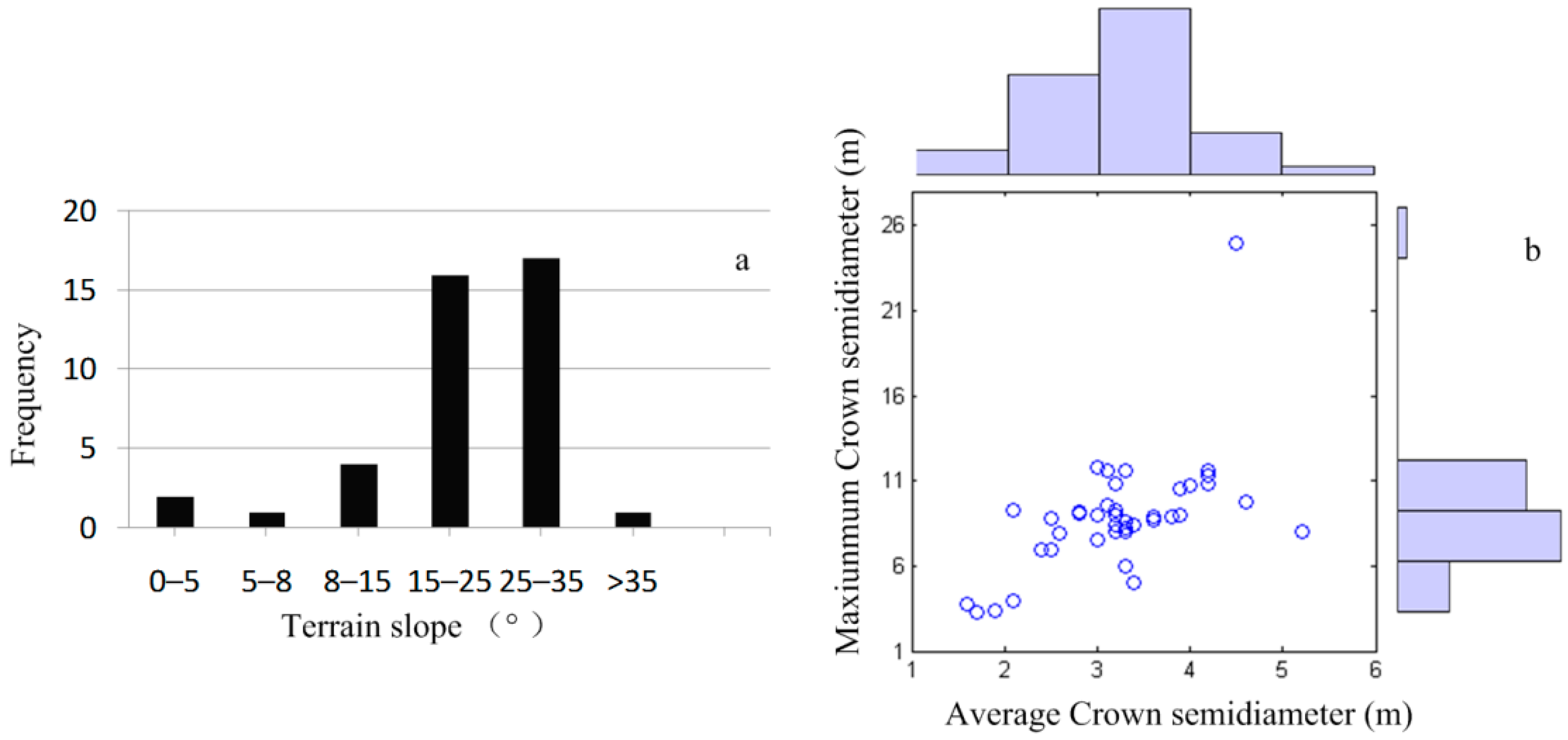
3. Methodology
3.1. Terrain Impacts on Canopy Height

| Slope i (°) | 5 | 10 | 20 | 30 | 40 | 50 | |
|---|---|---|---|---|---|---|---|
| Crown d (m) | |||||||
| 3 | 0.13 | 0.26 | 0.54 | 0.86 | 1.26 | 1.79 | |
| 5 | 0.22 | 0.44 | 0.91 | 1.44 | 2.10 | 2.97 | |
| 10 | 0.44 | 0.88 | 1.82 | 2.88 | 4.20 | 5.96 | |
| 15 | 0.66 | 1.32 | 2.73 | 4.33 | 6.29 | 8.94 | |
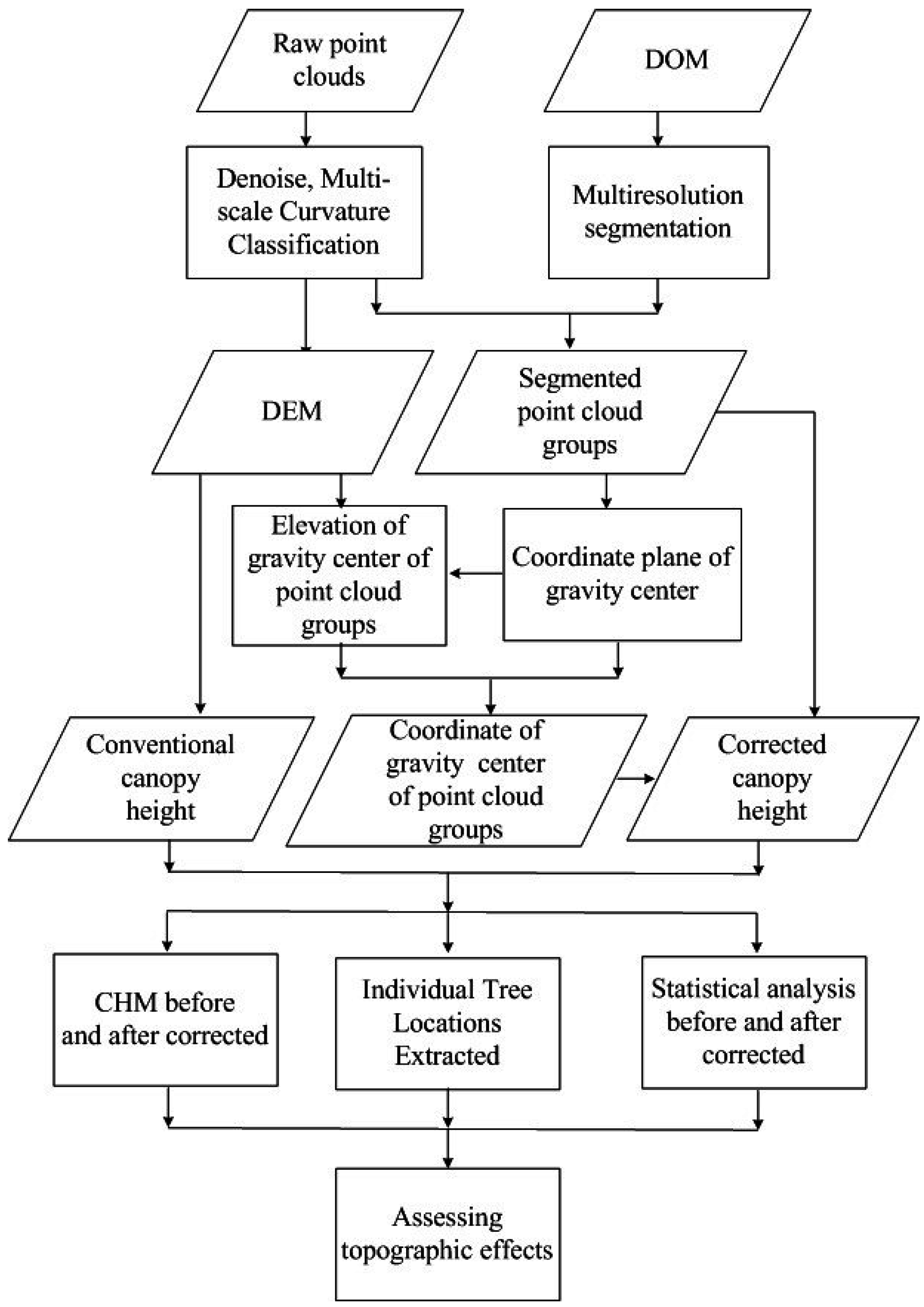
3.2. Processing of LiDAR Data
3.3. Crown Segmentation
3.4. Terrain Correction of Normalized Point Cloud
3.5. Individual Tree Locations Extracted
3.6. Assessing the Consequence of Correcting Topographic Effects
4. Results
4.1. CHMs before and after Correction


4.2. Impact of Topography on Individual Tree Extraction

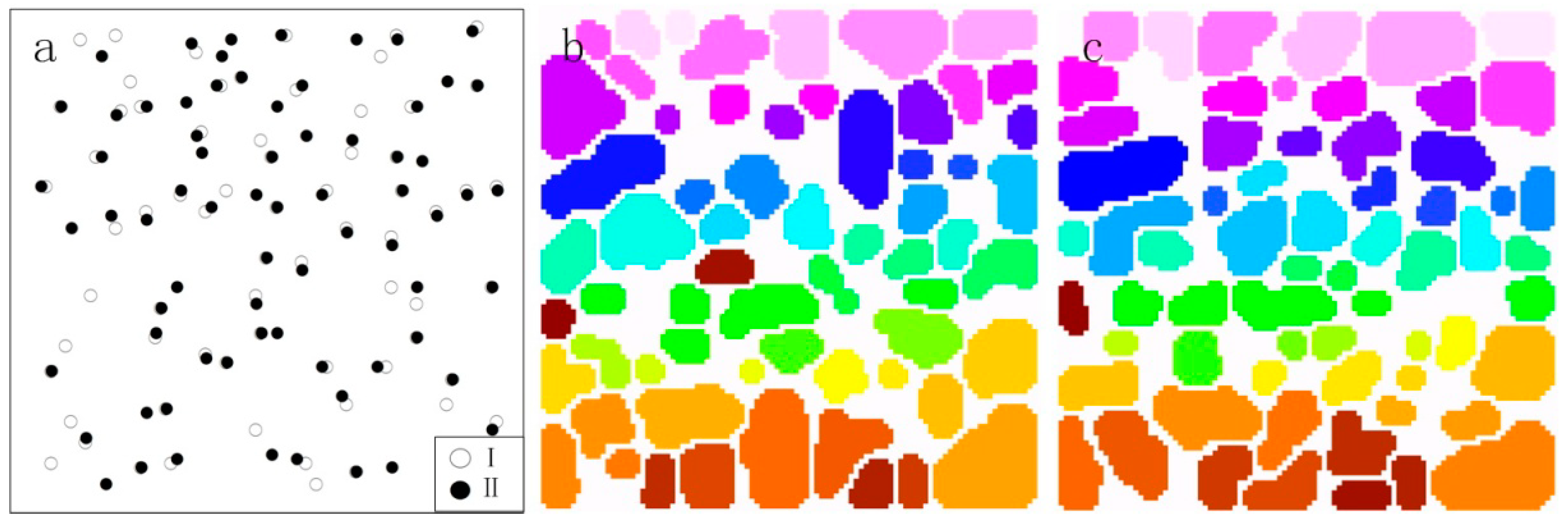
4.3. Impact of Topography on Plot-Level Canopy Height

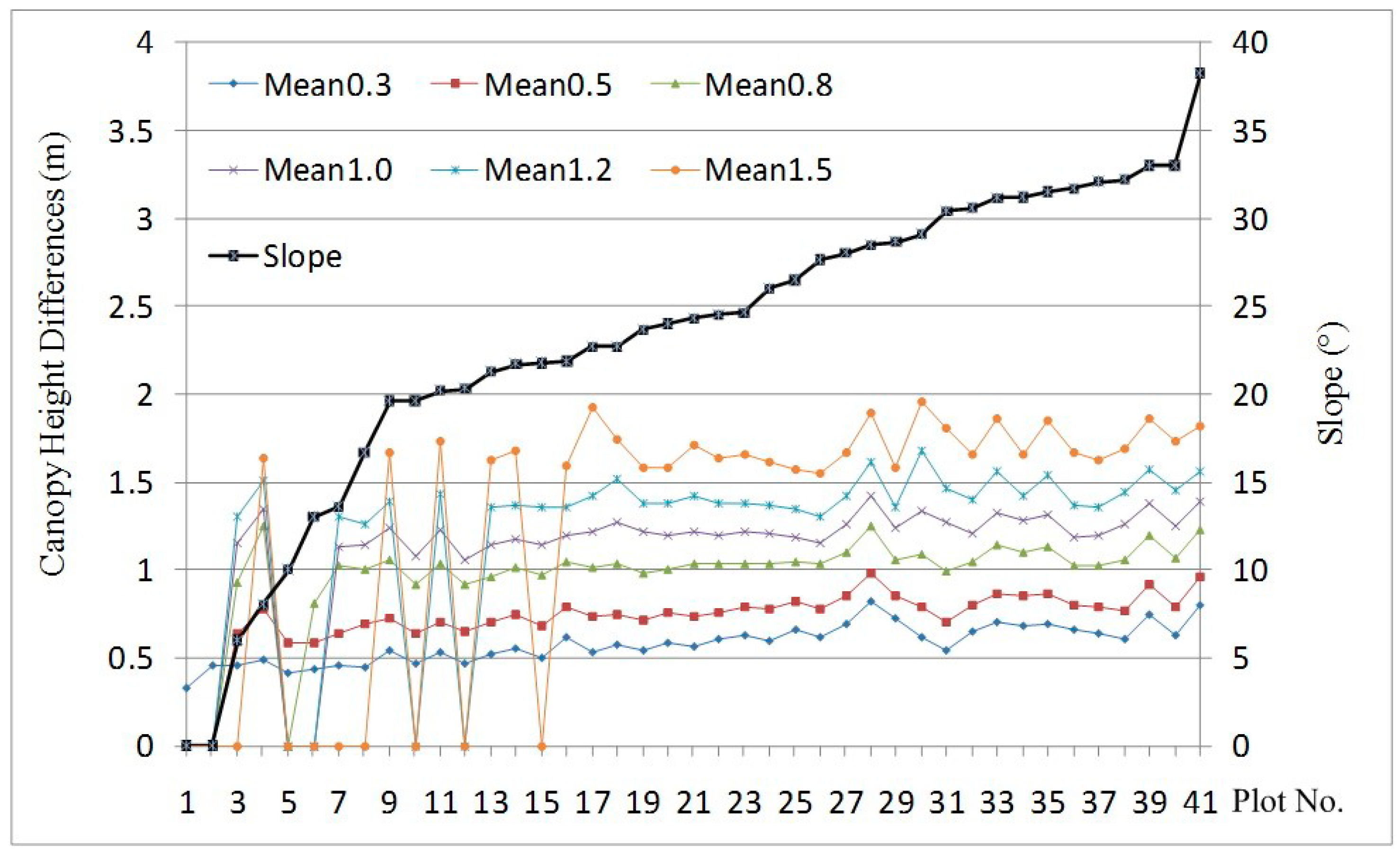


4.4. Consequence of Correcting Topographic Effects
| Coefficients | Model I ( n = 41) | Model II ( n = 41) | ||||||
|---|---|---|---|---|---|---|---|---|
| Estimated Values | Error Sum of Squares (SS) | F Ratio | P > F | Estimated Values | Error Sum of Squares (SS) | F Ratio | P > F | |
| β0 | 1.382009 | 0.0 | 0.000 | 1 | 0.814054 | 0.0 | 0.000 | 1 |
| β1 | −3.0262 | 36.37898 | 8.80696 | 0.005382 | −3.33578 | 34.44685 | 9.895305 | 0.003499 |
| β2 | ||||||||
| β3 | ||||||||
| β4 | 2.62023n7 | 34.95306 | 8.46176 | 0.006263 | 3.454034 | 26.42372 | 7.590555 | 0.009475 |
| β5 | −6.26901 | 21.0267 | 6.04019 | 0.019406 | ||||
| β6 | −3.33947 | 40.49818 | 9.804176 | 0.003506 | ||||
| β7 | 8.454889 | 19.45856 | 5.589724 | 0.024103 | ||||
| β8 | −7.81642 | 29.14998 | 8.37371 | 0.006697 | ||||
| β9 | 0.569135 | 23.73004 | 5.744788 | 0.022012 | 1.141369 | 52.70873 | 15.14126 | 0.000458 |
| β10 | 3.614737 | 19.06262 | 4.614855 | 0.038697 | 5.001009 | 25.61142 | 7.357213 | 0.010527 |
5. Discussion

6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Falkowski, M.J.; Wulder, M.A.; White, J.C.; Gillis, M.D. Supporting large-area, sample-based forest inventories with very high spatial resolution satellite imagery. Prog. Phys. Geogr. 2009, 33, 403–423. [Google Scholar] [CrossRef]
- Maltamo, M.; Packalén, P.; Yu, X.; Eerikäinen, K.; Hyyppä, J.; Pitkänen, J. Identifying and quantifying structural characteristics of heterogeneous boreal forests using laser scanner data. Forest Ecol. Manag. 2005, 216, 41–50. [Google Scholar] [CrossRef]
- Næsset, E.; Bjerknes, K.O. Estimating tree heights and number of stems in young forest stands using airborne laser scanner data. Remote Sens. Environ. 2001, 78, 328–340. [Google Scholar] [CrossRef]
- Evans, J.S.; Hudak, A.T.; Faux, R.; Smith, A. Discrete return LiDAR in natural resources: recommendations for project planning, data processing, and deliverables. Remote Sens. 2009, 1, 776–794. [Google Scholar] [CrossRef]
- Takahashi, T.; Yamamoto, K.; Senda, Y.; Tsuzuku, M. Estimating individual tree heights of sugi (Cryptomeria japonica D. Don) plantations in mountainous areas using small-footprint airborne LiDAR. J. Forest Res. 2005, 10, 135–142. [Google Scholar] [CrossRef]
- Hodgson, M.E.; Bresnahan, E. Accuracy of airborne LiDAR-derived elevation: Empirical assessment and error budget. Photogramm. Eng. Remote Sens. 2004, 70, 331–339. [Google Scholar] [CrossRef]
- Gatziolis, D.; Fried, J.S.; Monleon, V.S. Challenges to estimating tree height via LiDAR in closed-canopy forests: a parable from western Oregon. Forest Sci. 2010, 56, 139–155. [Google Scholar]
- Breidenbach, J.; Koch, B.; Kändler, G.; Kleusberg, A. Quantifying the influence of slope, aspect, crown shape and stem density on the estimation of tree height at plot level using LiDAR and InSAR data. Int. J. Remote Sens. 2008, 29, 1511–1536. [Google Scholar] [CrossRef]
- Gousie, M.B.; Franklin, W.R. Augmenting grid-based contours to improve thin plate DEM generation. Photogramm. Eng. Remote Sens. 2005, 71, 69–79. [Google Scholar] [CrossRef]
- Guo, Q.; Li, W.; Yu, H.; Alvarez, O. Effects of topographic variability and LiDAR sampling density on several DEM interpolation methods. Photogramm. Eng. Remote Sens. 2010, 76, 701–712. [Google Scholar] [CrossRef]
- Kraus, K.; Pfeifer, N. Determination of terrain models in wooded areas with airborne laser scanner data. ISPRS J. Photogramm. 1998, 53, 193–203. [Google Scholar] [CrossRef]
- Elmqvist, M.; Jungert, E.; Lantz, F.; Persson, A.; Soderman, U. Terrain modelling and analysis using laser scanner data. ISPRS Arch. 2001, 34, 219–226. [Google Scholar]
- Kobler, A.; Pfeifer, N.; Ogrinc, P.; Todorovski, L.; Oštir, K.; Džeroski, S. Repetitive interpolation: A robust algorithm for DTM generation from Aerial Laser Scanner Data in forested terrain. Remote Sens. Environ. 2007, 108, 9–23. [Google Scholar] [CrossRef]
- Axelsson, P. Processing of laser scanner data—Algorithms and applications. ISPRS J. Photogramm. 1991, 54, 138–147. [Google Scholar] [CrossRef]
- Vosselman, G. Slope based filtering of laser altimetry data. ISPRS Arch. 2000, 33, 935–942. [Google Scholar]
- Sithole, G. Filtering of laser altimetry data using a slope adaptive filter. ISPRS Arch. 2001, 34, 203–210. [Google Scholar]
- Chen, Q. Airborne LiDAR data processing and information extraction. Photogramm. Eng. Remote Sens. 2007, 73, 109–112. [Google Scholar]
- Evans, J.S; Hudak, A.T. A multiscale curvature algorithm for classifying discrete return LiDAR in forested environments. IEEE. Trans. Geosci. Remote Sens. 2007, 45, 1029–1038. [Google Scholar] [CrossRef]
- Popescu, S.C. Estimating biomass of individual pine trees using airborne LiDAR. Biomass Bioenergy 2007, 31, 646–655. [Google Scholar] [CrossRef]
- Zeng, Y.; Schaepman, M.E.; Wu, B.; Clevers, J.G.P.W.; Bregt, A.K. Scaling-based forest structural change detection using an inverted geometric-optical model in the three gorges region of china. Remote Sens. Environ. 2008, 112, 4261–4271. [Google Scholar] [CrossRef]
- Leckie, D.; Gougeon., F.; Hill, D.; Quinn, R.; Armstrong, L.; Shreenan, R. Combined high-density LiDAR and multispectral imagery for individual tree crown analysis. Can. J. Remote Sens. 2003, 29, 633–649. [Google Scholar] [CrossRef]
- Lim, K.; Treitz, P.; Baldwin, K.; Morrison, I.; Green, J. LiDAR remote sensing of biophysical properties of tolerant northern hardwood forests. Can. J. Remote Sens. 2003, 29, 658–678. [Google Scholar] [CrossRef]
- Koukoulas, S.; Blackburn, G.A. Quantifying the spatial properties of forest canopy gaps using LiDAR imagery and GIS. Int. J. Remote Sens. 2004, 25, 3049–3072. [Google Scholar] [CrossRef]
- Næsset, E.; Gobakken, T. Estimation of above-and below-ground biomass across regions of the boreal forest zone using airborne laser. Remote Sens. Environ. 2008, 112, 3079–3090. [Google Scholar] [CrossRef]
- Nilsson, M. Estimation of tree heights and stand volume using an airborne LiDAR system. Remote Sens. Environ. 1996, 56, 1–7. [Google Scholar] [CrossRef]
- Ferster, C.J.; Coops, N.C.; Trofymow, J.A. Aboveground large tree mass estimation in a coastal forest in British Columbia using plot-level metrics and individual tree detection from LiDAR. Can. J. Remote Sens. 2009, 35, 270–275. [Google Scholar] [CrossRef]
- Lefsky, M.A.; Cohen, W.B.; Acker, S.A.; Parker, G.G.; Spies, T.A.; Harding, D. LiDAR remote sensing of the canopy structure and biophysical properties of Douglas-fir western hemlock forests. Remote Sens. Environ. 1999, 70, 339–361. [Google Scholar] [CrossRef]
- Næsset, E. Predicting forest stand characteristics with airborne scanning laser using a practical two-stage procedure and field data. Remote Sens. Environ. 2002, 80, 88–99. [Google Scholar] [CrossRef]
- Brandtberg, T.; Walter, F. Automated delineation of individual tree crowns in high spatial resolution aerial images by multiple-scale analysis. Mach. Vis. Appl. 1998, 11, 64–73. [Google Scholar] [CrossRef]
- Dralle, K.; Rudemo, M. Stem number estimation by kernel smoothing of aerial photos. Can. J. Remote Sens. 1996, 26, 1228–1236. [Google Scholar]
- Wulder, M.; Niemann, K.O.; Goodenough, D. Local maximum filtering for the extraction of tree locations and basal area from high spatial resolution imagery. Remote Sens. Environ. 2000, 73, 103–114. [Google Scholar] [CrossRef]
- Pouliot, D.A.; King, D.J.; Bell, F.W.; Pitt, D.G. Automated tree crown detection and delineation in high-resolution digital camera imagery of coniferous forest regeneration. Remote Sens. Environ. 2002, 82, 322–334. [Google Scholar] [CrossRef]
- Schardt, M.; Ziegler, M.; Wimmer, A.; Wack, R.; Hyyppae, J. Assessment of Forest Parameters by Means of Laser Scanning. ISPRS Arch. 2002, 302–309. [Google Scholar]
- Wang, L.; Gong, P.; Biging, G.S. Individual Tree-Crown Delineation and Treetop Detection in High-Spatial-Resolution Aerial Imagery. Photogramm. Eng. Remote Sens. 2004, 70, 351–358. [Google Scholar] [CrossRef]
- Kravchenko, A.; Bullock, D.G. A comparative study of interpolation methods for mapping soil properties. Agron. J. 1999, 91, 393–400. [Google Scholar] [CrossRef]
- Zhao, D.; Pang, Y.; Li, Z.Y.; Sun, G.Q. Filling invalid values in a LiDAR-derived canopy height model with morphological crown control. Int. J. Remote Sens. 2013, 34, 4636–4654. [Google Scholar] [CrossRef]
- Avery, T.; Burkhart, H. Forest Measurments, 5th ed.; McGraw-Hill: New York, NY, USA, 2002. [Google Scholar]
- Hyyppä, J.; Kelle, O.; Lehikoinen, M.; Inkinen, M. A segmentation-based method to retrieve stem volume estimates from 3-D tree height models produced by laser scanners. IEEE. Trans. Geosci. Remote Sens. 2001, 39, 969–975. [Google Scholar] [CrossRef]
- Persson, Å.; Holmgren, J.; Söderman, U. Detecting and measuring individual trees using an airborne laser scanner. Photogramm. Eng. Remote Sens. 2002, 68, 925–932. [Google Scholar]
- Brandtberg, T.; Warner, T.A.; Landenberger, R.E.; McGraw, J.B. Detection and analysis of individual leaf-off tree crowns in small footprint, high sampling density Lidar data from eastern deciduous forest in North America. Remote Sens. Environ. 2003, 85, 290–303. [Google Scholar] [CrossRef]
- Duan, Z.G.; Zeng, Y.; Zhao, D.; Wu, B.F.; Zhao, Y.J.; Zhu, J.J. Method of removing pits of canopy height model from airborne LiDAR. Trans. CSAE 2014, 30, 209–217, (in Chinese with English abstract). [Google Scholar]
- Chen, Q.; Baldocchi, D.; Gong, P.; Kelly, M. Isolating individual trees in a savanna woodland using small footprint LiDAR data. Photogramm. Eng. Remote Sens. 2006, 72, 923–932. [Google Scholar] [CrossRef]
- Tiede, D.; Hochleitner, G.; Blaschke, T. A full GIS-based workflow for tree identification and tree crown delineation using laser scanning, CMRT, 2005, Vienna, Austria, 29–30 August 2005. 9–14.
- Koch, B.; Heyder, U.; Weinacker, H. Detection of Individual Tree Crowns in Airborne LiDAR Data. Photogramm. Eng. Remote Sens. 2006, 72, 357–363. [Google Scholar] [CrossRef]
- Lee, H.; Slatton, K.C.; Roth, B.E.; Cropper, W.P., JR. Adaptive clustering of airborne LiDAR data to segment individual tree crowns in managed pine forests. Int. J. Remote Sens. 2010, 31, 117–139. [Google Scholar] [CrossRef]
- Jakubowski, M.K.; Li, W.K.; Guo, Q.H.; Kelly, M. Delineating Individual Trees from LiDAR Data: A Comparison of Vector- and Raster-based Segmentation Approaches. Remote Sens. 2013, 5, 4163–4186. [Google Scholar] [CrossRef]
- Yang, W.; Ni-Meister, W.; Lee, S. Assessment of the impacts of surface topography, off-nadir pointing and vegetation structure on vegetation LiDAR waveforms using an extended geometric optical and radiative transfer model. Remote Sens. Environ. 2011, 115, 2810–2822. [Google Scholar] [CrossRef]
- Lee, S.; Ni-Meister, W.; Yang, W.Z.; Chen, Q. Physically based vertical vegetation structure retrieval from ICESat data: Validation using LVIS in White Mountain National Forest, New Hampshire, USA. Remote Sens. Environ. 2011, 115, 2776–2785. [Google Scholar] [CrossRef]
- Park, T.; Kennedy, R.E.; Choi, S.; Wu, J.; Lefsky, M.A.; Bi, J.; Mantooth, J.A.; Myneni, R.B.; Knyazikhin, Y. Application of Physically-Based Slope Correction for Maximum Forest Canopy Height Estimation Using Waveform Lidar across Different Footprint Sizes and Locations: Tests on LVIS and GLAS. Remote Sens. 2014, 6, 6566–6586. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duan, Z.; Zhao, D.; Zeng, Y.; Zhao, Y.; Wu, B.; Zhu, J. Assessing and Correcting Topographic Effects on Forest Canopy Height Retrieval Using Airborne LiDAR Data. Sensors 2015, 15, 12133-12155. https://doi.org/10.3390/s150612133
Duan Z, Zhao D, Zeng Y, Zhao Y, Wu B, Zhu J. Assessing and Correcting Topographic Effects on Forest Canopy Height Retrieval Using Airborne LiDAR Data. Sensors. 2015; 15(6):12133-12155. https://doi.org/10.3390/s150612133
Chicago/Turabian StyleDuan, Zhugeng, Dan Zhao, Yuan Zeng, Yujin Zhao, Bingfang Wu, and Jianjun Zhu. 2015. "Assessing and Correcting Topographic Effects on Forest Canopy Height Retrieval Using Airborne LiDAR Data" Sensors 15, no. 6: 12133-12155. https://doi.org/10.3390/s150612133







