A Graphene-Based Biosensing Platform Based on Regulated Release of an Aptameric DNA Biosensor
Abstract
:1. Introduction
2. Experimental Section
2.1. Materials and Reagents
2.2. Apparatus
2.3. Adsorption and Regulated Release of the DNA Biosensor from Graphene Oxide Surface
2.4. Detection of ATP by Using the Biosensing Platform
2.5. Selectivity Investigation of the Proposed Biosensing Platform
3. Results and Discussion
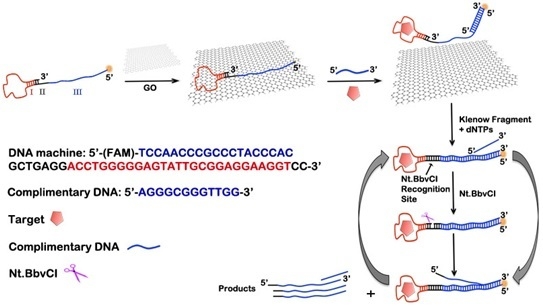
3.1. Design of the GO-Based DNA Biosensor and Polymerization/Nicking Enzyme Synergetic Isothermal Amplification Principle
3.2. Feasibility of the Biosensing Platform for ATP Detection

3.3. Absorption and Regulated Release of the DNA Biosensor
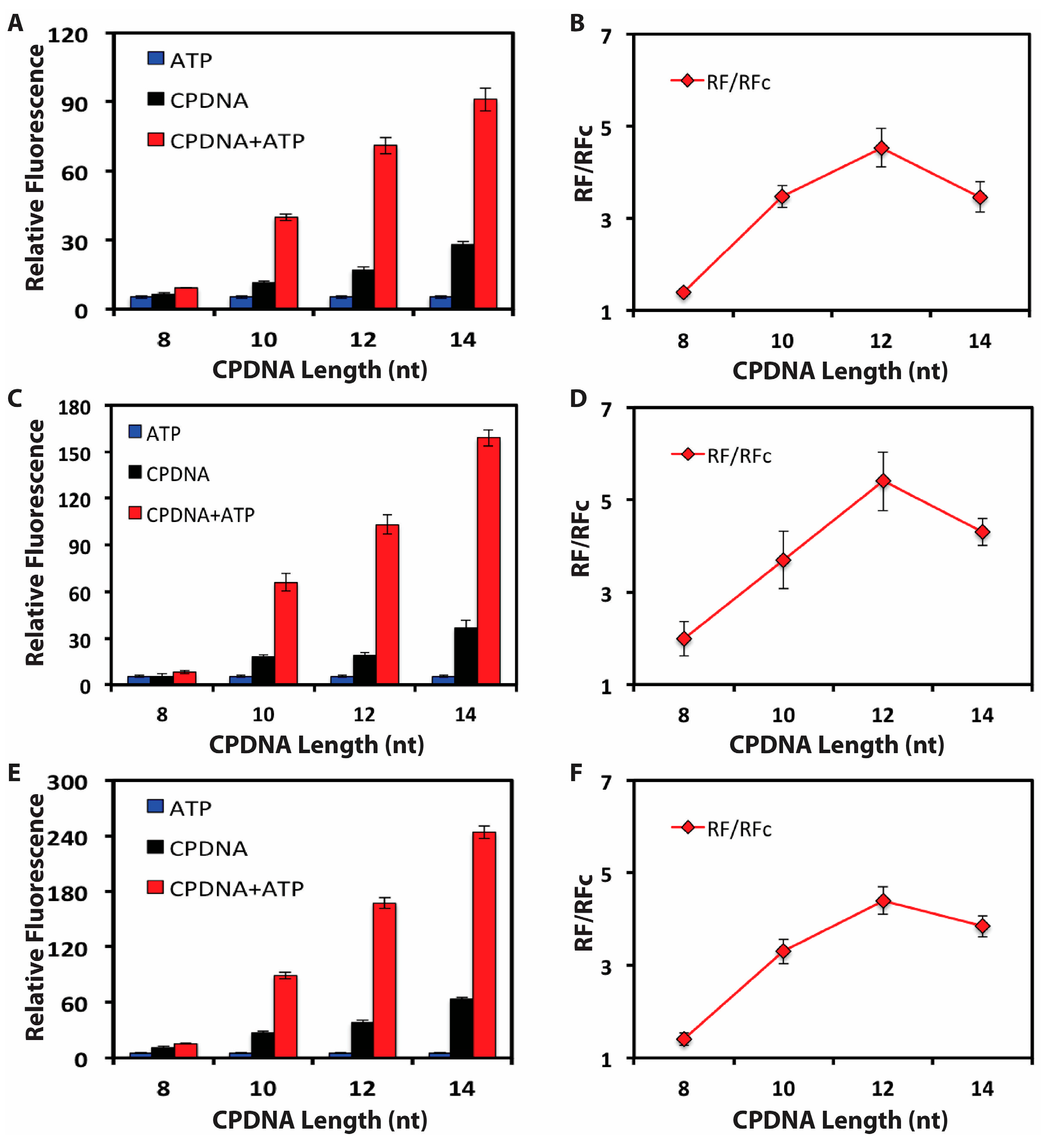
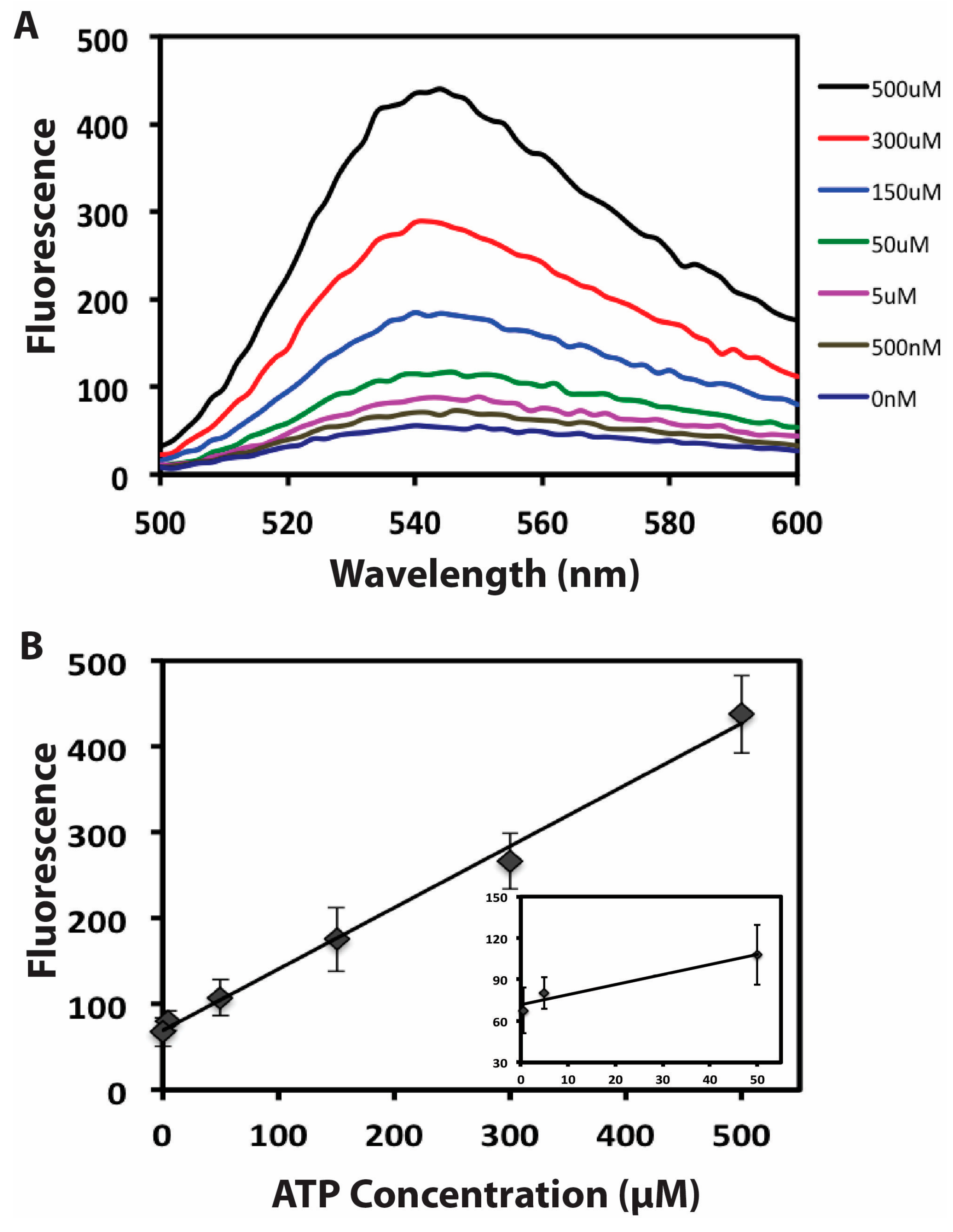
3.4. Quantitative Detection of ATP
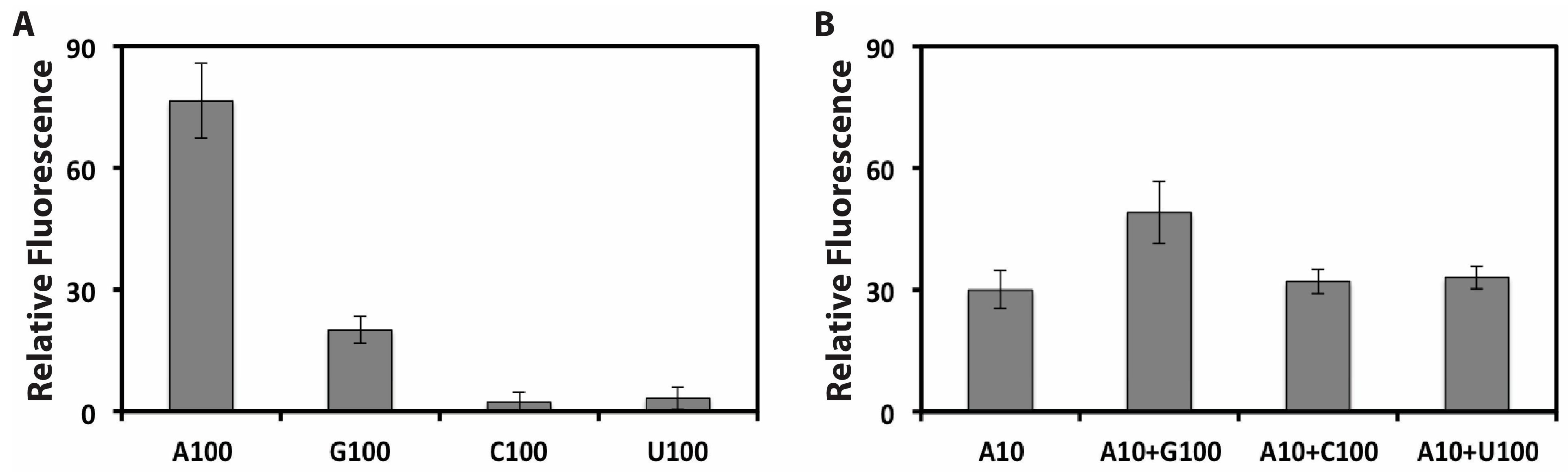
3.5. Selective Measurement of ATP
3.6. Application of the Biosensing Platform in Real Sample
4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chen, Y.; Lee, S.H.; Mao, C. A DNA nanomachine based on a duplex-triplex transition. Angew. Chem. Int. Ed. Engl. 2004, 43, 5335–5338. [Google Scholar] [CrossRef] [PubMed]
- Simmel, F.C.; Dittmer, W.U. DNA nanodevices. Small 2005, 1, 284–299. [Google Scholar] [CrossRef] [PubMed]
- Seeman, N.C. From genes to machines: DNA nanomechanical devices. Trends Biochem. Sci. 2005, 30, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.G.; Elbaz, J.; Willner, I. DNA machines: Bipedal walker and stepper. Nano Lett. 2011, 11, 304–309. [Google Scholar] [CrossRef] [PubMed]
- Modi, S.; Swetha, M.G.; Goswami, D.; Gupta, G.D.; Mayor, S.; Krishnan, Y. A DNA nanomachine that maps spatial and temporal pH changes inside living cells. Nat. Nanotechnol. 2009, 4, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Z.; Zhang, C.Y. Highly sensitive detection of protein with aptamer-based target-triggering two-stage amplification. Anal. Chem. 2012, 84, 1623–1629. [Google Scholar] [CrossRef] [PubMed]
- Freage, L.; Wang, F.; Orbach, R.; Willner, I. Multiplexed analysis of genes and of metal ions using enzyme/DNAzyme amplification machineries. Anal. Chem. 2014, 86, 11326–11333. [Google Scholar] [CrossRef] [PubMed]
- Shlyahovsky, B.; Li, D.; Weizmann, Y.; Nowarski, R.; Kotler, M.; Willner, I. Spotlighting of cocaine by an autonomous aptamer-based machine. J. Am. Chem. Soc. 2007, 129, 3814–3815. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Bock, L.C.; Griffin, L.C.; Latham, J.A.; Vermaas, E.H.; Toole, J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 1992, 355, 564–566. [Google Scholar] [CrossRef] [PubMed]
- And, R.N.; Li, Y. In vitro selection of structure-switching signaling aptamers. Angew. Chem. 2005, 44, 1085–1089. [Google Scholar]
- Shangguan, D.; Li, Y.; Tang, Z.; Cao, Z.C.; Chen, H.W.; Mallikaratchy, P.; Sefah, K.; Yang, C.J.; Tan, W. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl. Acad. Sci. USA 2006, 103, 11838–11843. [Google Scholar] [CrossRef] [PubMed]
- Patel, D.J. Structural analysis of nucleic acid aptamers. Curr. Opin. Chem. Biol. 1997, 1, 32–46. [Google Scholar] [CrossRef]
- Hermann, T.; Patel, D.J. RNA bulges as architectural and recognition motifs. Structure 2000, 8, R47–R54. [Google Scholar] [CrossRef]
- Xie, S.J.; Zhou, H.; Liu, D.; Shen, G.L.; Yu, R.; Wu, Z.S. In situ amplification signaling-based autonomous aptameric machine for the sensitive fluorescence detection of cocaine. Biosens. Bioelectron. 2013, 44, 95–100. [Google Scholar] [CrossRef] [PubMed]
- He, J.L.; Wu, Z.S.; Zhou, H.; Wang, H.Q.; Jiang, J.H.; Shen, G.L.; Yu, R.Q. Fluorescence aptameric sensor for strand displacement amplification detection of cocaine. Anal. Chem. 2010, 82, 1358–1364. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Pei, H.; Wan, Y.; Su, Y.; Huang, Q.; Song, S.; Fan, C. DNA nanostructure-decorated surfaces for enhanced aptamer-target binding and electrochemical cocaine sensors. Anal. Chem. 2011, 83, 7418–7423. [Google Scholar] [CrossRef] [PubMed]
- Dreyer, D.R.; Park, S.; Bielawski, C.W.; Ruoff, R.S. The chemistry of graphene oxide. Chem. Soc. Rev. 2010, 39, 228–240. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Feng, H.; Li, J. Graphene oxide: Preparation, functionalization, and electrochemical applications. Chem. Rev. 2012, 112, 6027–6053. [Google Scholar] [CrossRef] [PubMed]
- Morales-Narvaez, E.; Merkoci, A. Graphene oxide as an optical biosensing platform. Adv. Mater. 2012, 24, 3298–3308. [Google Scholar] [CrossRef] [PubMed]
- Loh, K.P.; Bao, Q.; Eda, G.; Chhowalla, M. Graphene oxide as a chemically tunable platform for optical applications. Nat. Chem. 2010, 2, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Z.; Wang, J.; Li, J.; Lin, Y. Graphene and graphene oxide: Biofunctionalization and applications in biotechnology. Trends Biotechnol. 2011, 29, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.; Kim, Y.K.; Shin, D.; Ryoo, S.R.; Hong, B.H.; Min, D.H. Biomedical applications of graphene and graphene oxide. Acc. Chem. Res. 2013, 46, 2211–2224. [Google Scholar] [CrossRef] [PubMed]
- Varghese, N.; Mogera, U.; Govindaraj, A.; Das, A.; Maiti, P.K.; Sood, A.K.; Rao, C.N. Binding of DNA nucleobases and nucleosides with graphene. ChemPhysChem 2009, 10, 206–210. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Sun, Z.; Zhang, X.; Liu, J. Mechanisms of DNA sensing on graphene oxide. Anal. Chem. 2013, 85, 7987–7993. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.H.; Yang, H.H.; Zhu, C.L.; Chen, X.; Chen, G.N. A graphene platform for sensing biomolecules. Angew. Chem. Int. Ed. Engl. 2009, 48, 4785–4787. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Ding, L.; Yan, F.; Ji, H.; Ju, H. The use of polyethylenimine-grafted graphene nanoribbon for cellular delivery of locked nucleic acid modified molecular beacon for recognition of microRNA. Biomaterials 2011, 32, 3875–3882. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.H.; Kong, R.M.; Zhang, X.B.; Meng, H.M.; Liu, W.N.; Tan, W.; Shen, G.L.; Yu, R.Q. Graphene-DNAzyme based biosensor for amplified fluorescence “turn-on” detection of Pb2+ with a high selectivity. Anal. Chem. 2011, 83, 5062–5066. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Z.; Hu, D.; Lin, C.T.; Li, J.; Lin, Y. Aptamer/Graphene Oxide Nanocomplex for in Situ Molecular Probing in Living Cells. J. Am. Chem. Soc. 2010, 132, 9274–9276. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Tang, L.; Wang, Y.; Jiang, J.; Li, J. Graphene fluorescence resonance energy transfer aptasensor for the thrombin detection. Anal. Chem. 2010, 82, 2341–2346. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Song, J.; Shuang, S.; Dong, C.; Brennan, J.D.; Li, Y. A Graphene-Based Biosensing Platform Based on the Release of DNA Probes and Rolling Circle Amplification. ACS Nano 2014, 8, 5564–5573. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.H.; Li, J.; Lin, M.H.; Wang, Y.W.; Yang, H.H.; Chen, X.; Chen, G.N. Amplified Aptamer-Based Assay through Catalytic Recycling of the Analyte 69. Angew. Chem. 2010, 122, 8632–8635. [Google Scholar] [CrossRef]
- Manohar, S.; Mantz, A.R.; Bancroft, K.E.; Hui, C.Y.; Jagota, A.; Vezenov, D.V. Peeling Single-Stranded DNA from Graphite Surface to Determine Oligonucleotide Binding Energy by Force Spectroscopy. Nano Lett. 2008, 8, 4365–4372. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Lau, P.S.; Liu, M.; Shuang, S.; Dong, C.; Li, Y. A general strategy to create RNA aptamer sensors using “regulated” graphene oxide adsorption. ACS Appl. Mater. Interface 2014, 6, 21806–21812. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.J.; Liu, J. Molecular beacon lighting up on graphene oxide. Anal. Chem. 2012, 84, 4192–4198. [Google Scholar] [CrossRef] [PubMed]
- Yi, M.; Yang, S.; Peng, Z.; Liu, C.; Li, J.; Zhong, W.; Yang, R.; Tan, W. Two-photon graphene oxide/aptamer nanosensing conjugate for in vitro or in vivo molecular probing. Anal. Chem. 2014, 86, 3548–3554. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Wang, Z.G.; Tang, H.W.; Pang, D.W. Low background signal platform for the detection of ATP: When a molecular aptamer beacon meets graphene oxide. Biosens. Bioelectron. 2011, 29, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, C.; Jiang, Y.; Hu, Y.; Li, J.; Yang, S.; Li, Y.; Yang, R.; Tan, W.; Huang, C.Z. Graphene signal amplification for sensitive and real-time fluorescence anisotropy detection of small molecules. Anal. Chem. 2013, 85, 1424–1430. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, F.; Aizen, R.; Yehezkeli, O.; Willner, I. Graphene oxide/nucleic-acid-stabilized silver nanoclusters: Functional hybrid materials for optical aptamer sensing and multiplexed analysis of pathogenic DNAs. J. Am. Chem. Soc. 2013, 135, 11832–11839. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Freeman, R.; Willner, I. Amplified fluorescence aptamer-based sensors using exonuclease III for the regeneration of the analyte. Chemistry 2012, 18, 2207–2211. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.J.; Yang, L.; Levy, M.; Ellington, A.D. Using a deoxyribozyme ligase and rolling circle amplification to detect a non-nucleic acid analyte, ATP. J. Am. Chem. Soc. 2005, 127, 2022–2023. [Google Scholar] [CrossRef] [PubMed]
- Gorman, M.W.; Feigl, E.O.; Buffington, C.W. Human plasma ATP concentration. Clin. Chem. 2007, 53, 318–325. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mao, Y.; Chen, Y.; Li, S.; Lin, S.; Jiang, Y. A Graphene-Based Biosensing Platform Based on Regulated Release of an Aptameric DNA Biosensor. Sensors 2015, 15, 28244-28256. https://doi.org/10.3390/s151128244
Mao Y, Chen Y, Li S, Lin S, Jiang Y. A Graphene-Based Biosensing Platform Based on Regulated Release of an Aptameric DNA Biosensor. Sensors. 2015; 15(11):28244-28256. https://doi.org/10.3390/s151128244
Chicago/Turabian StyleMao, Yu, Yongli Chen, Song Li, Shuo Lin, and Yuyang Jiang. 2015. "A Graphene-Based Biosensing Platform Based on Regulated Release of an Aptameric DNA Biosensor" Sensors 15, no. 11: 28244-28256. https://doi.org/10.3390/s151128244
APA StyleMao, Y., Chen, Y., Li, S., Lin, S., & Jiang, Y. (2015). A Graphene-Based Biosensing Platform Based on Regulated Release of an Aptameric DNA Biosensor. Sensors, 15(11), 28244-28256. https://doi.org/10.3390/s151128244