The Evolution of the Bacterial Luciferase Gene Cassette (lux) as a Real-Time Bioreporter
Abstract
: The bacterial luciferase gene cassette (lux) is unique among bioluminescent bioreporter systems due to its ability to synthesize and/or scavenge all of the substrate compounds required for its production of light. As a result, the lux system has the unique ability to autonomously produce a luminescent signal, either continuously or in response to the presence of a specific trigger, across a wide array of organismal hosts. While originally employed extensively as a bacterial bioreporter system for the detection of specific chemical signals in environmental samples, the use of lux as a bioreporter technology has continuously expanded over the last 30 years to include expression in eukaryotic cells such as Saccharomyces cerevisiae and even human cell lines as well. Under these conditions, the lux system has been developed for use as a biomedical detection tool for toxicity screening and visualization of tumors in small animal models. As the technologies for lux signal detection continue to improve, it is poised to become one of the first fully implantable detection systems for intra-organismal optical detection through direct marriage to an implantable photon-detecting digital chip. This review presents the basic biochemical background that allows the lux system to continuously autobioluminesce and highlights the important milestones in the use of lux-based bioreporters as they have evolved from chemical detection platforms in prokaryotic bacteria to rodent-based tumorigenesis study targets. In addition, the future of lux imaging using integrated circuit microluminometry to image directly within a living host in real-time will be introduced and its role in the development of dose/response therapeutic systems will be highlighted.1. Introduction
Bacterial bioluminescence, commonly known as the lux reaction, is the most widely distributed luminescent mechanism on the planet [1] and, although this process of bacterial light production has been observed for centuries, it was not until the mid 1900s that it began to be evaluated scientifically [2,3]. Beginning in the 1980s, after several decades of research, the understanding of this system became advanced enough that it was possible to exogenously express the full gene cassette, comprised of five genes (luxCDABE), in alternative host organisms such as Escherichia coli [4]. As researcher’s understanding of the biochemistry behind the lux reaction continued to be refined, and genetic manipulation techniques improved, it soon became possible to exploit this cassette as a reporter system across a wide variety of bacterial species for an extremely diverse set of monitoring objectives.
Following the success of these myriad lux-based bacterial bioreporters, attempts were made to incorporate the system into eukaryotic organisms in order to expand the lux system’s usefulness as a reporter. While initially expression of the bacterial genes was unsuccessful, through rearrangement of the lux cassette gene expression pattern and improvement of expression efficiency via codon-optimization and the addition of specialized linker regions, these hurdles were overcome and the lux reaction was demonstrated to occur in the lower eukaryote Saccharomyces cerevisiae [5]. Building upon this early success of eukaryotic expression, the luxAB genes were then further engineered to express in a human cell line, leading to the emergence of the lux system as a truly multifunctional reporter system similar to the more commonly employed firefly luciferase system [6].
Recently the lux system has undergone another substantial improvement, as it has been demonstrated that the full cassette can be optimized in a similar manner to the luxAB genes in order to promote fully autonomous bioluminescent production in a human cell line without the need to exogenously supplement a chemical substrate [7]. This review will highlight the development of the lux cassette from a curiosity observed in marine bacteria, through its extensive use as a bacterial bioreporter system and modification for expression in eukaryotic organisms, up to its recent demonstration as the only fully autonomous, substrate-free bioluminescent reporter system available in the eukaryotic host background. The unique, autonomous nature of the lux cassette will also be reviewed in light of the development of advanced photon detection hardware, detailing the future directions of lux development and its potential for biomedical as well as basic research applications.
Wild-Type lux Background
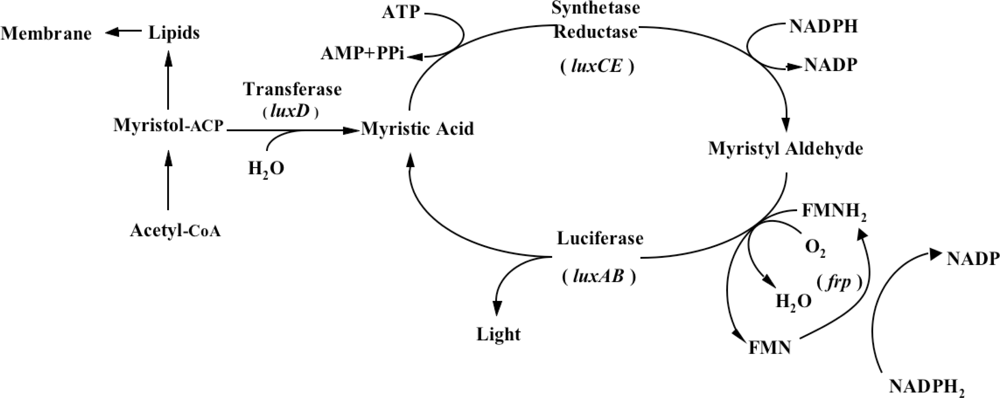
One does not have to look very far to see the glow of naturally bioluminescent organisms. On land bioluminescence is most commonly observed in the glow of fungi growing on decaying wood or from insects displaying their luminescent signal after dusk, while in marine environments bioluminescence is most commonly observed in single celled bacteria that are found either living freely or in symbiosis with larger hosts. It is these bioluminescent bacteria that are the most abundant and widely distributed of the light emitting organisms on Earth and they can be found in both aquatic (freshwater and marine) and terrestrial environments. Despite the widespread prevalence of bacterial bioluminescence, however, the majority of these organisms are classified into just three genera: Vibrio, Photobacterium, and Photorhabdus (Xenorhabdus) [1]. Although they are viable as free-living bacteria, these organisms are most commonly observed in symbiosis with a larger host. There is still no consensus as to the evolutionary benefit of bioluminescent production, however, in general it is theorized that the production of light can aid in the consumption of free living bacteria by higher trophic organisms, transferring them to a more controlled, nutrient rich habitat inside the host, or that, likewise, symbiotic bacteria can aid their hosts through the production of light that attracts prey, aids in camouflage, or attracts mates, in return for the shelter and nutrients provided by living within the body of the host organism [8]. Regardless of the reasons, the genetic system employed for the generation of bioluminescence is well conserved across all known bioluminescent bacteria. The luciferase protein is a heterodimer formed by the luxA and luxB gene products. The luxC, luxD, and luxE gene products encode for a reductase, transferase, and synthase respectively, that work together in a single complex to generate an aldehyde substrate for the bioluminescent reaction. In some species, there is an additional gene, frp, that functions as a flavin reductase to aid in the regeneration of the required FMNH2 substrate. Together with molecular oxygen, these components are all that are required to produce a bioluminescent signal [9] (Figure 1).
In addition, some marine species have additional genes that govern the expression of the remainder of the operon. The luxI and luxR genes function as an autoinducer and transcriptional activator (Figure 2), allowing the bioluminescent bacteria to participate in quorum sensing, therefore producing a bioluminescent signal only at high population densities [10]. This allows the production of light to be regulated and only expressed when it is beneficial to the organism, without unduly consuming metabolic energy.
2. Bacterial lux-Based Reporter Systems
2.1. First Examples of Transgenic lux Expression
Even before the mechanisms responsible for bioluminescence were completely explored in the organisms that naturally express it, the lux system had gained attention in regards to its potential as a reporter because of the facile detection and quantification of visible light produced by the Lux proteins. Work therefore quickly began to introduce the lux genes into non-luminescent bacterial species to determine if the modified organisms could acquire the light-emitting phenotype. These initial attempts to isolate and exogenously express the lux genes in non-native hosts such as E. coli were driven primarily by the desire to understand the organization, regulation, and function of the genetic components responsible for bioluminescence. The first major advancement in these investigations occurred in 1982 when Belas et al. [4] demonstrated bioluminescent production from recombinant E. coli containing the Vibrio harveyi luxA and luxB genes upon addition of exogenous aldehyde. In less than one year following this successful demonstration, the entire lux cassette (luxCDABE), together with its associated regulatory genes, was isolated from V. fischeri and introduced into E. coli [11], resulting in the first recombinant strain capable of emitting light without aldehyde supplementation. This process was soon repeated using the Photorhabdus luminescens genes, demonstrating the universal nature of the operon [12,13]. Although these efforts focused primarily at interrogating the functions of the individual lux genes, investigators began to appreciate the sensitivity, ease of detection, and non-destructive features the lux system offered as a bioluminescent reporter. The practical nonexistence of in vivo bioluminescence from bacteria not expressing the lux genes provides a high signal-to-noise ratio for the recombinant lux reporter system and the fast turnover rate of the bacterial luciferase enzyme allows for rapid light production. Taken together, these attributes have allowed the development of bacterial bioreporters that can be continuously monitored in a near real-time fashion. In addition, because the resulting bioluminescence is emitted in an autonomous manner, these reporters can be employed in a high throughput manner with very low cost, making them attractive options for a large number of biomonitoring applications.
2.2. Development of lux as a Method for Visualizing Gene Expression
The first use of lux as a biomonitoring technology came soon after its transgenic expression in E. coli, when Enbreghet et al. [14] fused the lux cassette to an inducible promoter that could be used to monitor gene expression in vivo. Using this experimental design it became possible to monitor autonomous bioluminescence as an indicator for the transcriptional activity of a promoter of interest. Using this method, the first major targets of study were the E. coli lac and ara promoters and it was discovered that upon IPTG or arabinose induction, light production in hosts expressing lux fusions increased between 600 to 1,000-fold. Following these reports the lux system was used to monitor regulation of the lateral flagella genes in Vibrio parahaemolyticus [14,15], providing its first demonstration in a previously uncharacterized system. These applications represented a significant shift in the way gene expression was investigated because, unlike traditional biochemical assays using enzymatic reporters, the bioluminescent signal from the lux genes could be easily detected and measured with high sensitivity without cell perturbation. This allowed the same sample to be continuously monitored, thus revealing the dynamics of gene expression through changes in bioluminescence over time. This new method was therefore capable of generating data that could not previously be generated.
2.3. lux-Based Bioluminescence as a Tool for Cellular Population Monitoring
While the Lux proteins do not require exogenous substrate addition, their function does require continued access to the molecular oxygen, FMNH2, and aldehyde co-substrates. For this reason, their bioluminescence can only be detected in actively growing cells. This knowledge, combined with the discovery that lux bioluminescent output is proportionally correlated to the number of cells present, has therefore been used as a simple, sensitive, and non-destructive means for in situ bacterial monitoring. This was first demonstrated by Shaw et al. [16] in 1986 when constitutively expressed V. fischeri luxCDABE genes were introduced into the phytopathogen Xanthomonas campestris, and their subsequent invasion of a cauliflower leaf was visualized. Similarly, de Weger and colleagues [17] were successfully able to detect luxCDABE-labeled Pseudomonas fluorescens in the rhizosphere of soybean roots using the same technique. Additionally, through the use of a lux-based system rather than an enzymatic reporter, it was possible for these researchers to achieve detection limits three orders of magnitude lower than what was previously possible, leading to improved signal detection. These early examples highlighted the application of lux-based bioluminescence as a rapid, simple and sensitive tool for in situ detection of living bacteria and established the foundation for future research using lux to monitor genetically engineered microorganisms. In perhaps the most notable use of the lux genes for tracking a cellular population, a P. fluorescens strain was transformed with the lux genes and used for the first bioremediation-related environmental field release of a genetically engineered microorganism in 1996.
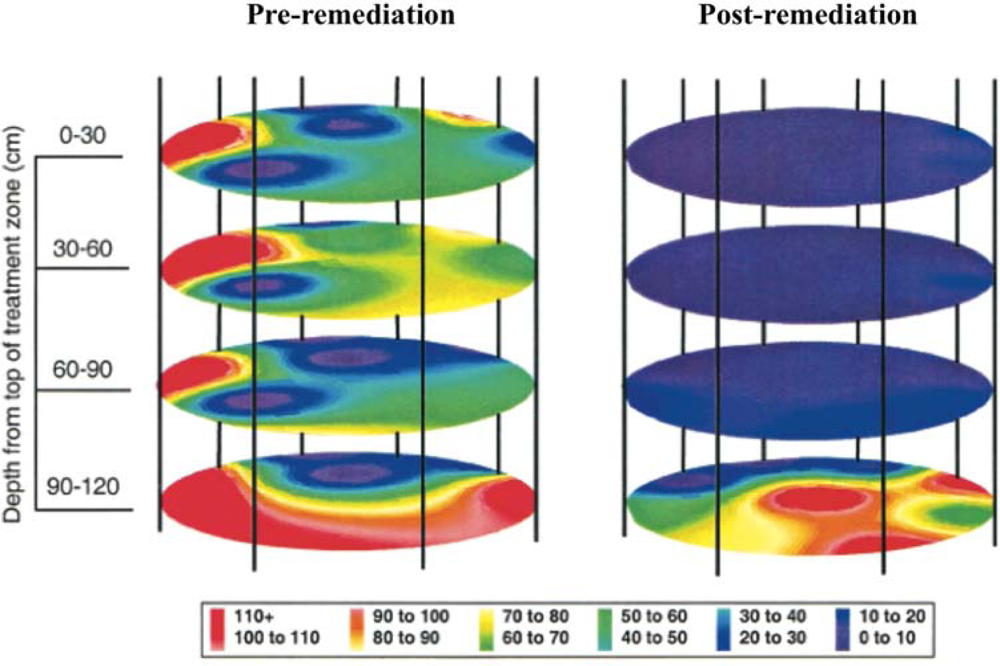
This release was approved by the Environmental Protection Agency to determine the efficiency of bioremediation process monitoring through inoculation of the bioluminescent strain directly into contaminated soil and to determine its ability to monitor the bioremediation of polycyclic aromatic hydrocarbons [18]. By placing the lux genes under the control of promoters in the naphthalene degradation pathway, it was possible to monitor their bioluminescent output as a measure of naphthalene contamination in the soil [19]. Using a combination of bioluminescent and traditional culture based detection methods, the release area was monitored for two years after the release of bioluminescent P. fluorescens. Over this time, regular sampling was performed to track the amount of bacteria present in the soil, as well as the amount of bioluminescence produced, which were indicative of organism presence and naphthalene degradation, respectively (Figure 3). Based on culture detection methods, the bioluminescent P. fluorescens persisted in both contaminated and non-contaminated soils, decaying at similar rates and producing similar colony counts [18]. The long term nature and difficulty in remote monitoring of bacterial populations presented in this study illustrates how the unique properties of the lux operon can provide it with an advantage over its substrate requiring bioluminescent or UV stimulation requiring fluorescent counterparts. Because of its autonomous nature, the lux-tagged P. fluorescens could be continually surveyed for bioluminescent production, without the need for repeated stimulation to induce a reporter signal.
2.4. The Use of lux for Exogenous Target Detection
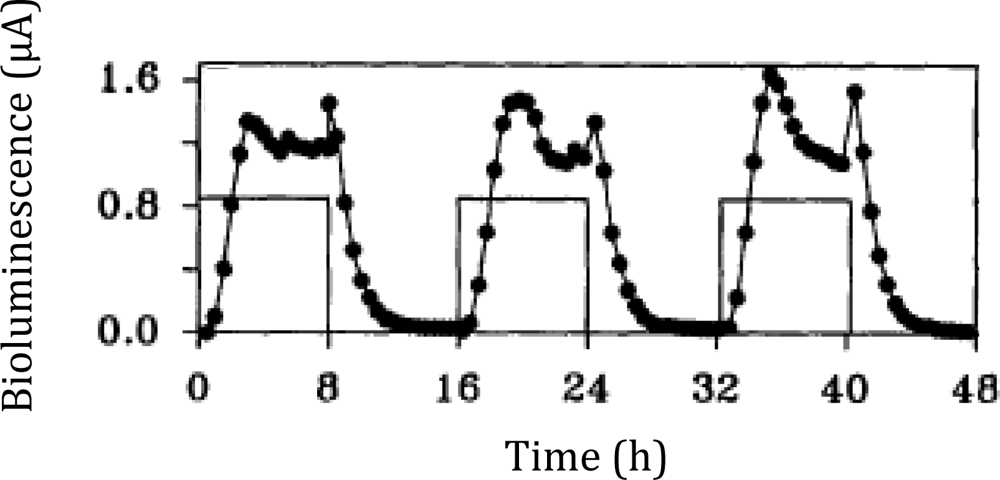
Following the work that demonstrated how the lux cassette could be used as a tool for visualizing gene expression, it soon became clear that these genes could be adapted for use as a traditional bioreporter target through activation under specific, predetermined conditions as well. By expressing the lux cassette under the control of a promoter with a known inducer, the resultant bioluminescent emission could be used as an indicator for the presence of the given stimulus, and fluctuation of the bioluminescent signal could be interpreted as changes in the bioavailable concentration of the inducer compound. Building upon these ideas, the first use of bioluminescence for monitoring metabolic activity was demonstrated in Pseudomonas putida by Burlage et al. in 1990 [20]. Here, naphthalene degradation was monitored using a transcriptional fusion of the salicylate inducible nah promoter and the luxCDABE genes. Salicylate is an intermediate metabolite of naphthalene, which is eventually degraded to acetaldehyde and pyruvate in Pseudomonas. Therefore, naphthalene degradation could be correlated to the light emission upon induction with naphthalene-derived salicylate. The nondestructive nature of the lux system allowed for this analysis to occur in real time in a growing culture, providing continuous monitoring of naphthalene metabolism across various stages of growth. It was later determined by King et al. [19] that the bioluminescent signal was controlled in a dose/response fashion (Figure 4), therefore demonstrating its usefulness in determining contaminant levels in mixed environmental samples. This opened the door for a multitude of environmental bioreporters featuring lux, such as that developed by Applegate and colleagues that was used to monitor for water soluble benzene, toluene, ethylbenzene, and xylene (BTEX) compounds indicative of petroleum spills. This reporter, constructed by linking expression of the lux cassette to the toluene dioxygenase promoter, was capable of detecting as little as 30 μg of toluene/L in as quickly as 2 h and maintained its detection ability for over 100 generations without antibiotic selection [21].
Another common target for lux-based environmental sensing has been phenol. Notably, Abd-El-Haleem et al. [22] constructed one of the first lux-based phenol biosensors by inserting a mopR-like promoter fused to the V. fischeri lux cassette genes into Acinetobacter sp DF4. This reporter was capable of demonstrating a lower detection limit of 2.5 ppm in 4 h when exposed to phenol, and was only responsive to three of the ten phenol derivatives tested, suggesting that it was relatively specific as well. This is, however, not by any means the only lux-based phenol reporter to be developed. Davidov et al. [23] made extensive use of recA promoters fused to lux cassettes, with each of the reporters containing a slight variation in its promoter sequence, that were expressed either in E. coli or Salmonella typhimurium and using lux genes from either V. fischeri or P. luminescens. The most sensitive of these reporters was that expressing the V. fischeri lux genes in E. coli, which was capable of detecting 0.008 mg phenol/L in 2 h. This same construct, when expressed in S. typhimurium was also capable of detecting phenol in 2 h but required a minimum concentration of 16 mg phenol/L, demonstrating the differences in host phenol bioavailability.
2.5. Further Uses of lux as a Bacterial Bioreporter
As the popularity of the lux system has grown over the years, an increasing number of bacterial reporters have been leveraged for the detection of a wide variety of contaminants. While this review focuses only on the seminal examples of lux’s growth as a reporter system, a larger list of target compounds and detection limits of various bioreporters can be found in recent reviews [24,25] and Table 1.
3. Eukaryotic Expression of the lux Cassette
Despite its success as a bacterial bioreporter, widespread use of the lux system faced a major hurdle in that it was initially believed to be capable of expression only in prokaryotes. Although several attempts were made to express the lux genes in eukaryotic hosts, none of these made significant headway [74–76]. It would not be until 2003 that the first major achievement was documented with the demonstration of autonomous bioluminescence from the yeast Saccharomyces cerevisiae [5]. Following this breakthrough, the lux genes continued to be modified and improved for eukaryotic expression, later being developed into a reliable yeast-based bioassay tool and, eventually, becoming capable of expression in a human cell line [7], opening the door for continued development in the future.
3.1. lux Expression in Yeast
It was the demonstration of lux function in S. cerevisiae by Gupta et al. [5] in 2003 that marked the first time a eukaryotic organism successfully produced levels of bioluminescence comparable to prokaryotic lux-based bioreporters (Figure 5). To achieve this, Gupta and colleagues chose to express the lux genes from the terrestrial bacterium P. luminescens rather than those from the traditional marine organisms V. harveyi or V. fischeri. This was done because the resulting luciferase proteins from P. luminescens exhibit a higher thermal stability than those of their marine counterparts. To mimic the organization and expression of the lux genes found in prokaryotic organisms, the luxA and luxB genes were expressed from a single promoter and linked by an internal ribosomal entry site (IRES). Under this expression strategy it was determined that bioluminescence was 20 times greater than that reported for fused luciferases upon exposure to an n-decanal substrate. Building upon these findings, the remainder of the lux genes were incorporated using the same strategy, with a pair of genes linked by an IRES element and driven by a unique promoter. When expressed concurrently this design was capable of producing an easily detectable bioluminescent signal.
Using the lessons learned from creation of the bioluminescent yeast strain, work was then begun to develop the eukaryotic lux system into a functional bioreporter for the detection of estrogenic compounds—a task that was not possible using prokaryotic hosts. Sanseverino et al. [45] built upon the lux plasmids developed by Gupta, creating a second set that constitutively expressed the luxC, luxD, luxE, and frp genes while regulating expression of the remaining luxA and luxB genes through insertion of human estrogen response elements (Figure 6). Upon exposure to estrogenic compounds, yeast expressing these regulated lux genes would produce a bioluminescent signal in as quickly as 1 h. This improved significantly over the colorimetric yeast estrogen screen, which could take as long as five days to produce results under identical conditions. Within two years of this successful demonstration of lux-based bioluminescent yeast as estrogen reporters, the same group had expanded the functionality of the assay by incorporating detection of androgenic compounds as well. This was accomplished by replacing the estrogen response elements that controlled lux expression with palindromic androgen response elements. However, this simple change allowed for parallel detection of estrogenic and androgenic compounds when combined with the existing lux-based estrogen reporter. Just as before, the lux-based system was shown to be more effective, faster, and more specific than existing screening methods [32].
3.2. lux Expression in Mammalian Cells
Continuing the development of the lux system as a functional eukaryotic reporter system, it was further refined and optimized for expression in human cell lines. This process was undertaken because the lux system possesses some unique characteristics compared to the alternative fluorescent and bioluminescent reporter systems that are available for use in human cell lines. Because the lux system is capable of producing continuous bioluminescence without addition of an exogenous luciferin, it is the only bioluminescent reporter available for human cell imaging that can function independently, and therefore produce real-time data in vivo [7,77]. In addition, because it is bioluminescent rather than fluorescent, it is not subject to the same high levels of background produced by some fluorescent reporters such as GFP [78]. These traits make it an attractive option for imaging in the human cellular background.
Despite the potential benefits of human cell line expression, it would still be five years from the first published report of human optimization until full cassette expression was finally reported [6,7]. Despite the success achieved in expressing the full lux cassette in S. cerevisiae, it was not possible to transfect human cells with the same constructs and produce a bioluminescent signal. In 2005, Patterson et al. [6] were the first to demonstrate that efficient expression of the luxA and luxB genes required optimization of the DNA sequence of each gene to more closely match the human codon preference. This optimization served to increase the efficiency of transcription/translation and increase the probability that the full length of the open reading frame would be recognized and expressed by the human ribosome (Table 2). These results mirrored those demonstrated in the prokaryotic arena by Craney et al. [79], who reported that codon modification of the AT-rich P. luminescens lux operon allowed for improved expression in GC-rich bacteria such as Streptomyces coelicolor. However, unlike the work of Craney et al., Patterson had to include additional modifications such as the replacement of the yeast IRES sequence with a mammalian optimized IRES element to provide for improved expression of the downstream luxB gene in the mammalian cellular environment. Through incorporation of these modifications, Patterson et al. were able to demonstrate constitutive expression of the lux luciferase heterodimer, providing the framework for future optimization of the full lux cassette and creating a substrate dependent lux reporter system that could be used in a manner similar to firefly luciferase, only with addition of the inexpensive aldehyde n-decanal rather than D-luciferin.
Following the inroads made by Patterson et al., it was not long before the full lux cassette was optimized for expression in human cell lines. This was accomplished by re-engineering each of the lux genes to more closely match the human codon preference, separating them by human optimized IRES elements, and then dividing their expression across three sequences in two separate plasmids. The original construct created by Patterson was retained, but co-expressed with a second plasmid containing the luxC and luxE genes (separated by an IRES element) under the control of the human elongation factor 1-α promoter and the luxD and frp genes (also separated by an IRES element) under the control of the cytomegalovirus immediate early promoter (Figure 7).
It is possible that this two plasmid expression system could itself contribute to the production of bioluminescence in human cells since Yagur-Kroll and Belkin [80] have recently reported that splitting the five lux genes into two smaller units (luxAB and luxCDE) resulted in improved bioreporter performance in E. coli. This increased bioluminescent production is hypothesized to be due to the associated enhanced transcriptional and/or translational efficiency achieved through the expression of smaller open reading frames, which theoretically could serve the same function in eukaryotic cells as well.
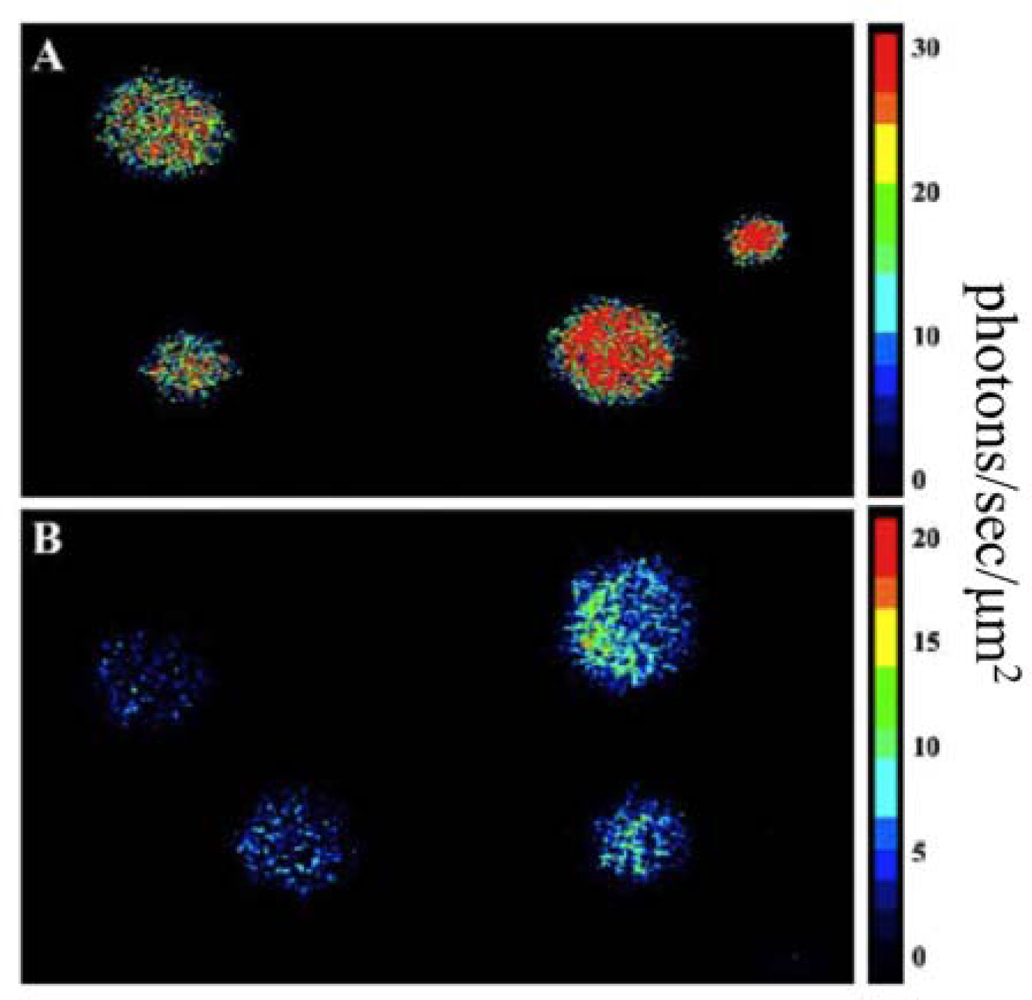
Using this expression strategy, it was demonstrated that these changes were both necessary and sufficient for autonomous production of a bioluminescent signal when expressed in a human kidney cell line (Figure 8) [7]. It should be noted that bioluminescent production from the human-optimized lux cassette was demonstrated to be several orders of magnitude lower than that of the more common firefly luciferase reporter and therefore greater numbers of bioluminescent cells were required to produce a significantly detectable signal in both cell culture (15,000 lux-expressing cells vs. 50 firefly luciferase-expressing cells) and small animal imaging experiments (25,000 lux-expressing cells vs. 2,500 firefly luciferase cells). However, due to the autonomous nature of the lux system, bioluminescent production was maintained over a longer period and produced less variability than did the firefly luciferase system [78].
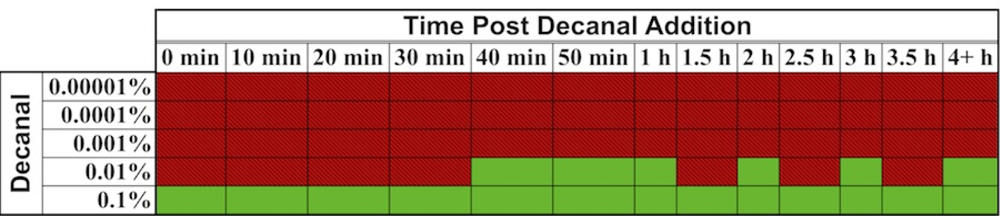
In one interesting experiment that took advantage of the autonomous nature of lux bioluminescence, constitutively bioluminescent human cells expressing the lux genes were used to evaluate the cytotoxicity of the aldehyde n-decanal [77]—the same aldehyde that was used by Patterson et al. [6] to stimulate bioluminescent production in cell extracts containing optimized luxA and luxB genes. By monitoring the changes in bioluminescent production following aldehyde treatment, it was possible to evaluate not only which concentrations were cytotoxic to the cells, but also at what time following exposure the effects began to take place, how long the cells were able to continue functioning under a diminished capacity following introduction of the aldehyde, and at what point cells succumbed to treatment and died. It was demonstrated that treatment with 0.00001%, 0.0001%, and 0.001% volumes of aldehyde did not show any changes in bioluminescence [77], despite the fact that this compound has been shown previously to function as a substrate for the lux reaction [6]. However, while treatment with a 0.1% volume of aldehyde quickly diminished bioluminescent production, treatment with 0.01% allowed the investigators to visualize a 3.5 h period where the bioluminescent output from cells oscillated between a significant decline and the negative control value, followed by a final, constant departure to significantly reduced bioluminescent production (Figure 9) [77]. The monitoring of these oscillating changes highlights the utility of the autonomous lux system, because if individual readings were taken at set time points rather than the near continuous monitoring performed in the investigation, it would have been possible to completely omit this period of activity from the resultant data.
4. Future of lux Imaging
With the extensive use of lux as a bioreporter in bacteria, and the recent proof-in-principal demonstration in eukaryotes, the majority of development that remains for this unique reporter system will most likely revolve around its further optimization and expansion in the eukaryotic cellular background. Close et al. [7] have reported that the availability of the FMNH2 co-substrate is currently the limiting reagent in the lux reaction in human cells, and increases in the efficiency of expressing the Frp protein, which reduces FMN to FMNH2, could lead to bioluminescent increases as large as 151-fold. Increases in the availability of the aldehyde co-substrate were also shown to enhance bioluminescent production, however, not to as great an extent and came with possible concerns over increased cytotoxicity. It is also important to note that, unlike alternate reporter systems such as GFP and firefly luciferase, the eukaryotic-optimized lux system has only recently become available for use. As the system continues to be used and is exposed to a wider variety of investigators, it can be further optimized and bioluminescent output can be improved. This would enhance the usability of the lux system and give it a broader appeal than it has currently.
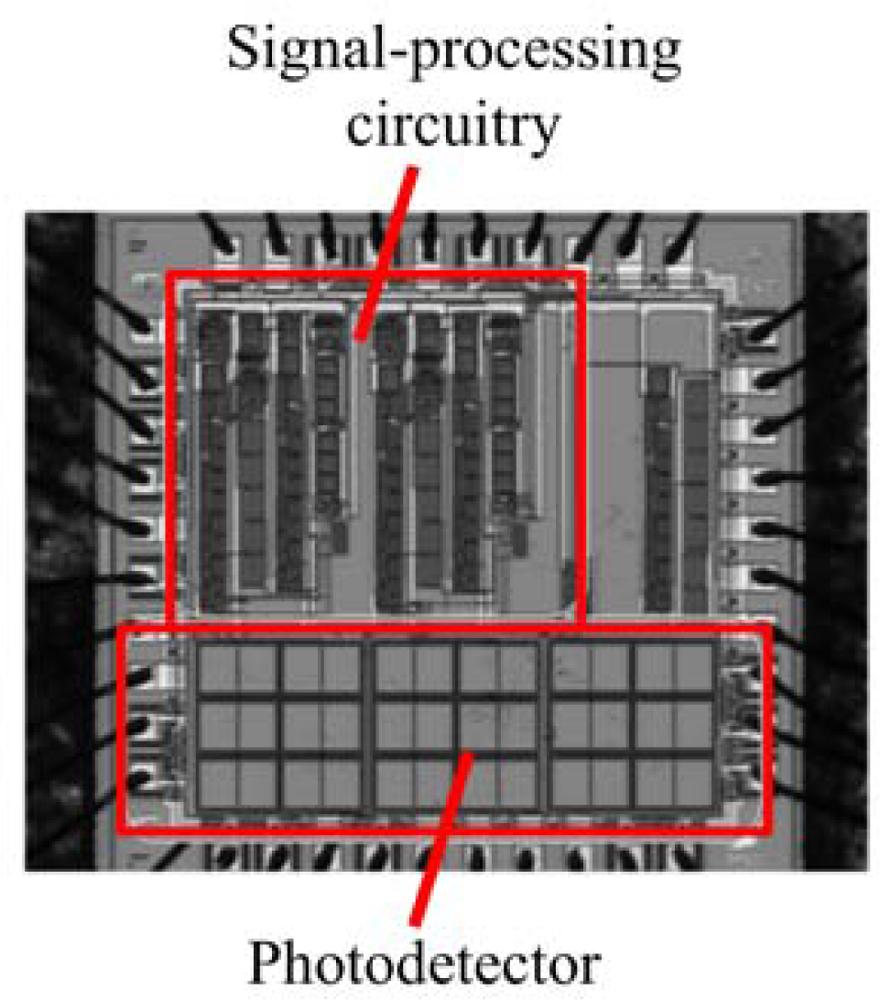
Separate from the biological modifications that could be engineered into the lux system, its autonomous nature already presents unique opportunities for merging it directly with existing detection technologies. In several instances, investigators lead by Sayler et al. [81–83] have integrated bacteria expressing the full lux cassette onto small footprint (∼1 cm3), low power (3.3 milliwatt) integrated circuit transducers. These miniaturized devices, called bioluminescent bioreporter integrated circuits (BBICs), can provide for the detection of the bioreporter optical signal, the distinguishing of this signal from noise, the digital processing of the signal, and local communication of the result within a single, self contained package (Figure 10).
When bioluminescent bacterial cells are interfaced to the BBIC, as few as 5,000 can be detected and distinguished from background [81] and it is sensitive enough to differentiate bioluminescent output levels stemming from changes in the concentration of the exposed stimulating analyte [81,82]. These devices could be paired with lux-expressing eukaryotic cells and then implanted into small animal models for direct internal imaging of reporter signals without the need to anesthetize or remove the animal from its natural habitat, offering unparalleled opportunities for studying changes in physiology and compound bioavailability under a wide range of conditions. Similarly, these cells could be complexed with microcircuitry capable of initiating hormone or therapeutic compound dosage. In this fashion, the lux-expressing cells could be programed to continuously monitor the body for specific target compounds, acting as real-time biosentinels that detect changes in physiology and whose resultant changes in bioluminescent output could trigger the release of counteractive compounds. This would allow the development of fully autonomous, implantable dose/response therapeutic devices.
5. Conclusions
While the use of bacterial bioluminescence as a reporter system has been employed for quite a long time, it is still a continually developing reporter system. There are many examples from the recent literature that demonstrate new and creative lux-based bacterial biosensors that are employed for myriad sensing applications, and there are also recent examples showing how modification of these genes has expanded their usage to new applications that were previously thought impossible. The unique ability of the lux system to produce a bioluminescent signal without exogenous substrate input has ensured that it will continue to find use in basic and applied scientific research for years to come. Whether or not it continues to be improved for function in eukaryotic cells may well decide the true extent of lux usage in the future, however, for the time being it remains both an interesting and practical example of the benefits available from an optical reporter system.
Acknowledgments
Portions of this review reflecting work by the authors was supported by the the National Institutes of Health, National Cancer Institute, Cancer Imaging Program, award number CA127745-01, the University of Tennessee Research Foundation Technology Maturation Funding program, National Science Foundation Division of Chemical, Bioengineering, Environmental, and Transport Systems (CBET) under award number CBET-0853780, and the Army Defense University Research Instrumentation Program.
References
- Close, D.M.; Ripp, S.; Sayler, G.S. Reporter proteins in whole-cell optical bioreporter detection systems, biosensor integrations, and biosensing applications. Sensors 2009, 9, 9147–9174. [Google Scholar]
- McElroy, W.; Hastings, J.; Sonnenfeld, V.; Coulombre, J. The requirement of riboflavin phosphate for bacterial luminescence. Science 1953, 118, 385–386. [Google Scholar]
- Strehler, B.; Harvey, E.; Chang, J.; Cormier, M. The luminescent oxidation of reduced riboflavin or reduced riboflavin phosphate in the bacterial luciferin-luciferase reaction. Proc. Natl. Acad. Sci. USA 1954, 40, 10–12. [Google Scholar]
- Belas, R.; Mileham, A.; Cohn, D.; Hilmen, M.; Simon, M.; Silverman, M. Bacterial bioluminescence: Isolation and expression of the luciferase genes from Vibrio harveyi. Science 1982, 218, 791–793. [Google Scholar]
- Gupta, R.K.; Patterson, S.S.; Ripp, S.; Sayler, G.S. Expression of the Photorhabdus luminescens lux genes (luxA, B, C, D, and E) in Saccharomyces cerevisiae. FEMS Yeast Res 2003, 4, 305–313. [Google Scholar]
- Patterson, S.S.; Dionisi, H.M.; Gupta, R.K.; Sayler, G.S. Codon optimization of bacterial luciferase (lux) for expression in mammalian cells. J. Ind. Microbiol. Biotechnol 2005, 32, 115–123. [Google Scholar]
- Close, D.M.; Patterson, S.S.; Ripp, S.; Baek, S.J.; Sanseverino, J.; Sayler, G.S. Autonomous bioluminescent expression of the bacterial luciferase gene cassette (lux) in a mammalian cell line. PLoS One 2010, 5, e12441. [Google Scholar]
- Nealson, K.H.; Hastings, J. Bacterial bioluminescence: Its control and ecological significance. Microbiol. Mol. Biol. Rev 1979, 43, 496–518. [Google Scholar]
- Meighen, E.A. Molecular biology of bacterial bioluminescence. Microbiol. Rev 1991, 55, 123–142. [Google Scholar]
- Swartzman, E.; Silverman, M.; Meighen, E. The luxR gene product of Vibrio harveyi is a transcriptional activator of the lux promoter. J. Bacteriol 1992, 174, 7490–7493. [Google Scholar]
- Engebrecht, J.; Nealson, K.; Silverman, M. Bacterial bioluminescence: Isolation and genetic analysis of functions from Vibrio fischeri. Cell 1983, 32, 773–781. [Google Scholar]
- Fernandezpinas, F.; Wolk, C. Expression of luxCD-E in Anabeaena sp. can replace the use of exogenous aldehyde for in vivo localization of transcription by luxAB. Gene 1994, 150, 169–174. [Google Scholar]
- Frackman, S.; Anhalt, M.; Nealson, K. Cloning, organization, and expression of the bioluminescence genes of Xenorhabdus luminescens. J. Bacteriol 1990, 172, 5767–5773. [Google Scholar]
- Engebrecht, J.; Simon, M.; Silverman, M. Measuring gene expression with light. Science 1985, 227, 1345–1347. [Google Scholar]
- Belas, R.; Simon, M.; Silverman, M. Regulation of lateral flagella gene transcription in Vibrio parahaemolyticus. J. Bacteriol 1986, 167, 210–218. [Google Scholar]
- Shaw, J.J.; Kado, C.I. Development of a Vibrio bioluminescence gene set to monitor phytopathogenic bacteria during the ongoing disease process in a nondisruptive manner. Nat. Biotechnol 1986, 4, 560–564. [Google Scholar]
- de Weger, L.; Dunbar, P.; Mahafee, F.; Lugtenberg, B.; Sayler, G. Use of bioluminescence markers to detect Pseudomonas spp. in the rhizosphere. Appl. Environ. Microbiol 1991, 57, 3641–3644. [Google Scholar]
- Ripp, S.; Nivens, D.E.; Ahn, Y.; Werner, C.; Jarrell, J.; Easter, J.P.; Cox, C.D.; Burlage, R.S.; Sayler, G.S. Controlled field release of a bioluminescent genetically engineered microorganism for bioremediation process monitoring and control. Environ. Sci. Technol 2000, 34, 846–853. [Google Scholar]
- King, J.M.H.; Digrazia, P.M.; Applegate, B.; Burlage, R.; Sanseverino, J.; Dunbar, P.; Larimer, F.; Sayler, G.S. Rapid, sensitive bioluminescent reporter technology for naphthalene exposure and biodegradation. Science 1990, 249, 778–781. [Google Scholar]
- Burlage, R.S.; Sayler, G.S.; Larimer, F. Monitoring of naphthalene catabolism by bioluminescence with nah-lux transcriptional fusions. J. Bacteriol 1990, 172, 4749–4757. [Google Scholar]
- Applegate, B.M.; Kehrmeyer, S.R.; Sayler, G.S. A chromosomally based tod-luxCDABE whole-cell reporter for benzene, toluene, ethybenzene, and xylene (BTEX) sensing. Appl. Environ. Microbiol 1998, 64, 2730–2735. [Google Scholar]
- Abd-El-Haleem, D.; Ripp, S.; Scott, C.; Sayler, G.S. A luxCDABE-based bioluminescent bioreporter for the detection of phenol. J. Ind. Microbiol. Biotechnol 2002, 29, 233–237. [Google Scholar]
- Davidov, Y.; Rozen, R.; Smulski, D.R.; Van Dyk, T.K.; Vollmer, A.C.; Elsemore, D.A.; LaRossa, R.A.; Belkin, S. Improved bacterial SOS promoter::lux fusions for genotoxicity detection. Mutat. Res 2000, 466, 97–107. [Google Scholar]
- Belkin, S.; Gu, M. Whole Cell Sensing Systems II: Applications; Springer-Verlag: Berlin, Germany, 2010. [Google Scholar]
- Ripp, S.; Layton, A.C.; Sayler, G.S. The microbe as a reporter: Microbial bioreporter sensing technologies for chemical and biological detection. In Environmental Microbiology: Current Technology and Water Applications; Sen, K., Ashbolt, N.J., Eds.; Caister Academic Press: Norfolk, UK, 2011; pp. 281–305. [Google Scholar]
- Hay, A.G.; Rice, J.F.; Applegate, B.M.; Bright, N.G.; Sayler, G.S. A bioluminescent whole-cell reporter for detection of 2,4-dichlorophenoxyacetic acid and 2,4-dichlorophenol in soil. Appl. Environ. Microbiol 2000, 66, 4589–4594. [Google Scholar]
- Toba, F.A.; Hay, A.G. A simple solid phase assay for the detection of 2,4-D in soil. J. Microbiol. Methods 2005, 62, 135–143. [Google Scholar]
- Burlage, R.S. Organic contaminant detection and biodegradation characteristics. In Methods in Molecular Biology/Bioluminescence; LaRossa, R., Ed.; Humana Press: Totowa, NJ, USA, 1998; pp. 259–268. [Google Scholar]
- Rozen, Y.; Nejidat, A.; Gartemann, K.H.; Belkin, S. Specific detection of p-chlorobenzoic acid by Escherichia coli bearing a plasmid-borne fcbA'::lux fusion. Chemosphere 1999, 38, 633–641. [Google Scholar]
- Wallace, W.H.; Fleming, J.T.; White, D.C.; Sayler, G.S. An algD-lux bioluminescent reporter plasmid to monitor alginate production in biofilms. Microb. Ecol 1994, 27, 225–239. [Google Scholar]
- Simpson, M.L.; Paulus, M.J.; Jellison, G.E.; Sayler, G.S.; Nivens, D.E.; Dionisi, H.M.; Ripp, S.; Applegate, B.; Patterson, G.; Bolton, E.; et al. Bioluminescent bioreporter integrated circuits (BBICs): Whole-cell environmental monitoring devices. Proceedings of the 30th International Conference on Environmental Systems (Society of Automotive Engineers), Toulouse, France, 10–13 July 2000.
- Eldridge, M.L.; Sanseverino, J.; Layton, A.C.; Easter, J.P.; Schultz, T.W.; Sayler, G.S. Saccharomyces cerevisiae BLYAS, a new bioluminescent bioreporter for detection of androgenic compounds. Appl. Environ. Microbiol 2007, 73, 6012–6018. [Google Scholar]
- Francis, K.P.; Joh, D.; Bellinger-Kawahara, C.; Hawkinson, M.J.; Purchio, T.F.; Contag, P.R. Monitoring bioluminescent Staphylococcus aureus infections in mice using a novel luxABCDE construct. Infect. Immun 2000, 68, 3594–3600. [Google Scholar]
- Flynn, H.C.; Meharg, A.A.; Bowyer, P.K.; Paton, G.I. Antimony bioavailability in mine soils. Environ. Pollut 2003, 124, 93–100. [Google Scholar]
- Ivask, A.; Green, T.; Polyak, B.; Mor, A.; Kahru, A.; Virta, M.; Marks, R. Fibre-optic bacterial biosensors and their application for the analysis of bioavailable Hg and As in soils and sediments from Aznalcollar mining area in Spain. Biosens. Bioelectron 2007, 22, 1396–1402. [Google Scholar]
- Corbisier, P.; Thiry, E.; Diels, L. Bacterial biosensors for the toxicity assessment of solid waste. Environ. Toxicol. Water Qual 1996, 11, 171–177. [Google Scholar]
- Peitzsch, N.; Eberz, G.; Nies, D.H. Alcaligenes eutrophus as a bacterial chromate sensor. Appl. Environ. Microbiol 1998, 64, 453–458. [Google Scholar]
- Tibazarwa, C.; Wuertz, S.; Mergeay, M.; Wyns, L.; van der Lelie, D. Regulation of the cnr cobalt and nickel resistance determinant of Ralstonia eutropha (Alcaligenes eutrophus) CH34. J. Bacteriol 2000, 182, 1399–1409. [Google Scholar]
- Riether, K.B.; Dollard, M.A.; Billard, P. Assessment of heavy metal bioavailability using Escherichia coli zntAp::lux and copAp::lux based biosensors. Appl. Microbiol. Biotechnol 2001, 57, 712–716. [Google Scholar]
- Lopes, N.; Hawkins, S.; Jegier, P.; Menn, F.-M.; Sayler, G.; Ripp, S. Detection of dichloromethane with a bioluminescent (lux) bacterial bioreporter. J. Ind. Microbiol. Biotechnol 2012, 39, 45–53. [Google Scholar]
- Belkin, S.; Smulski, D.R.; Dadon, S.; Vollmer, A.C.; Van Dyk, T.K.; LaRossa, R.A. A panel of stress-responsive luminous bacteria for the detection of selected classes of toxicants. Wat. Res 1997, 31, 3009–3016. [Google Scholar]
- Vollmer, A.C.; Belkin, S.; Smulski, D.R.; Van Dyk, T.K.; LaRossa, R.A. Detection of DNA damage by use of Escherichia coli carrying recA'-lux, uvrA'-lux, or alkA'-lux reporter plasmids. Appl. Environ. Microbiol 1997, 63, 2566–2571. [Google Scholar]
- Hwang, E.T.; Ahn, J.M.; Kim, B.C.; Gu, M.B. Construction of a nrdA::luxCDABE fusion and its use in Escherichia coli as a DNA damage biosensor. Sensors 2008, 8, 1297–1307. [Google Scholar]
- van Dyk, T.K.; DeRose, E.J.; Gonye, G.E. LuxArray, a high-density, genomewide transcription analysis of Escherichia coli using bioluminescent reporter strains. J. Bacteriol 2001, 183, 5496–5505. [Google Scholar]
- Sanseverino, J.; Gupta, R.K.; Layton, A.C.; Patterson, S.S.; Ripp, S.A.; Saidak, L.; Simpson, M.L.; Schultz, T.W.; Sayler, G.S. Use of Saccharomyces cerevisiae BLYES expressing bacterial bioluminescence for rapid, sensitive detection of estrogenic compounds. Appl. Environ. Microbiol 2005, 71, 4455–4460. [Google Scholar]
- Min, J.; Lee, C.W.; Moon, S.H.; LaRossa, R.A.; Gu, M.B. Detection of radiation effects using recombinant bioluminescent Escherichia coli strains. Radiat. Environ. Biophys 2000, 39, 41–45. [Google Scholar]
- Rupani, S.P.; Gu, M.B.; Konstantinov, K.B.; Dhurjati, P.S.; Van Dyk, T.K.; Larossa, R.A. Characterization of the stress response of a bioluminescent biological sensor in batch and continuous cultures. Biotechnol. Prog 1996, 12, 387–392. [Google Scholar]
- van Dyk, T.K.; Reed, T.R.; Vollmer, A.C.; LaRossa, R.A. Synergistic induction of the heat shock response in Escherichia coli by simultaneous treatment with chemical inducers. J. Bacteriol 1995, 177, 6001–6004. [Google Scholar]
- Jouanneau, S.; Durand, M.J.; Courcoux, P.; Blusseau, T.; Thouand, G. Improvement of the identification of four heavy metals in environmental samples by using predictive decision tree models coupled with a set of five bioluminescent bacteria. Environ. Sci. Technol 2011, 45, 2925–2931. [Google Scholar]
- Bang, Y.B.; Lee, S.E.; Rhee, J.H.; Choi, S.H. Evidence that expression of the Vibrio vulnificus hemolysin gene is dependent on cyclic AMP and cyclic AMP receptor protein. J. Bacteriol 1999, 181, 7639–7642. [Google Scholar]
- Belkin, S.; Smulski, D.R.; Vollmer, A.C.; Van Dyk, T.K.; LaRossa, R.A. Oxidative stress detection with Escherichia coli harboring a katG'::lux fusion. Appl. Environ. Microbiol 1996, 62, 2252–2256. [Google Scholar]
- Contag, C.H.; Contag, P.R.; Mullins, J.I.; Spilman, S.D.; Stevenson, D.K.; Benaron, D.A. Photonic detection of bacterial pathogens in living hosts. Mol. Microbiol 1995, 18, 593–603. [Google Scholar]
- Khang, Y.H.; Yang, Z.K.; Burlage, R.S. Measurement of iron-dependence of pupA promoter activity by a pup-lux bioreporter. J. Microbiol. Biotechnol 1997, 7, 352–355. [Google Scholar]
- Selifonova, O.V.; Eaton, R.W. Use of an ipb-lux fusion to study regulation of the isopropylbenzene catabolism operon of Pseudomonas putida RE204 and to detect hydrophobic pollutants in the environment. Appl. Environ. Microbiol 1996, 62, 778–783. [Google Scholar]
- Magrisso, S.; Belkin, S.; Erel, Y. Lead bioavailability in soil and soil components. Water Air Soil Pollut 2009, 202, 315–323. [Google Scholar]
- Larose, C.; Dommergue, A.; Marusczak, N.; Coves, J.; Ferrari, C.P.; Schneider, D. Bioavailable mercury cycling in polar snowpacks. Environ. Sci. Technol 2011, 45, 2150–2156. [Google Scholar]
- Yan, L.; Allen, M.S.; Simpson, M.L.; Sayler, G.S.; Cox, C.D. Direct quantification of N-(3-oxo-hexanoyl)-L-homoserine lactone in culture supernatant using a whole-cell bioreporter. J. Microbiol. Methods 2007, 68, 40–45. [Google Scholar]
- Heitzer, A.; Webb, O.F.; Thonnard, J.E.; Sayler, G.S. Specific and quantitative assessment of naphthalene and salicylate bioavailability by using a bioluminescent catabolic reporter bacterium. Appl. Environ. Microbiol 1992, 58, 1839–1846. [Google Scholar]
- Everhart, J.L.; McNear, D.; Peltier, E.; van der Lelie, D.; Chaney, R.L.; Sparks, D.L. Assessing nickel bioavailability in smelter-contaminated soils. Sci. Total Environ 2006, 367, 732–744. [Google Scholar]
- Abd-El-Haleem, D.; Ripp, S.; Zaki, S.; Sayler, G.S. Detection of nitrate/nitrite bioavailability in wastewater using a luxCDABE-based Klebsiella oxytoca bioluminescent bioreporter. J. Microbiol. Biotechnol 2007, 17, 1254–1261. [Google Scholar]
- Lee, H.J.; Gu, M.B. Construction of a sodA::luxCDABE fusion Escherichia coli: Comparison with a katG fusion strain through their responses to oxidative stresses. Appl. Microbiol. Biotechnol 2003, 60, 577–580. [Google Scholar]
- Layton, A.C.; Muccini, M.; Ghosh, M.M.; Sayler, G.S. Construction of a bioluminescent reporter strain to detect polychlorinated biphenyls. Appl. Environ. Microbiol 1998, 64, 5023–5026. [Google Scholar]
- Ripp, S.; Applegate, B.; Nivens, D.E.; Simpson, M.L.; Sayler, G.S. Advances in whole-cell bioluminescent bioreporters for environmental monitoring and chemical sensing. Proceedings of the AIChE Annual Meeting, Los Angeles, CA, USA, 12–17 November 2000.
- Shimizu, T.; Ohta, Y.; Tsutsuki, H.; Noda, M. Construction of a novel bioluminescent reporter system for investigating Shiga toxin expression of enterohemorrhagic Escherichia coli. Gene 2011, 478, 1–10. [Google Scholar]
- Hansen, L.H.; Sorensen, S.J. Detection and quantification of tetracyclines by whole cell biosensors. FEMS Microbiol. Lett 2000, 190, 273–278. [Google Scholar]
- Elad, T.; Benovich, E.; Magrisso, S.; Belkin, S. Toxicant identification by a luminescent bacterial bioreporter panel: Application of pattern classification algorithms. Environ. Sci. Technol 2008, 42, 8486–8491. [Google Scholar]
- Elad, T.; Lee, J.H.; Gu, M.B.; Belkin, S. Microbial cell arrays. In Whole Cell Sensing Systems I: Reporter Cells and Devices; Belkin, S., Gu, M.B., Eds.; Springer: Berlin, Germany, 2010; Volume 117; pp. 85–108. [Google Scholar]
- Shingleton, J.T.; Applegate, B.M.; Nagel, A.C.; Bienkowski, P.R.; Sayler, G.S. Induction of the tod operon by trichloroethylene in Pseudomonas putida TVA8. Appl. Environ. Microbiol 1998, 64, 5049–5052. [Google Scholar]
- Burlage, R.S.; Patek, D.R.; Everman, K.R. Method for Detection of Buried Explosives Using a Biosensor. U.S. Patent 5,972,638, October 1999.
- Vollmer, A.C.; Kwakye, S.; Halpern, M.; Everbach, E.C. Bacterial stress response to 1-megahertz pulsed ultrasound in the presence of microbubbles. Appl. Environ. Microbiol 1998, 64, 3927–3931. [Google Scholar]
- Elasri, M.O.; Miller, R.V. A Pseudomonas aeruginosa biosensor responds to exposure to ultraviolet radiation. Appl. Microbiol. Biotechnol 1998, 50, 455–458. [Google Scholar]
- Bakhrat, A.; Eltzov, E.; Finkelstein, Y.; Marks, R.S.; Raveh, D. UV and arsenate toxicity: A specific and sensitive yeast bioluminescence assay. Cell Biol. Toxicol 2011, 27, 227–236. [Google Scholar]
- Erbe, J.L.; Adams, A.C.; Taylor, K.B.; Hall, L.M. Cyanobacteria carrying an smt-lux transcriptional fusion as biosensors for the detection of heavy metal cations. J. Ind. Microbiol 1996, 17, 80–83. [Google Scholar]
- Almashanu, S.; Musafia, B.; Hadar, R.; Suissa, M.; Kuhn, J. Fusion of luxA and luxB and its expression in Escherichia coli, Saccharomyces cerevisiae and Drosophila melanogaster. J. Biolumin. Chemilumin 1990, 5, 89–97. [Google Scholar]
- Kirchner, G.; Roberts, J.L.; Gustafson, G.D.; Ingolia, T.D. Active bacterial luciferase from a fused gene: Expression of a Vibrio harveyi luxAB translational fusion in bacteria, yeast and plant cells. Gene 1989, 81, 349–354. [Google Scholar]
- Olsson, O.; Koncz, C.; Szalay, A.A. The use of the luxA gene of the bacterial luciferase operon as a reporter gene. Mol. Gen. Genet 1988, 215, 1–9. [Google Scholar]
- Close, D.; Hahn, R.; Ripp, S.; Sayler, G.S. Determining toxicant bioavailability using a constitutively bioluminescent human cell line. Proceedings for the 3rd Annual Biomedical Science and Engineering Conference, Oak Rige, TN, USA, 15–17 March 2011.
- Close, D.M.; Hahn, R.; Patterson, S.S.; Ripp, S.; Sayler, G.S. Comparison of human optimized bacterial luciferase, firefly luciferase, and green fluorescent protein for continuous imaging of cell culture and animal models. J. Biomed. Opt 2011, 16, e12441. [Google Scholar]
- Craney, A.; Hohenauer, T.; Xu, Y.; Navani, N.K.; Li, Y.F.; Nodwell, J. A synthetic luxCDABE gene cluster optimized for expression in high-GC bacteria. Nucleic Acids Res 2007. [Google Scholar] [CrossRef]
- Yagur-Kroll, S.; Belkin, S. Upgrading bioluminescent bacterial bioreporter performance by splitting the lux operon. Anal. Bioanal. Chem 2011, 400, 1071–1082. [Google Scholar]
- Nivens, D.E.; McKnight, T.E.; Moser, S.A.; Osbourn, S.J.; Simpson, M.L.; Sayler, G.S. Bioluminescent bioreporter integrated circuits: Potentially small, rugged and inexpensive whole-cell biosensors for remote environmental monitoring. J. Appl. Microbiol 2004, 96, 33–46. [Google Scholar]
- Ripp, S.; DiClaudio, M.; Sayler, G. Biosensors as environmental monitors. In Environmental Microbiology, 2nd ed; Mitchell, R., Gu, J., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2010; pp. 213–233. [Google Scholar]
- Vijayaraghavan, R.; Islam, S.; Zhang, M.; Ripp, S.; Caylor, S.; Bull, N.D.; Moser, S.; Terry, S.; Blalock, B.; Sayler, G. A bioreporter bioluminescent integrated circuit for very low-level chemical sensing in both gas and liquid environments. Sens. Actuat. B-Chem 2007, 123, 922–928. [Google Scholar]
- Simpson, M.L.; Sayler, G.S.; Applegate, B.M.; Ripp, S.; Nivens, D.E.; Paulus, M.J.; Jellison, G.E. Bioluminescent-bioreporter integrated circuits form novel whole-cell biosensors. Trends Biotechnol 1998, 16, 332–338. [Google Scholar]




| Analyte | Time for induction | Concentration | Reference |
|---|---|---|---|
| 2,3 Dichlorophenol | 2 h | 50 mg/L | [23] |
| 2,4,6 Trichlorophenol | 2 h | 10 mg/L | [23] |
| 2,4-D | 20–60 min | 0.44 mg/L | [26,27] |
| 3-Xylene | Hours | 3 μM | [28] |
| 4-Chlorobenzoate | 1 h | 380 μM–6.5 mM | [29] |
| 4-Nitrophenol | 2 h | 0.25 mg/L | [23] |
| Alginate production | 1 h | 50–150 mM NaCl | [30] |
| Ammonia | 30 min | 20 μM | [31] |
| Androgenic chemicals | 3–4 h | 10−9–10−10 M | [32] |
| Antibiotic effectiveness against Staphylococcus aureus infections in mice | 4 h | 100 CFU | [33] |
| Antimony (antimonate and antimonite) | 3–4 h | 0.1 mg/L | [34] |
| Arsenic | 3–4 h | 80 μg/L As(V); 8 μg/L As(III) | [35] |
| BTEX (benzene, toluene, ethylbenzene, xylene) | 1–4 h | 0.03–50 mg/L | [21] |
| Cadmium | 4 h | 19 mg/kg | [36] |
| Chlorodibromomethane | 2 h | 20 mg/L | [23] |
| Chloroform | 2 h | 300 mg/L | [23] |
| Chromate | 1 h | 10 μM | [37] |
| Cobalt | 9 μM | 2.0 mM | [38] |
| Copper | 1 h | 0.05 mg/L | [39] |
| Dichloromethane | 1–2 h | ∼0.01 mg/L | [40] |
| DNA damage (cumene hydroperoxide) | 50 min | 6.25 mg/mL | [41] |
| DNA damage (mitomycin) | 1 h | 0.032 μg/mL | [42] |
| Not specified | 0.31 μg/mL | [43] | |
| DNA damage and other cell stressors/activators | A library of luxCDABE-based fusions with 689 E. coli gene promoters | [44] | |
| Estrogenic chemicals | 1 h | 10−11 M | [45] |
| Gamma-irradiation | 1.5 h | 1.5–200 Gy | [46] |
| Heat shock | 20 min | Various, depending on chemical inducer used | [47,48] |
| Heavy metals | A multi-bioreporter panel for detecting and identifying multiple heavy metal contaminants in a single sample | [49] | |
| Hemolysin production | Not specified | 5 mM cAMP | [50] |
| Hydrogen peroxide | 20 min | 0.1 mg/L | [51] |
| in vivo monitoring of Salmonella typhimurium infections in living mice | 4 h | 100 CFU | [52] |
| Iron | Hours | 10 nM–1 μM | [53] |
| Isopropyl benzene | 1–4 h | 1–100 μM | [54] |
| Lead | 1 h | 0.33 mg/L | [39,55] |
| Mercury | 2 h | 0.5 ng/L | [56] |
| N-acyl homoserine lactones (3-oxo-C6-HSL) | 200 min | 3 nM | [57] |
| Naphthalene | 8–24 min | 12–120 μM | [58] |
| Nickel | 4–6 h | 0.1 μM | [38,59] |
| Nitrate | ∼1 h | 1 mg/L | [60] |
| Organic peroxides | 20 min | Not specified | [51] |
| Oxidative stress | Not specified | 0.015 ppm (paraquat) | [61] |
| PCBs | 1–3 h | 0.8 μM | [62] |
| p-chlorobenzoic acid | 40 min | 0.06 g/L | [29] |
| p-cymene | <30 min | 60 ppb | [63] |
| Pentachlorophenol | 2 h | 0.008 mg/L | [23] |
| Phenol | 2 h | 16 mg/L | [23] |
| Salicylate | 15 min | 36 μM | [58] |
| Shiga toxin expression in E. coli | Gene expression profiling in enterohemorrhagic E. coli | [64] | |
| Tetracycline | 40 min | 5 ng/mL | [65] |
| Toxicity monitoring | Use of multi-bioreporter arrays to survey and identify multiple chemicals within single samples | [66,67] | |
| Trichloroethylene | 1–1.5 h | 5–80 μM | [68] |
| Trinitrotoluene | Not specified | Not specified | [69] |
| Ultrasound | 1 h | 500 W/cm2 | [70] |
| Ultraviolet light (bacterial) | 1 h | 2.5–20 J/m2 | [71] |
| Ultraviolet light (yeast) | 1 h | 7 mJ/cm2 | [72] |
| Zinc | 4 h | 0.5–4 μM | [73] |
| Gene | Type | Begin | End | Length | I a | T b | CodRg c | P d | Translated e |
|---|---|---|---|---|---|---|---|---|---|
| luxA (wt) | 1 | 61 | 1,083 | 1,023 | 45 | 42 | 791 | 0.7 | 67.01 |
| luxA (op) | 1 | 1 | 1,083 | 1,083 | 66 | 42 | 1,910 | 0.88 | 181.78 |
| luxB (wt) | 1 | 1 | 984 | 984 | 51 | 38 | 585 | 0.97 | 46.37 |
| luxB (op) | 1 | 1 | 984 | 984 | 66 | 41 | 1,952 | 0.99 | 185.60 |
aInitiaion signal;bTermination;cCoding region score;dProbability of an exon;eExon score.
© 2012 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Close, D.; Xu, T.; Smartt, A.; Rogers, A.; Crossley, R.; Price, S.; Ripp, S.; Sayler, G. The Evolution of the Bacterial Luciferase Gene Cassette (lux) as a Real-Time Bioreporter. Sensors 2012, 12, 732-752. https://doi.org/10.3390/s120100732
Close D, Xu T, Smartt A, Rogers A, Crossley R, Price S, Ripp S, Sayler G. The Evolution of the Bacterial Luciferase Gene Cassette (lux) as a Real-Time Bioreporter. Sensors. 2012; 12(1):732-752. https://doi.org/10.3390/s120100732
Chicago/Turabian StyleClose, Dan, Tingting Xu, Abby Smartt, Alexandra Rogers, Robert Crossley, Sarah Price, Steven Ripp, and Gary Sayler. 2012. "The Evolution of the Bacterial Luciferase Gene Cassette (lux) as a Real-Time Bioreporter" Sensors 12, no. 1: 732-752. https://doi.org/10.3390/s120100732
APA StyleClose, D., Xu, T., Smartt, A., Rogers, A., Crossley, R., Price, S., Ripp, S., & Sayler, G. (2012). The Evolution of the Bacterial Luciferase Gene Cassette (lux) as a Real-Time Bioreporter. Sensors, 12(1), 732-752. https://doi.org/10.3390/s120100732




