Are the Genes nadA and norB Involved in Formation of Aflatoxin G1?
Abstract
:1. Introduction
2. Results
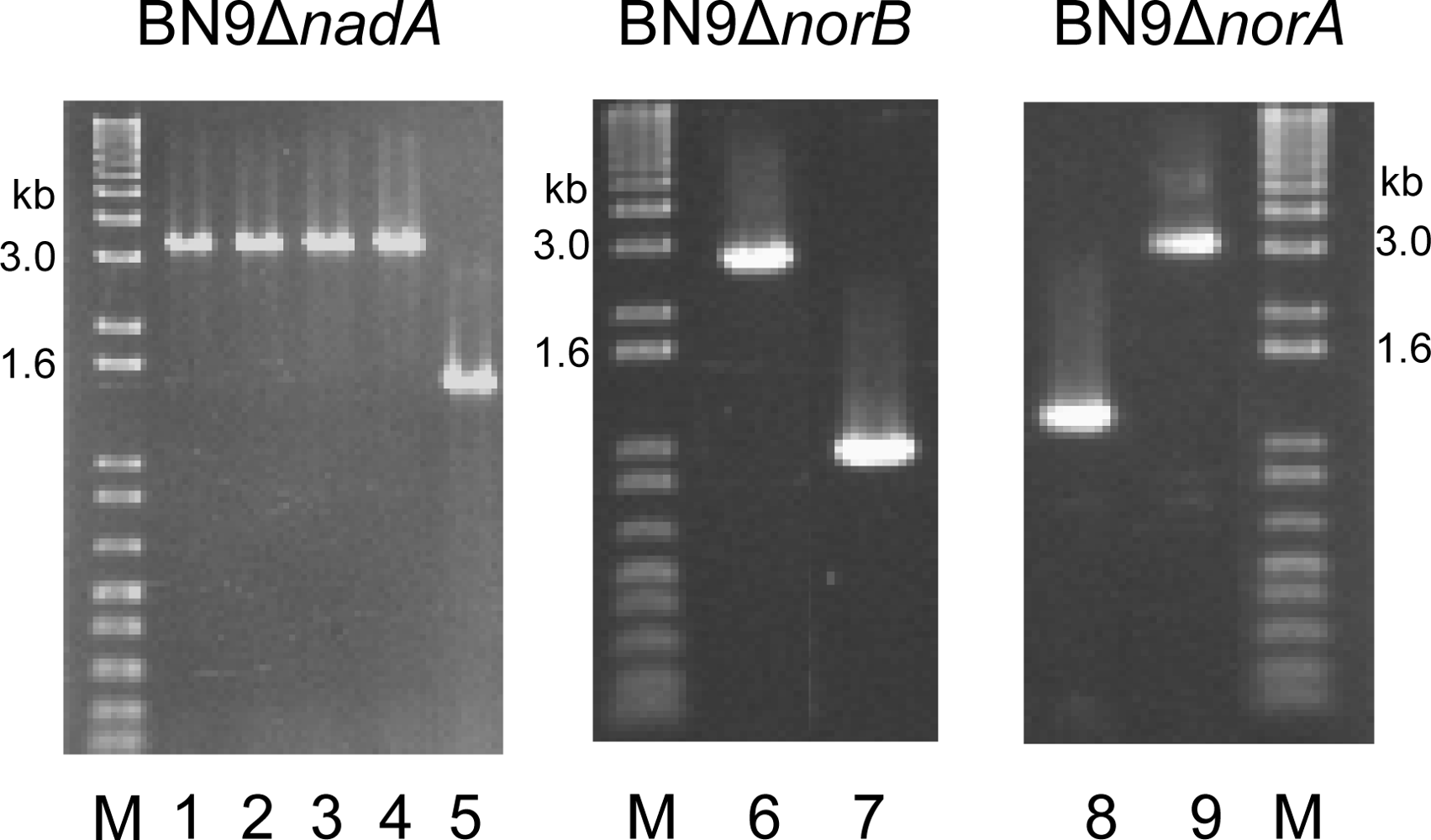
2.1. Production of gene disruptants
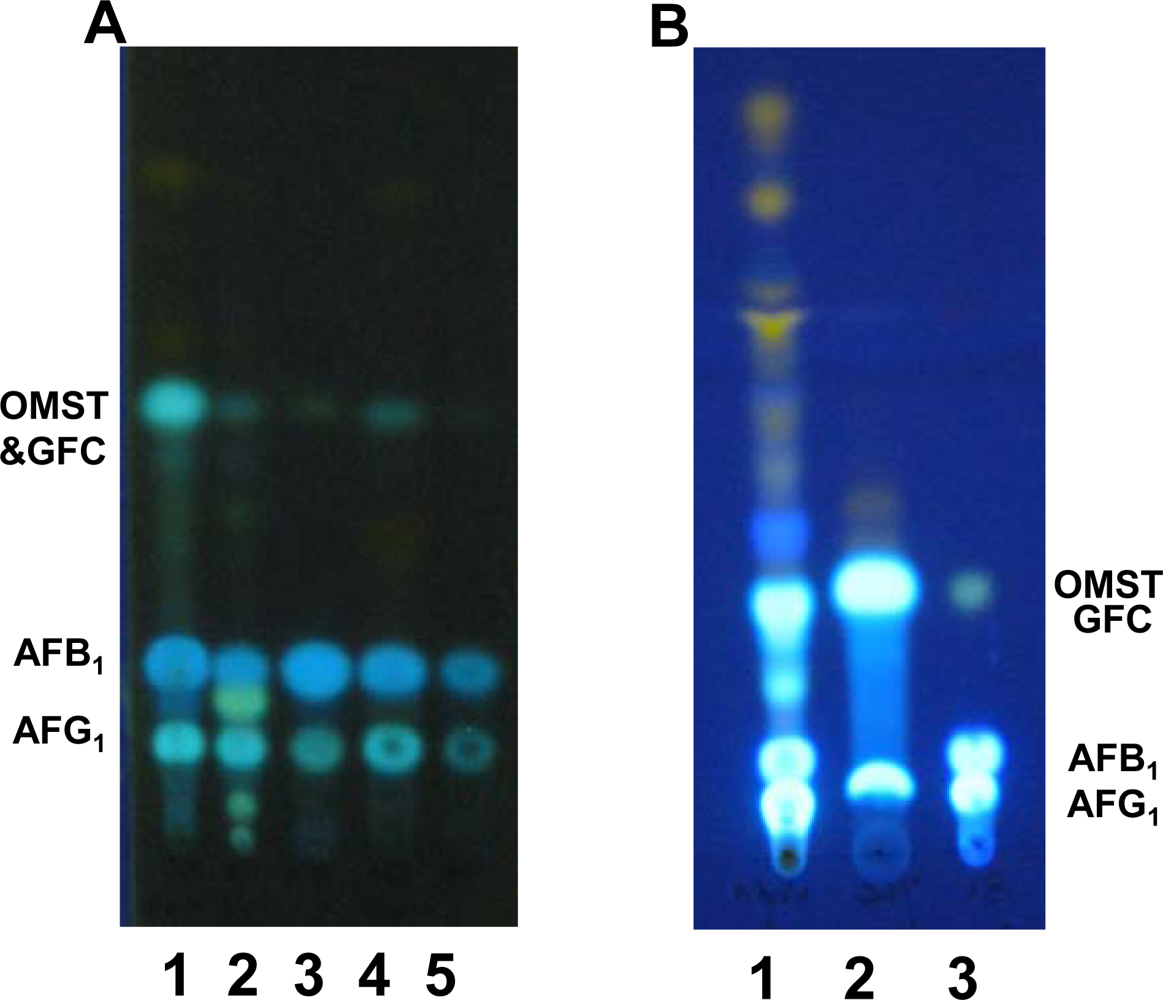
2.2. TLC analyses of extracts of BN9ΔnadA, BN9ΔnorA and BN9ΔnorB
2.3. LC/MS analysis of transformant extracts
2.4. LC/MS analysis of BN9ΔnorA and BN9ΔnorB transformant extracts
2.5. Comparsion of NadA in A. parasiticus and A. flavus
3. Discussion
4. Experimental Section
4.1. Construction of gene knockout vectors
4.2. Thin layer chromatography (TLC) and liquid chromatography mass spectrometry (LC/MS)
4.3. Sequence comparison
5. Conclusions
Acknowledgments
References
- Bhatnagar, D; Yu, J; Ehrlich, KC. Toxins of filamentous fungi. In Fungal Allergy and Pathogenicity; Breitenbach, M, Crameri, R, Lehrer, SB, Eds.; Karger: Basel, 2002; pp. 167–206. [Google Scholar]
- Brown, BJ; Hyun, JW; Duvvuri, S; Karplus, PA; Massey, V. The role of glutamine-114 in old yellow enzyme. J. Biol. Chem 2002, 277, 2138–2145. [Google Scholar]
- Cai, JJ; Zeng, H; Shima, Y; Hatabayashi, H; Nakagawa, H; Ito, Y; Nakajima, H; Yabe, K. Involvement of the nadA gene in formation of G-group aflatoxins in Aspergillus parasiticus. Fungal. Genet. Biol 2008, 45, 1081–1093. [Google Scholar]
- Cary, JW; Bhatnagar, D; Linz, JE. Aflatoxins: biological significance and regulation of biosynthesis. In Microbial Foodborne Diseases: Mechanisms of Pathogenesis and Toxin Synthesis; Cary, JW, Linz, JE, Bhatnagar, D, Eds.; Technomic Publishing Company: Lancaster, PA, 2000; pp. 317–361. [Google Scholar]
- Chang, P-K. A highly efficient gene-targeting system for Aspergillus parasiticus. Lett. Appl. Microbiol 2008, 13, 13. [Google Scholar]
- Chang, P-K; Ehrlich, KC; Linz, JE; Bhatnagar, D; Cleveland, TE; Bennett, JW. Characterization of the Aspergillus parasiticus niaD and niiA gene cluster. Curr. Genet 1996, 30, 68–75. [Google Scholar]
- Ehrlich, KC; Chang, P-K; Yu, J; Cotty, PJ. Aflatoxin biosynthesis cluster gene cypA is required for G aflatoxin formation. Appl. Environ. Microbiol 2004, 70, 6518–6524. [Google Scholar]
- Fox, KM; Karplus, PA. Old yellow enzyme at 2 A resolution: overall structure, ligand binding, and comparison with related flavoproteins. Structure 1994, 2, 1089–1105. [Google Scholar]
- Henderberg, A; Bennett, JW; Lee, LS. Biosynthetic origin of aflatoxin G1: Confirmation of sterigmatocystin and lack of confirmation of aflatoxin B1 as precursors. J. Gen. Microbiol 1988, 134, 661–667. [Google Scholar]
- Kubodera, T; Yamashita, N; Nishimura, A. Transformation of Aspergillus sp. and Trichoderma reesei using the pyrithiamine resistance gene (ptrA) of Aspergillus oryzae. Biosci. Biotechnol. Biochem 2002, 66, 404–406. [Google Scholar]
- Price, MS; Yu, J; Nierman, WC; Kim, HS; Pritchard, B; Jacobus, CA; Bhatnagar, D; Cleveland, TE; Payne, GA. The aflatoxin pathway regulator AFLR induces gene transcription inside and outside of the aflatoxin biosynthetic cluster. Fems. Micro. Biol. Lett 2006, 255, 275–279. [Google Scholar]
- Stubblefield, RD; Shotwell, OL; Shannon, GM. Aflatoxins M1 and M2 and parasiticol: thin layer chromatography and physical and chemical properties. J. Assoc. Off. Anal. Chem 1972, 55, 762–767. [Google Scholar]
- Udwary, DW; Casillas, LK; Townsend, CA. Synthesis of 11-hydroxy O-methylsterigmatocystin and the role of a cytochrome P-450 in the final step of aflatoxin biosynthesis. J. Am. Chem. Soc 2002, 124, 5294–5303. [Google Scholar]
- Williams, RE; Bruce, NC. New uses for an Old Enzyme--the Old Yellow Enzyme family of flavoenzymes. Microbiology 2002, 148, 1607–1614. [Google Scholar]
- Yu, J; Chang, P-K; Bhatnagar, D; Cleveland, TE. Cloning of a sugar utilization gene cluster in Aspergillus parasiticus. Biochim. Biophys. Acta 2000, 1493, 211–214. [Google Scholar]
- Yu, J; Chang, P-K; Ehrlich, KC; Cary, JW; Montalbano, B; Dyer, JM; Bhatnagar, D; Cleveland, TE. Characterization of the critical amino acids of an Aspergillus parasiticus cytochrome P-450 monooxygenase encoded by ordA that is involved in the biosynthesis of aflatoxins B1, G1, B2, and G2. Appl. Environ. Microbiol 1998, 64, 4834–4841. [Google Scholar]
- Yu, J; Chang, PK; Ehrlich, KC; Cary, JW; Bhatnagar, D; Cleveland, TE; Payne, GA; Linz, JE; Woloshuk, CP; Bennett, JW. Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol 2004, 70, 1253–1262. [Google Scholar]



| LC Peak | Retention time, min (relative peak height) | Mass/charge [M+H+] (relative peak intensity) | |
|---|---|---|---|
| A. BN9ΔnadA | Peak 1 | Peak 2 | |
| 1 | 3.7 (4) | 371 (100) | 363 (6) |
| 2 | 4.3 (69) | 329 (100) | 355 (15) |
| 3 | 4.5 (100) | 313 (100) | 329 (39) |
| 4
| 5.3 (2)
| 387 (100)
| 339 (24)
|
| B. BN9ΔnadA-TLC | |||
| 1 | 4.1 (50) | 329 (100) | 387 (15) |
| 2 | 4.3 (100) | 329 (100) | 383 (7) |
| 3 | 5.0 (7) | 339 (100) | 291 (46) |
| 4
| 5.4 (20)
| 387 (100)
| |
| C. BN9ΔnorB | |||
| 1 | 3.7 (23) | 363 (100) | 361 (18) |
| 2 | 4.2 (5) | 355 (100) | 371 (6) |
| 3 | 4.3 (23) | 329 (100) | 311 (7) |
| 4 | 4.5 (100) | 313 (100) | 327 (37) |
| 5
| 5.0 (3)
| 339 (100)
| |
| D. BN9ΔnorA | |||
| 3 | 4.3 (100) | 329 (100) | 311 (9) |
| 4 | 4.4 (82) | 313 (100) | 329 (25) |
| 5
| 5.0 (20)
| 339 (100)
| 295 (5)
|
| E. BN9control | |||
| 1 | 4.3 (100) | 329 (100) | 311 (6) |
| 2 | 4.5 (94) | 313 (100) | 285 (5) |
| 3 | 5.0 (16) | 339 (100) | 295 (2) |
© 2008 by MDPI This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ehrlich, K.C.; Scharfenstein, L.L., Jr.; Montalbano, B.G.; Chang, P.-K. Are the Genes nadA and norB Involved in Formation of Aflatoxin G1? Int. J. Mol. Sci. 2008, 9, 1717-1729. https://doi.org/10.3390/ijms9091717
Ehrlich KC, Scharfenstein LL Jr., Montalbano BG, Chang P-K. Are the Genes nadA and norB Involved in Formation of Aflatoxin G1? International Journal of Molecular Sciences. 2008; 9(9):1717-1729. https://doi.org/10.3390/ijms9091717
Chicago/Turabian StyleEhrlich, Kenneth C., Leslie L. Scharfenstein, Jr., Beverly G. Montalbano, and Perng-Kuang Chang. 2008. "Are the Genes nadA and norB Involved in Formation of Aflatoxin G1?" International Journal of Molecular Sciences 9, no. 9: 1717-1729. https://doi.org/10.3390/ijms9091717




