Molecular, Genetic and Agronomic Approaches to Utilizing Pulses as Cover Crops and Green Manure into Cropping Systems
Abstract
:1. Introduction
2. Cover Crops, Advantages and Disadvantages
3. Focus on Leguminous Cover Crops
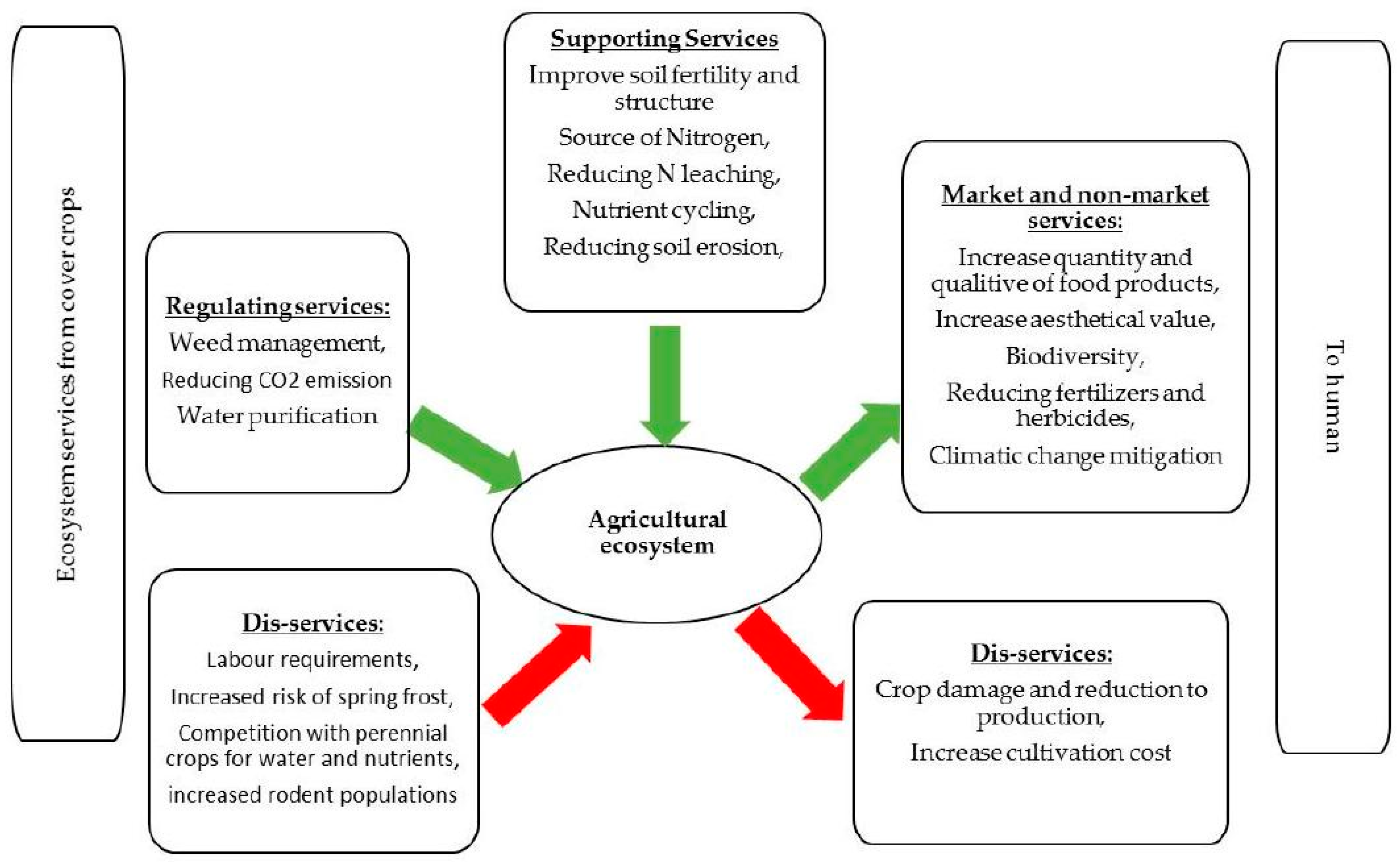
4. Regulating Services from the Leguminous Cover Crops to Agro-Ecosystems
4.1. Weed Management
4.2. Reducing CO2 Emission
- (a)
- In almost all cases, the no-till systems are highly dependent on herbicide use such as the non-selective glyphosate or other contact herbicides that would have higher environmental impact such as terrestrial ecotoxicity.
- (b)
- Yields are often maintained with higher mineral nitrogen input due to tillage operations especially in the first years of continuous use of reduced tillage in temperate climates [50].
- (c)
- Control of weeds (annual or perennial) tends to depend on tillage operations.
- (d)
- Launch of appropriate soil conservation technologies similar to what has been successfully used with soybean for decades in mainstream agriculture [51].
- (e)
- Integration of mulch seeding or direct seeding of leguminous cover crops in systems where grasses have been used as a superior mulch (e.g., oats for annual weeds control due to competition and allelopathy).
5. The Most Common Grain Legumes as Cover Crops
5.1. Cowpea (Vigna unguiculata L. Walp.) as a Cover Crop
5.1.1. Origin and Adaptation
5.1.2. Genetic Diversity
5.1.3. Important Traits for Breeding and Molecular Approaches
5.2. Faba Bean (Vicia faba L.) as a Cover Crop
5.2.1. Origin and Adaptation
5.2.2. Genetic Diversity
5.2.3. Important Traits for Breeding
5.2.4. Molecular Approach for Breeding
5.3. Pea (Pisum sativum L.) as a Cover Crop
5.3.1. Origin and Adaptation
5.3.2. Genetic Diversity
5.3.3. Important Traits for Breeding
5.3.4. Molecular Approach for Breeding
6. Conclusions
- To fully exploit their benefits, proper integration into agro-ecosystems should be safeguarded. In this context, the interaction of the effects with various components such as soil types, wet or dry growing conditions, or lodging should be validated.
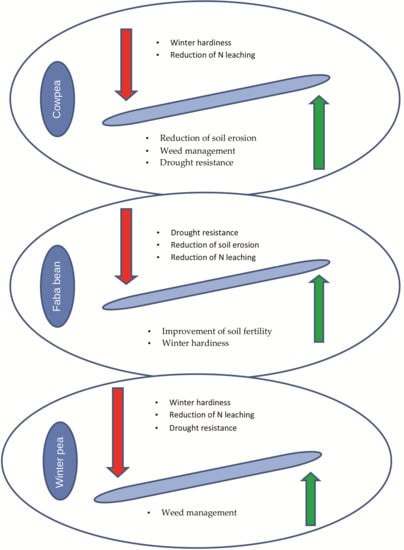
- Different roles for major grain legumes have been attributed. Cowpea could be an excellent summer cover crop that contributes to the weed suppression, reduction of soil erosion and increase of soil N. On the other hand, the importance of faba bean, as winter cover crop, related to its ability of N fixation has been monitored. Finally, peas are related to their enhanced ability to compete with weeds.
- Generally, high level of genetic diversity exists in the germplasm of the aforementioned species.
- Recent developments into whole genome sequencing (e.g., cowpeas) would allow significant contributions into crop improvements.
- Despite the progress in breeding programs, the existing genetic diversity and molecular tools have not been completely exploited.
7. Prospects for Future Research
- Sound empirical data (from field trials) are needed to quantify the positive effects of legumes on major agronomic characteristics such as yields, weed competition, nitrogen use, P and K availability and general crop health. For example, clear differences of its effects on the succeeding crops should be manifested as related to sowing time (autumn vs. spring sowing times).
- Grain legume crop improvements would be documented in the near future. Those crop improvements would be based upon gene expression information and other resources that are rapidly accumulated. In cowpeas, for example, breeding programs have been starting to exploit these resources for molecular breeding, especially for MARS and MABC. The new publicly available resources and knowledge would help the breeding of cowpea in order to become an ideal summer cover crop. In the post genomic era, the ultimate achievement for a breeder will be the simple detection of the chromosome sections that are inherited from each parent in order to assist the selection procedure and diminish the need for widespread field trials.
Acknowledgments
Conflicts of Interest
References
- Wittwer, R.A.; Dorn, B.; Jossi, W.; van der Heijden, M.G.A. Cover crops support ecological intensification of arable cropping systems. Sci. Rep. 2017. [Google Scholar] [CrossRef] [PubMed]
- Travlos, I.S. Legumes as cover crops or components of intercropping systems and their effects on weed populations and crop productivity. In Progress in Food Science and Technology; Greco, A.J., Ed.; Nova Science Publishers Inc.: Hauppauge, NY, USA, 2010; pp. 151–164. [Google Scholar]
- Smith, S.M.; Frye, W.W.; Varco, J.J. Legume winter cover crops. Adv. Soil Sci. 1987, 7, 95–139. [Google Scholar]
- Singh, Y.; Singh, B.; Ladha, J.K.; Khind, C.S.; Gupta, R.K.; Meelu, O.P.; Pasuquin, E. Long-term effects of organic inputs on yield and soil fertilityin the rice-wheat rotation. Soil Sci. Soc. Am. J. 2004, 63, 845–853. [Google Scholar]
- Meisinger, J.J.; Hargrove, W.L.; Mikkelsen, R.L.; Williams, J.R.; Benson, V.W. Effects of cover crops on groundwater quality. In Cover Crops for Clean Water; Meisinger, J.J., Hargrove, W.L., Eds.; SWCS: Ankeny, IA, USA, 1991. [Google Scholar]
- Batary, P.; Baldi, A.; Kleijn, D.; Tscharntke, T. Landscape-moderated biodiversity effects of agri-environmental management: A meta-analysis. Proc. R. Soc. Biol. Sci. 2011, 278, 1894–1902. [Google Scholar] [CrossRef] [PubMed]
- Christ, K.L.; Burritt, R.L. Critical environmental concerns in wine production: An integrative review. J. Clean. Prod. 2013, 53, 232–242. [Google Scholar] [CrossRef]
- Török, P.; Vida, E.; Deák, B.; Lengyel, B.; Tóthmérész, S.Z. Grassland restoration on former croplands in Europe: An assessment of applicability of techniques and costs. Biodivers. Conserv. 2011, 20, 2311–2332. [Google Scholar] [CrossRef]
- Hartwig, N.L.; Ammon, H.U. 50th Anniversary—Invited article—Cover crops and living mulches. Weed Sci. 2002, 50, 688–699. [Google Scholar] [CrossRef]
- Miglecz, T.; Valko, O.; Torok, P.; Deak, B.; Kelemen, A.; Donko, A.; Drexler, D.; Tothmeresz, B. Establishment of three cover crop mixtures in vineyards. Sci. Hortic. 2015, 197, 117–123. [Google Scholar] [CrossRef]
- Ingels, C.A.; Scow, K.M.; Whisson, D.A.; Drenovsky, R.E. Effects of cover crops on grapevines, yield, juice composition, soil microbial ecology, and gopher activity. Am. J. Enol. Vitic. 2005, 56, 19–29. [Google Scholar]
- Volaire, F.; Lelievre, F. Role of Summer Dormant Perennial Grasses as Intercrops in Rainfed Mediterranean Vineyards. Crop Sci. 2010, 50, 2046–2054. [Google Scholar] [CrossRef]
- Hatch, T.A.; Hickey, C.C.; Wolf, T.K. Cover Crop, Rootstock, and Root Restriction Regulate Vegetative Growth of Cabernet Sauvignon in a Humid Environment. Am. J. Enol. Vitic. 2011, 62, 298–311. [Google Scholar] [CrossRef]
- Tesic, D.; Keller, M.; Hutton, R.J. Influence of vineyard floor management practices on grapevine vegetative growth, yield, and fruit composition. Am. J. Enol. Vitic. 2007, 58, 1–11. [Google Scholar]
- Giese, G.; Wolf, T.K. Root Pruning and Groundcover to Optimize Vine Vigor and Berry Composition in Cabernet Sauvignon Grapevines. Am. J. Enol. Vitic. 2009, 60, 551a. [Google Scholar]
- Monteiro, A.; Lopes, C.M. Influence of cover crop on water use and performance of vineyard in Mediterranean Portugal. Agric. Ecosyst. Environ. 2007, 121, 336–342. [Google Scholar] [CrossRef]
- Hua, L.; Xi, Z.-M.; Fang, Y.-l.; Zhang, Z.-W. Effects of grass cover in vineyard onthe vine growth and wine quality. J. Fruit Sci. 2005, 22, 697–701. [Google Scholar]
- Ingels, C.A.; Bugg, R.L.; McGourty, G.T.; Christensen, L.P. Cropping in Vineyards: A Growers’ Handbook; University of California Agriculture and Natural Resources: Oakland, CA, USA, 1998. [Google Scholar]
- Monteiro, A.; Lopes, C.M.; Machado, J.P.; Fernandes, N.; Araujo, A.; Moreira, I. Cover cropping in a sloping, non-irrigated vineyard: 1-Effects on weed composition and dynamics. Cienc. Tec. Vitivinic. 2008, 23, 29–36. [Google Scholar]
- Morlat, R.; Jacquet, A. Grapevine root system and soil characteristics in a vineyard maintained long-term with or without interrow sward. Am. J. Enol. Vitic. 2003, 54, 1–7. [Google Scholar]
- Smith, R.; Bettiga, L.; Cahn, M.; Baumgartner, K.; Jackson, L.E.; Bensen, T. Vineyard floor management affects soil, plant nutrition, and grape yield and quality. Calif. Agric. 2008, 62, 184–190. [Google Scholar] [CrossRef]
- Dabney, S.M.; Delgado, J.A.; Reeves, D.W. Using winter cover crops to improve soil and water quality. Commun. Soil Sci. Plant Anal. 2001, 32, 1221–1250. [Google Scholar] [CrossRef]
- Dorn, B.; Jossi, W.; Heijden, M.G.A. Weed suppression by cover crops: Comparative on-farm experiments under integrated and organic conservation tillage. Weed Res. 2015, 55, 586–597. [Google Scholar] [CrossRef]
- Thorup-Kristensen, K.; Dresboll, D.B. Incorporation time of nitrogen catch crops influences the N effect for the succeeding crop. Soil Use Manag. 2010, 26, 27–35. [Google Scholar] [CrossRef]
- Celette, F.; Gaudin, R.; Gary, C. Spatial and temporal changes to the water regime of a Mediterranean vineyard due to the adoption of cover cropping. Eur. J. Agron. 2008, 29, 153–162. [Google Scholar] [CrossRef]
- McGourty, G. Cover cropping systems for organically farmed vineyards. Prac. Winery Vineyard Sep. Oct. 2004, 7, 38. [Google Scholar]
- Alonso-Ayuso, M.; Gabriel, J.L.; Quemada, M. The Kill Date as a Management Tool for Cover Cropping Success. PLoS ONE 2014, 9, e109587. [Google Scholar] [CrossRef] [PubMed]
- Fageria, N.K. Green manuring in crop production. J. Plant Nutr. 2007, 30, 691–719. [Google Scholar] [CrossRef]
- McGourty, G.T.; Reganold, J.P. Managing vineyard soil organic matter with cover crops. In Proceedings of the Soil Environment and Vine Mineral Nutrition Symposium; Christensen, P., Smart, D.R., Eds.; American Society for Enology and Viticulture Davis: Bellevue, Davis, CA, USA, 2005; pp. 145–151. [Google Scholar]
- Sullivan, P. Overview of Cover Crops and Green Manures; National Sustainable Agriculture Information Service: Butte, MT, USA, 2003; p. 16. [Google Scholar]
- Madge, D. Organic Viticulture: An Australian Manual; Department of Primary Industries: Victoria, Australia, 2005. [Google Scholar]
- Gabriel, J.L.; Quemada, M. Replacing bare fallow with cover crops in a maize cropping system: Yield, N uptake and fertiliser fate. Eur. J. Agron. 2011, 34, 133–143. [Google Scholar] [CrossRef]
- Liebman, M.; Graef, R.L.; Nettleton, D.; Cambardella, C.A. Use of legume green manures as nitrogen sources for corn production. Renew. Agric. Food Syst. 2012, 27, 180–191. [Google Scholar] [CrossRef]
- Miguez, F.E.; Bollero, G.A. Review of corn yield response under winter cover cropping systems using meta-analytic methods. Crop Sci. 2005, 18, 2318–2329. [Google Scholar] [CrossRef]
- King, A.P.; Berry, A.M. Vineyard delta N-15, nitrogen and water status in perennial clover and bunch grass cover crop systems of California’s central valley. Agric. Ecosyst. Environ. 2005, 109, 262–272. [Google Scholar] [CrossRef]
- Bugg, R.L.; McGourty, G.; Sarrantonio, M.; Lanini, W.T.; Bartolucci, R. Comparison of 32 cover crops in an organic vineyard on the north coast of California. Biol. Agric. Hortic. 1996, 13, 63–81. [Google Scholar] [CrossRef]
- McGourty, G.; Nosera, J.; Tylicki, S.; Toth, A. Self-reseeding annual legumes evaluated as cover crops for untilled vineyards. Calif. Agric. 2008, 63, 191–194. [Google Scholar] [CrossRef]
- Mischler, R.A.; Curran, W.S.; Duiker, S.W.; Hyde, J.A. Use of a Rolled-rye Cover Crop for Weed Suppression in No-Till Soybeans. Weed Technol. 2010, 24, 253–261. [Google Scholar] [CrossRef]
- Teasdale, J.R. Contribution of cover crops to weed management in sustainable agricultural systems. J. Prod. Agric. 1996, 9, 475–479. [Google Scholar] [CrossRef]
- Teasdale, J.R. Reduced-Herbicide Weed Management-Systems for No-Tillage Corn (Zea-mays) in a Hairy Vetch (Vicia-villosa) Cover Crop. Weed Technol. 1993, 7, 879–883. [Google Scholar]
- Facelli, J.M.; Pickett, S.T.A. Plant Litter—Its Dynamics and Effects on Plant Community Structure. Bot. Rev. 1991, 57, 1–32. [Google Scholar] [CrossRef]
- Fuji, Y. Allelopathy of hairy vetch and Mucuna: Their application for sustainableagriculture. In Biodiversity and Allelopathy: From Organisms to Ecosystemsin the Pacific; Chou, C.H., Waller, G.R., Reinhardt, C., Eds.; Academia Sinica: Taipei, Taiwan, 1999. [Google Scholar]
- Caamal-Maldonado, J.A.; Jimenez-Osornio, J.J.; Torres-Barragan, A.; Anaya, A.L. The use of allelopathic legume cover and mulch species for weed control in cropping systems. Agron. J. 2001, 93, 27–36. [Google Scholar] [CrossRef]
- Lopez-Bellido, R.J.; Lopez-Bellido, L.; Lopez-Bellido, F.J.; Castillo, J.E. Faba bean (Vicia faba L.) response to tillage and soil residual nitrogen in a continuous rotation with wheat (Triticum aestivum L.) under rainfed Mediterranean conditions. Agron. J. 2003, 95, 1253–1261. [Google Scholar] [CrossRef]
- Lopez-Bellido, L.; Lopez-Bellido, R.J.; Redondo, R.; Benitez, J. Faba bean nitrogen fixation in a wheat-based rotation under rainfed Mediterranean conditions: Effect of tillage system. Field Crop. Res. 2006, 99, 172. [Google Scholar] [CrossRef]
- De Giorgio, D.; Fornaro, F. Nitrogen effects on five durum wheat genotypes (Triticumdurum Desf.) in a semiarid Mediterranean environment. Agric. Mediterr. 2004, 134, 201–215. [Google Scholar]
- Schaller, B.; Nemecek, T.; Streit, B.; Zihlmann, U.; Chervet, A.; Sturny, W.G. Life cycle assessment of a system under no-tillage and ploughing. AGRARForsch. Schweiz 2006, 13, 482–487. [Google Scholar]
- Köpke, U.; Schulte, H. Direct Seeding of Faba Beans in Organic Agriculture (poster). Cultivating the Future Based on Science. In Proceedings of the 2nd Conference of the International Society of Organic Agriculture Research (ISOFAR), Modena, Italy, 18–20 June 2008. [Google Scholar]
- Nemecek, T.; Erzinger, S. Modelling representative life cycle inventories for Swiss arable crops. Int. J. Life Cycle Assess. 2005, 10, 68–76. [Google Scholar] [CrossRef]
- Bäumer, K.; Köpke, U. Effects of Nitrogen Fertilization; EUR 11258; Commission of the European Communities: Brussels, Belgium, 1989; pp. 145–162. [Google Scholar]
- Derpsch, R. No-tillage and conservation agriculture: A progress report. In No-Till Farming Systems Book. Special Publication III of the World Association of Soil and Water Conservation; Goddard, T., Zoebisch, M., Gan, Y., Ellis, W., Watson, A., Sombatpanit, S., Eds.; Bangkok, Thailand, 2008; pp. 7–39. [Google Scholar]
- The State of Food Insecurity in the World. Available online: http://www.fao.org/3/a-i4646e (accessed on 5 June 2017).
- Pasquet, R.S.; Baudoin, J.P. Cowpea. In Tropical Plant Breeding; Charrier, A., Jacquot, M., Eds.; Enfield:Science: Baca Raton, FL, USA, 2001; pp. 177–198. [Google Scholar]
- Ehlers, J.D.; Hall, A.E. Cowpea (Vigna unguiculata L. Walp). Field Crop. Res. 1997, 53, 187–204. [Google Scholar] [CrossRef]
- Singh, B.B. Cowpea (Vigna unguiculata (L.) Walp). In Genetic Resources, Chromosome Engineering and Crop Improvement; Singh, R., Jauhar, P., Eds.; CRC Press: Boca Raton, FL, USA, 2005; pp. 117–162. [Google Scholar]
- Harrison, H.F.; Thies, J.A.; Fery, R.L.; Smith, J.P. Evaluation of cowpea genotypes for use as a cover crop. Hortscience 2006, 41, 1145–1148. [Google Scholar]
- Carsky, R.; Vanlauwe, B.; Lyasse, O. Cowpea rotation as a resource management technology for cereal-based systems in the savannas of West Africa. In Challenges and Opportunities for Enhancing Sustainable Cowpea Production; Fatokun, C., Tarawali, S., Singh, B.B., Kormawa, P.M., Tabo, R., Eds.; Intl. Inst. Tropical. Agric.: Ibadan, Nigeria, 2002; pp. 252–266. [Google Scholar]
- Sanginga, N.; Dashiell, K.E.; Diels, J.; Vanlauwe, B.; Lyasse, O.; Carsky, R.J.; Tarawali, S.; Asafo-Adjei, B.; Menkir, A.; Schulz, S.; et al. Sustainable resource management coupled to resilient germplasm to provide new intensive cereal–grain–legume–livestock systems in the dry savanna. Agric. Ecosyst. Environ. 2003, 100, 305–314. [Google Scholar] [CrossRef]
- Tarawali, S.A.; Singh, B.B.; Gupta, S.C.; Tabo, R.; Harris, F.; Nokoe, S.; Ferandez-Rivera, S.; Bationo, A.; Manyong, V.M.; Makinde, K.; et al. Cowpea as a key factor for a new approach to integrated crop–livestock systems research in the dry savannas of West Africa. In Challenges and Opportunities for Enhancing Sustainable Cowpea Production; Fatokun, C., Tarawali, S., Eds.; Intl. Inst. Tropical. Agric.: Ibadan, Nigeria, 2002; pp. 233–251. [Google Scholar]
- Wang, K.H.; McGovern, R.J.; McSorley, R.; Gallaher, R.N. Cowpea cover crop and solarization for managing root-knot and other plant-parasitic nematodes in herb and vegetable crops. Soil Crop Sci. Soc. Fla Proc. 2004, 63, 99–104. [Google Scholar]
- Calegari, A.; Ashburner, J.; Fowler, R. Conservation Agriculture in Africa. FAO Reg. Off. Afr. Accra Ghana 2005, 91. [Google Scholar]
- Creamer, N.G.; Baldwin, K.R. An evaluation of summer cover crops for use in vegetable production systems in North Carolina. Hortscience 2000, 35, 600–603. [Google Scholar]
- Vollmer, E.R.; Creamer, N.; Reberg-Horton, C.; Hoyt, G. Evaluating Cover Crop Mulches for No-till Organic Production of Onions. Hortscience 2010, 45, 61–70. [Google Scholar]
- Harrison, H.E.; Jackson, D.M.; Keinath, A.P.; Marino, P.C.; Pullaro, T. Broccoli production in cowpea, soybean,and velvetbean cover crop mulches. Horttechnology 2004, 14, 484–487. [Google Scholar]
- Adler, M.J.; Chase, C.A. Comparison of the allelopathic potential of leguminous summer cover crops: Cowpea, sunn hemp, and velvetbean. Hortscience 2007, 42, 289–293. [Google Scholar]
- Webber, C.L., III; White, P.M., Jr.; Dalley, C.; Petrie, E.C.; Viator, R.P.; Shrefler, J.W. Kenaf (Hibiscus cannabinus) and Cowpea (Vigna unguiculata) as Sugarcane Cover Crops. J. Agric. Sci. 2016, 8, 13. [Google Scholar]
- Summerfield, R.; Huxley, P.; Steele, N. Cowpea (Vigna unquiculata (L) walp.). Field Crop. Abstr. 1974, 27, 301–312. [Google Scholar]
- Xiong, H.Z.; Shi, A.N.; Mou, B.Q.; Qin, J.; Motes, D.; Lu, W.G.; Ma, J.B.; Weng, Y.J.; Yang, W.; Wu, D.X. Genetic Diversity and Population Structure of Cowpea (Vigna unguiculata L. Walp). PLoS ONE 2016, 11, e0160941. [Google Scholar] [CrossRef] [PubMed]
- Marechal, R.; Mascherpa, J.; Stainier, F. Etude taxonomique d’un groupe d’especes des genres Phaseolus et Vigna (Papilonaceae) sur la base des donnees morphologiques et polliques, traitees pour l’analyse informatique. Boissiera 1978, 28, 273. [Google Scholar]
- Boukar, O.; Fatokun, C.A.; Huynh, B.L.; Roberts, P.A.; Close, T.J. Genomic Tools in Cowpea Breeding Programs: Status and Perspectives. Front. Plant Sci. 2016, 7, 757. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.F.; Laudeman, T.W.; Rushton, P.J.; Spraggins, T.A.; Timko, M.P. CGKB: An annotation knowledge base for cowpea (Vigna unguiculata L.) methylation filtered genomic genespace sequences. BMC Bioinform. 2007, 8, 129. [Google Scholar] [CrossRef] [PubMed]
- Menancio-Hautea, D.; Fatokun, C.A.; Kumar, L.; Danesh, D.; Young, N.D. Comparative genome analysis of mungbean (Vigna radiata L. Wilczek) and cowpea (V. unguiculata L. Walpers) using RFLP mapping data. Theor. Appl. Genet. 1993, 86, 797–810. [Google Scholar] [CrossRef] [PubMed]
- Vasconcelos, E.V.; Fonseca, A.F.D.; Pedrosa-Harand, A.; Bortoleti, K.C.D.; Benko-Iseppon, A.M.; da Costa, A.F.; Brasileiro-Vidal, A.C. Intra- and interchromosomal rearrangements between cowpea (Vigna unguiculata (L.) Walp.) and common bean (Phaseolus vulgaris L.) revealed by BAC-FISH. Chromosom. Res. 2015, 23, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Lucas, M.R.; Diop, N.N.; Wanamaker, S.; Ehlers, J.D.; Roberts, P.A.; Close, T.J. Cowpea-Soybean Synteny Clarified through an Improved Genetic Map. Plant Genome 2011, 4, 218–225. [Google Scholar] [CrossRef]
- Boukar, O.; Fatokun, C.A.; Roberts, P.A.; Abberton, M.; Huynh, B.L.; Close, T.J.; Kyei-Boahen, S.; Higgins, T.J.V.; Ehlers, J.D. Cowpea. In Grain Legumes; De Ron, A.M., Ed.; Springer: New York, NY, USA, 2015; pp. 219–250. [Google Scholar]
- Ferry, R.L.; Singh, B.B. Cowpea genetics: A review of the recent literature. In Advances in Cowpea Research; Singh, B.B., Mohan-Raj, D.R., Eds.; Copublication of International Institute of Tropical Agriculture (UTA) and Japan International Research Center for Agricultural Sciences (JIRCAS): Ibadan, Nigeria, 1997. [Google Scholar]
- Singh, B.B.; Ehlers, J.D.; Sharma, B.; Freire-Filho, F.R. Recent progress in cowpea breeding. In Challenges and Opportunities for Enhancing Sustainable Cowpea Production; Fatokun, C.Α., Tarawali, S.A., Eds.; IITA: Ibadan, Nigeria, 2002; pp. 22–40. [Google Scholar]
- Muchero, W.; Diop, N.N.; Bhat, P.R.; Fenton, R.D.; Wanamaker, S.; Pottorff, M.; Hearne, S.; Cisse, N.; Fatokun, C.; Ehlers, J.D.; et al. A consensus genetic map of cowpea (Vigna unguiculata (L) Walp.) and synteny based on EST-derived SNPs. Proc. Natl. Acad. Sci. USA 2009, 106, 18159–18164. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Amatriain, M.; Mirebrahim, H.; Xu, P.; Wanamaker, S.I.; Luo, M.C.; Alhakami, H.; Alpert, M.; Atokple, I.; Batieno, B.J.; Boukar, O.; et al. Genome resources for climate-resilient cowpea, an essential crop for food security. Plant J. 2017, 89, 1042–1054. [Google Scholar] [CrossRef] [PubMed]
- Ehlers, J.D.; Ferry, R.L.; Hall, A.G. Cowpea breeding in the USA: New varieties and improved germplasm. In Challenges and Opportunities for Enhancing Sustainable Cowpea Production; Fatokun, C., Tarawali, S., Eds.; IITA: Ibadan, Nigeria, 2002; pp. 287–300. [Google Scholar]
- Hall, A.E.; Cisse, N.; Thiaw, S.; Elawad, H.O.A.; Ehlers, J.D.; Ismail, A.M.; Fery, R.L.; Roberts, P.A.; Kitch, L.W.; Murdock, L.L.; et al. Development of cowpea cultivars and germplasm by the Bean/Cowpea CRSP. Field Crop. Res. 2003, 82, 103–134. [Google Scholar] [CrossRef]
- Ishiyaku, M.F.; Singh, Β.Β. Inheritance of shortday-induced dwarfing in photosensitive cowpea. Afr. Crop Sci. J. 2001, 9, 1–8. [Google Scholar] [CrossRef]
- Roberts, P.; Ehlers, J.; Hall, A.; Matthews, W. Characterization of new resistance to rootknot nematodes in cowpea. In Advances in Cowpea Research; Singh, B., Mohan Raj, D., Dashiell, K., Jackai, L., Eds.; Copublication Intl. Inst. Tropical. Agric. (IITA) and Japan Intl. Res. Center Agric. Sci. (JIRCAS): Sayce, Devon, UK, 1997; pp. 207–214. [Google Scholar]
- Huynh, B.L.; Matthews, W.C.; Ehlers, J.D.; Lucas, M.R.; Santos, J.R.P.; Ndeve, A.; Close, T.J.; Roberts, P.A. A major QTL corresponding to the Rk locus for resistance to root-knot nematodes in cowpea (Vigna unguiculata L. Walp.). Theor. Appl. Genet. 2016, 129, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Pottorff, M.; Wanamaker, S.; Ma, Y.Q.Q.; Ehlers, J.D.; Roberts, P.A.; Close, T.J. Genetic and Physical Mapping of Candidate Genes for Resistance to Fusarium oxysporum f.sp tracheiphilum Race 3 in Cowpea (Vigna unguiculata (L.) Walp). PLoS ONE 2012, 7, e41600. [Google Scholar] [CrossRef] [PubMed]
- Souleymane, A.; Aken’Ova, M.; Fatokun, C.; Alabi, O. Screening for resistance to cowpea aphid (Aphis craccivora Koch) in wild and cultivated cowpea (Vigna unguiculata L.Walp.) accessions. Int. J. Sci. Environ. Technol. 2013, 2, 611–621. [Google Scholar]
- Huynh, B.L.; Ehlers, J.D.; Ndeve, A.; Wanamaker, S.; Lucas, M.R.; Close, T.J.; Roberts, P.A. Genetic mapping and legume synteny of aphid resistance in African cowpea (Vigna unguiculata L. Walp.) grown in California. Mol. Breed. 2015, 35, 36. [Google Scholar] [CrossRef] [PubMed]
- Li, J.X.; Lis, K.E.; Timko, M.P. Molecular genetics of race-specific resistance of cowpea to Striga gesnerioides (Wilid.). Pest Manag. Sci. 2009, 65, 520–527. [Google Scholar] [CrossRef] [PubMed]
- Timko, M.P.; Ehlers, J.D.; Roberts, P.A. Cowpea. In Genome Mapping and Molecular Breeding in Plants, Volume 3, Pulses, Sugar and Tuber Crops; Kole, C., Ed.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 49–67. [Google Scholar]
- Li, G.J.; Liu, Y.H.; Ehlers, J.D.; Zhu, Z.J.; Wu, X.H.; Wang, B.G.; Lu, Z.F. Identification of an AFLP fragment linked to rust resistance in asparagus bean and its conversion to a SCAR marker. Hortscience 2007, 42, 1153–1156. [Google Scholar]
- Mohammed, M.S.; Russom, Z.; Abdul, S.D. Inheritance of hairiness and pod shattering, heritability and correlation studies in crosses between cultivated cowpea (Vigna unguiculata (L.) Walp.) and its wild (var. pubescens) relative. Euphytica 2010, 171, 397–407. [Google Scholar] [CrossRef]
- Suanum, W.; Somta, P.; Kongjaimun, A.; Yimram, T.; Kaga, A.; Tomooka, N.; Takahashi, Y.; Srinives, P. Co-localization of QTLs for pod fiber content and pod shattering in F-2 and backcross populations between yardlong bean and wild cowpea. Mol. Breed. 2016, 36, 1–11. [Google Scholar] [CrossRef]
- Crépona, K.; Marget, P.; Peyronnet, C.; Carrouéea, B.; Arese, P.; Duc, G. Nutritional value of faba bean (Vicia faba L.) seeds for feed and food. Field Crop. Res. 2010, 115, 329–339. [Google Scholar] [CrossRef]
- Mihailović, V.; Pataki, I.; Mikić, A.; Katić, S.; Vasilević, S. Achievements in breeding annual forage crops in Serbia. Period. Sci. Res. Field Veg. Crop. 2007, 44, 79–86. [Google Scholar]
- Kopke, U.; Nemecek, T. Ecological services of faba bean. Field Crop. Res. 2010, 115, 217–233. [Google Scholar] [CrossRef]
- Ćupina, B.; Erić, P.; Kristić, Ð.; Vučković, S. Forage catch crops in sustainable agriculture and organing farming. Acta Agric. Serbica 2004, 17, 451–459. [Google Scholar]
- Turpin, J.E.; Herridge, D.F.; Robertson, M.J. Nitrogen fixation and soil nitrate interactions in field-grown chickpea (Cicer arietinum) and fababean (Vicia faba). Aust. J. Agric. Res. 2002, 53, 599–608. [Google Scholar] [CrossRef]
- Wang, Q.; Li, Y.; Alva, A. Growing Cover Crops to Improve Biomass Accumulation and Carbon Sequestration: A Phytotron Study. J. Environ. Prot. 2010, 1, 73. [Google Scholar] [CrossRef]
- Founíe, J.C. Soil Management in the Breede River Valley Wine Grape Region, South Africa. 1. Cover Crop Performance and Weed Control. S. Afr. J. Enol. Vitic. 2010, 31, 14–21. [Google Scholar]
- Sincik, M.; Turan, Z.M.; Goksoy, A.T. Responses of potato (Solanum tuberosum L.) to green manure cover crops and nitrogen fertilization rates. Am. J. Potato Res. 2008, 85, 150–158. [Google Scholar] [CrossRef]
- Lenzi, A.; Antich, D.; Bigongiali, F.; Mazzoncini, M.; Migliorini, P.; Tesi, R. Effect of different cover crops on organic tomato production. Renew. Agric. Food Syst. 2009, 24, 92–101. [Google Scholar] [CrossRef]
- Rose, T.J.; Damon, P.; Rengel, Z. Phosphorus-efficient faba bean (Vicia faba L.) genotypes enhance subsequent wheat crop growth in an acid and an alkaline soil. Crop Pasture Sci. 2010, 61, 1009–1016. [Google Scholar] [CrossRef]
- Grafton-CardwelI, E.E.; Ouyang, Y.; Bugg, R.L. Leguminous Cover Crops to Enhance Population Development of Euseius tularensis (Acari: Phytoseiidae) in Citrus. Biol. Control 1999, 16, 73–80. [Google Scholar] [CrossRef]
- Founíe, J.C.; Agenbag, G.A.; Louw, P.J.E. Cover Crop Management in a Chardonnay/99 Richter Vineyard in the Coastal Region, South Africa. 3. Effect of Different Cover Crops and Cover Crop Management Practices on Organic Matter and Macro-nutrient Content of a Medium-textured Soil. S. Afr. J. Enol. Vitic. 2007, 28, 61. [Google Scholar]
- Shirtliffe, S.J.; Johnson, E.N. Progress towards no-till organic weed control in western Canada. Renew. Agric. Food Syst. 2012, 27, 60–67. [Google Scholar] [CrossRef]
- Brennan, E.B.; Boyd, N.S.; Smith, R.F.; Foster, P. Seeding Rate and Planting Arrangement Effects on Growth and Weed Suppression of a Legume-Oat Cover Crop for Organic Vegetable Systems. Agron. J. 2009, 101, 979–988. [Google Scholar] [CrossRef]
- Novara, A.; Gristina, L.; Saladino, S.S.; Santoro, A.; Cerda, A. Soil erosion assessment on tillage and alternative soil managements in a Sicilian vineyard. Soil Tillage Res. 2011, 117, 140–147. [Google Scholar] [CrossRef]
- Barthès, B.; Roose, E. Aggregate stability as an indicator of soil susceptibility to runoff and erosion; validation at several levels. Catena 2002, 47, 133–149. [Google Scholar] [CrossRef]
- Six, J.; Bossuyt, H.; Degryze, S.; Denef, K. A history of research on the link between (micro)aggregates, soil biota, and soil organic matter dynamics. Soil Tillage Res. 2004, 79, 7–31. [Google Scholar] [CrossRef]
- Gao, Y.; Yu, G.R.; He, N.P. Equilibration of the terrestrial water, nitrogen, and carbon cycles: Advocating a health threshold for carbon storage. Ecol. Eng. 2013, 57, 366–374. [Google Scholar] [CrossRef]
- García-Díaz, A.; Bienes Allas, R.; Gristina, L.; Cerdà, A.; Pereira, P.; Novara, A. Carbon input threshold for soil carbon budget optimization in eroding vineyards. Geoderma 2016, 271, 144–149. [Google Scholar] [CrossRef]
- Cubero, J.I. Evolution of Vicia-Faba L. Theor. Appl. Genet. 1974, 45, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Zeven, A.C.; Zhukovsky, P.M. Dictionary of Cultivated Plants and Their Centres of Diversity; Centre for Agricultural Publishing and Documentation: Wageningen, The Netherlands, 1975; p. 219. [Google Scholar]
- Tanno, K.; Willcox, G. The origins of cultivation of Cicer arietinum L. and Vicia faba L.: Early finds from Tell el-Kerkh, north-west Syria, late 10th millennium BP. Veg. Hist. Archaeobotany 2006, 15, 197–204. [Google Scholar] [CrossRef]
- Abdel, L.Y.I. Effect of seed size and plant spacing on yield and yield components of Faba bean (Vicia fava L.). Res. J. Agric. Biolog. Sci. 2008, 4, 146–148. [Google Scholar]
- Gasim, S.; Link, W. Agronomic performance and the effect of soil fertilization on German winter faba bean. J. Central Eur. Agric. 2007, 8, 121–128. [Google Scholar]
- Singh, A.K.; Bharati, R.C.; Manibhushan, N.C.; Pedpati, A. An assessment of faba bean (Vicia faba L.) current status and future prospect. Afr. J. Agric. Res. 2013, 8, 6634–6641. [Google Scholar]
- Rajan, K.; Singh, A.K.; Pandey, A.K. Faba bean soils and their management for higher productivity. In Faba Bean (Vicia faba L): A Potential Leguminous Crop of India; ICAR, RC for ER: Patna, India, 2012. [Google Scholar]
- Singh, A.K.; Chandra, N.; Bharati, R.C.; Dimree, S.K. Effect of seed size and seeding depth on Fava bean (Vicia fava L.) productivity. Environ. Ecol. 2010, 28, 1722–1727. [Google Scholar]
- Gnanasambandam, A.; Paull, J.; Torres, A.; Kaur, S.; Leonforte, T.; Li, H.; Zong, X.; Yang, T.; Materne, M. Impact of Molecular Technologies on Faba Bean (Vicia faba L.) Breeding Strategies. Agronomy 2012, 2, 132–166. [Google Scholar] [CrossRef]
- Satovic, Z.; Avila, C.M.; Cruz-Izquierdo, S.; Diaz-Ruiz, R.; Garcia-Ruiz, G.M.; Palomino, C.; Gutierrez, N.; Vitale, S.; Ocana-Moral, S.; Gutierrez, M.V.; et al. A reference consensus genetic map for molecular markers and economically important traits in faba bean (Vicia faba L.). BMC Genom. 2013, 14, 932. [Google Scholar] [CrossRef] [PubMed]
- Cubero, J.I. Bean improvement: Proceedings of the Faba Bean conference ICARDA/IFAD Nile Valley Project. In Interespecific Hybridization in Vicia; Hawting, G., Webb, C., Eds.; Martinus Nihjoff: The Hague, The Netherlands, 1982; pp. 91–108. [Google Scholar]
- Van de Ven, M.; Powell, W.; Ramsay, G.; Waugh, R. Restriction fragment length polymorphisms as genetic markers in Vicia. Heredity 1990, 65, 329–342. [Google Scholar] [CrossRef]
- Link, W.; Dixkens, C.; Singh, M.; Schwall, M.; Melchinger, A.E. Genetic Diversity in European and Mediterranean Faba Bean Germ Plasm Revealed by Rapd Markers. Theor. Appl. Genet. 1995, 90, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Alghamdi, S.S.; Al-Faifi, S.A.; Migdadi, H.M.; Ammar, M.H.; Siddique, K.H.M. Inter-simple sequence repeat (ISSR)-based diversity assessment among faba bean genotypes. Crop Pasture Sci. 2011, 62, 755–760. [Google Scholar] [CrossRef]
- Terzopoulos, P.J.; Bebeli, P.J. Genetic diversity analysis of Mediterranean faba bean (Vicia faba L.) with ISSR markers. Field Crop. Res. 2008, 108, 39–44. [Google Scholar] [CrossRef]
- Zeid, M.; Schon, C.C.; Link, W. Genetic diversity in recent elite faba bean lines using AFLP markers. Theor. Appl. Genet. 2003, 107, 1304–1314. [Google Scholar] [CrossRef] [PubMed]
- Zong, X.X.; Liu, X.J.; Guan, J.P.; Wang, S.M.; Liu, Q.C.; Paull, J.G.; Redden, R. Molecular variation among Chinese and global winter faba bean germplasm. Theor. Appl. Genet. 2009, 118, 971–978. [Google Scholar] [CrossRef] [PubMed]
- Zong, X.; Ren, J.; Guan, J.; Wang, S.; Liu, Q.; Paull, J.G.; Redden, R. Molecular variation among Chinese and global germplasm in spring faba bean areas. Plant Breed. 2010, 129, 508–513. [Google Scholar] [CrossRef]
- Tribouillois, H.; Fort, F.; Cruz, P.; Charles, R.; Flores, O.; Garnier, E.; Justes, E. A Functional Characterisation of a Wide Range of Cover Crop Species: Growth and Nitrogen Acquisition Rates, Leaf Traits and Ecological Strategies. PLoS ONE 2015, 10, e0122156. [Google Scholar] [CrossRef] [PubMed]
- Duc, G.; Bao, S.Y.; Baum, M.; Redden, B.; Sadiki, M.; Suso, M.J.; Vishniakova, M.; Zong, X.X. Diversity maintenance and use of Vicia faba L. genetic resources. Field Crop. Res. 2010, 115, 270–278. [Google Scholar] [CrossRef]
- Sillero, J.C.; Villegas-Fernandez, A.M.; Thomas, J.; Rojas-Molina, M.M.; Emeran, A.A.; Fernandez-Aparicio, M.; Rubiales, D. Faba bean breeding for disease resistance. Field Crop. Res. 2010, 115, 297–307. [Google Scholar] [CrossRef]
- Khazaei, H.; O’Sullivan, D.M.; Sillanpaa, M.J.; Stoddard, F.L. Use of synteny to identify candidate genes underlying QTL controlling stomatal traits in faba bean (Viciafaba L.). Theor. Appl. Genet. 2014, 127, 2371–2385. [Google Scholar] [CrossRef] [PubMed]
- Arbaoui, M.; Balko, C.; Link, W. Study of faba bean (Vicia faba L.) winter-hardiness and development of screening methods. Field Crop. Res. 2008, 106, 60–67. [Google Scholar] [CrossRef]
- Link, W.; Balko, C.; Stoddard, F.L. Winter hardiness in faba bean: Physiology and breeding. Field Crop. Res. 2010, 115, 287–296. [Google Scholar] [CrossRef]
- Flores, F.; Nadal, S.; Solis, I.; Winkler, J.; Sass, O.; Stoddard, F.L.; Link, W.; Raffiot, B.; Muel, F.; Rubiales, D. Faba bean adaptation to autumn sowing under European climates. Agron. Sustain. Dev. 2012, 32, 727–734. [Google Scholar] [CrossRef]
- Nasrullahzadeh, S.; Ghassemi-Golezani, K.; Javanshir, A.; Valizade, M.; Shakiba, M.R. Effects of shade stress on ground cover and grain yield of faba bean (Vicia faba L.). J. Food Agric. Environ. 2007, 5, 337–340. [Google Scholar]
- Tavakkoli, E.; Paull, J.; Rengasamy, P.; McDonald, G.K. Comparing genotypic variation in faba bean (Vicia faba L.) in response to salinity in hydroponic and field experiments. Field Crop. Res. 2012, 127, 99–108. [Google Scholar] [CrossRef]
- Link, W.; Ederer, W.; Gumber, R.K.; Melchinger, A.E. Detection and characterization of two new CMS systems in faba bean (Vicia faba). Plant Breed. 1997, 116, 158–162. [Google Scholar] [CrossRef]
- Ghaouti, L.; Link, W. Local vs. formal breeding and inbred line vs. synthetic cultivar for organic farming: Case of Vicia faba L. Field Crop. Res. 2009, 110, 167–172. [Google Scholar] [CrossRef]
- Torres, A.M.; Avila, C.M.; Gutierrez, N.; Palomino, C.; Moreno, M.T.; Cubero, J.I. Marker-assisted selection in faba bean (Vicia faba L.). Field Crop. Res. 2010, 115, 243–252. [Google Scholar] [CrossRef]
- Kaur, S.; Kimber, R.B.E.; Cogan, N.O.I.; Materne, M.; Forster, J.W.; Paull, J.G. SNP discovery and high-density genetic mapping in faba bean (Vicia faba L.) permits identification of QTLs for ascochyta blight resistance. Plant Sci. 2014, 217, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Nakamura, Y.; Asamizu, E.; Isobe, S.; Tabata, S. Genome sequencing and genome resources in model legumes. Plant Physiol. 2007, 144, 588–593. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Izquierdo, S.; Avila, C.M.; Satovic, Z.; Palomino, C.; Gutierrez, N.; Ellwood, S.R.; Phan, H.T.T.; Cubero, J.I.; Torres, A.M. Comparative genomics to bridge Vicia faba with model and closely-related legume species: Stability of QTLs for flowering and yield-related traits. Theor. Appl. Genet. 2012, 125, 1767–1782. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, A.F.; Christensen, D.A.; McKinnon, J.J. Effects of pea, barley, and alfalfa silage on ruminal nutrient degradability and performance of dairy cows. J. Dairy Sci. 2000, 83, 2859–2865. [Google Scholar] [CrossRef]
- Spies, J.M.; Warkentin, T.D.; Shirtliffe, S.J. Variation in Field Pea (Pisum sativum) Cultivars for Basal Branching and Weed Competition. Weed Sci. 2011, 59, 218–223. [Google Scholar] [CrossRef]
- Hattab, S.; Dridi, B.; Chouba, L.; Kheder, M.B.; Bousetta, H. Photosynthesis and growth responses of pea Pisum sativum L. under heavy metals stress. J. Environ. Sci. 2009, 21, 1552–1556. [Google Scholar] [CrossRef]
- Murungu, F.S.; Chiduza, C.; Muchaonyerwa, P.; Mnkeni, P.N.S. Decomposition, Nitrogen, and Phosphorus Mineralization from Residues of Summer-Grown Cover Crops and Suitability for a Smallholder Farming System in South Africa. Commun. Soil Sci. Plant Anal. 2011, 42, 2461–2472. [Google Scholar] [CrossRef]
- Turgut, I.; Bilgili, U.; Duman, A.; Acikgoz, E. Effect of green manuring on the yield of sweet corn. Agron. Sustain. Dev. 2005, 25, 433–438. [Google Scholar] [CrossRef]
- Decker, A.M.; Clark, A.J.; Meisinger, J.J.; Mulford, F.R.; Mcintosh, M.S. Legume Cover Crop Contributions to No-Tillage Corn Production. Agron. J. 1994, 86, 126–135. [Google Scholar] [CrossRef]
- Brennan, E.B.; Boyd, N.S.; Smith, R.F.; Foster, P. Comparison of Rye and Legume-Rye Cover Crop Mixtures for Vegetable Production in California (vol 103, pg 449, 2011). Agron. J. 2011, 103, 1563–1564. [Google Scholar] [CrossRef]
- Akemo, M.C.; Regnier, E.E.; Bennett, M.A. Weed suppression in spring-sown rye (Secale cereale)-pea (Pisum sativum) cover crop mixes. Weed Technol. 2000, 14, 545–549. [Google Scholar] [CrossRef]
- Creamer, N.G.; Bennett, M.A.; Stinner, B.R. Evaluation of cover crop mixtures for use in vegetable production systems. Hortscience 1997, 32, 866–870. [Google Scholar]
- Kato-Noguchi, H. Isolation and identification of an allelopathic substance in Pisum sativum. Phytochemistry 2003, 62, 1141–1144. [Google Scholar] [CrossRef]
- Price, A.J.; Stoll, M.E.; Bergtold, J.S.; Arriaga, F.J.; Balkcom, K.S.; Kornecki, T.S.; Raper, R.L. Effect of cover crop extracts on cotton and radish radicleelongation. Int. J. Fac. Agric. Biol. 2008, 3, 60–66. [Google Scholar]
- Blackshaw, R.E.; Molnar, L.J.; Moyer, J.R. Suitability of legume cover crop-winter wheat intercrops on the semi-arid Canadian prairies. Can. J. Plant Sci. 2010, 90, 479–488. [Google Scholar] [CrossRef]
- Isik, D.; Kaya, E.; Ngouajio, M.; Mennan, H. Summer cover crops for weed management and yield improvement in organic lettuce (Lactuca sativa) production. Phytoparasitica 2009, 37, 193–203. [Google Scholar] [CrossRef]
- Baggs, E.M.; Watson, C.A.; Rees, R.M. The fate of nitrogen from incorporated cover crop and green manure residues. Nutr. Cycl. Agroecosyst. 2000, 56, 153–163. [Google Scholar] [CrossRef]
- Smýkal, P.; Jovanović, Ž.; Stanisavljević, N.; Zlatković, B.; Ćupina, B.; Đorđević, V.; Mikić, A.; Medović, A. A comparative study of ancient DNA isolated from charred pea (Pisum sativum L.) seeds from an Early Iron Age settlement in southeast Serbia: Inference for pea domestication. Genet. Res. Crop Evol. 2014, 61, 1533–1544. [Google Scholar] [CrossRef]
- Cousin, R. Peas (Pisum sativum L). Field Crop. Res. 1997, 53, 111–130. [Google Scholar] [CrossRef]
- Willcox, G.; Fornite, S.; Herveux, L. Early Holocene cultivation before domestication in northern Syria. Veg. Hist. Archaeobotany 2008, 17, 313–325. [Google Scholar] [CrossRef]
- Riehl, S.; Zeidi, M.; Conard, N.J. Emergence of Agriculture in the Foothills of the Zagros Mountains of Iran. Science 2013, 341, 65–67. [Google Scholar] [CrossRef] [PubMed]
- McPhee, K. Dry pea production and breeding—A mini-review. J. Food Agric. Environ. 2003, 1, 64–69. [Google Scholar]
- Smýkal, P.; Aubert, G.; Burstin, J.; Coyne, C.J.; Ellis, N.T.H.; Flavell, A.J.; Ford, R.; Hýbl, M.; Macas, J.; Neumann, P.; et al. Pea (Pisum sativum L.) in the Genomic Era. Agronomy 2012, 2, 74–115. [Google Scholar] [CrossRef]
- Smýkal, P.; Coyne, C.; Redden, R.; Maxted, N.P. Genetic and Genomic Resources of Grain Legume Improvement; Singh, M., Upadhyaya, H., Eds.; Elsevier: Amsterdam, The Netherlands, 2013. [Google Scholar]
- Jing, R.C.; Vershinin, A.; Grzebyta, J.; Shaw, P.; Smykal, P.; Marshall, D.; Ambrose, M.J.; Ellis, T.H.N.; Flavell, A.J. The genetic diversity and evolution of field pea (Pisum) studied by high throughput retrotransposon based insertion polymorphism (RBIP) marker analysis. BMC Evolut. Biol. 2010, 10, 44. [Google Scholar] [CrossRef] [PubMed]
- Leino, M.W.; Bostrom, E.; Hagenblad, J. Twentieth-century changes in the genetic composition of Swedish field pea metapopulations. Heredity 2013, 110, 338–346. [Google Scholar] [CrossRef] [PubMed]
- Abi-Ghanem, R.; Bodah, E.T.; Wood, M.; Braunwart, K. Potential breeding for high nitrogen fixation in Pisum sativum L.: Germplasm phenotypic characterization and genetic investigation. Am. J. Plant. Sci. 2013. [Google Scholar] [CrossRef]
- Santalla, M.; Amurrio, J.M.; De Ron, A.M. Symbiotic Interactions between Rhizobium leguminosarum Strains and Elite Cultivars of Pisum sativum L. J. Agron. Crop Sci. 2001, 187, 59–68. [Google Scholar] [CrossRef]
- Valentine, A.J.; Benedito, V.A.; Kang, Y. Legume Nitrogen Fixation and Soil Abiotic Stress: From Physiology to Genomics and Beyond. In Annual Plant Reviews; Wiley-Blackwell: Hoboken, NJ, USA, 2010; pp. 207–248. [Google Scholar]
- McMurray, L.S.; Davidson, J.A.; Lines, M.D.; Leonforte, A.; Salam, M.U. Combining management and breeding advances to improve field pea (Pisum sativum L.) grain yields under changing climatic conditions in south-eastern Australia. Euphytica 2011, 180, 69–88. [Google Scholar] [CrossRef]
- Alvino, A.; Leone, A. Response to Low Soil-Water Potential in Pea Genotypes (Pisum-Sativum L.) with Different Leaf Morphology. Sci. Hortic. 1993, 53, 21–34. [Google Scholar] [CrossRef]
- Sanchez, F.J.; Manzanares, M.; de Andres, E.F.; Tenorio, J.L.; Ayerbe, L. Turgor maintenance, osmotic adjustment and soluble sugar and proline accumulation in 49 pea cultivars in response to water stress. Field Crop. Res. 1998, 59, 225–235. [Google Scholar] [CrossRef]
- Stoddard, F.L.; Balko, C.; Erskine, W.; Khan, H.R.; Link, W.; Sarker, A. Screening techniques and sources of resistance to abiotic stresses in cool-season food legumes. Euphytica 2006, 147, 167–186. [Google Scholar] [CrossRef]
- Auld, D.L.; Ditterline, R.L.; Murray, G.A.; Swensen, J.B. Screening Peas for Winterhardiness under Field and Laboratory Conditions. Crop Sci. 1983, 23, 85–88. [Google Scholar] [CrossRef]
- Zhang, X.; Wan, S.; Hao, J.; Hu, J.; Yang, T.; Zong, X. Large-scale evaluation of pea (Pisum sativum L.) germplasm for cold tolerance in the field during winter in Qingdao. Crop J. 2016, 4, 377–383. [Google Scholar] [CrossRef]
- Fowler, D.B.; Breton, G.; Limin, A.E.; Mahfoozi, S.; Sarhan, F. Photoperiod and temperature interactions regulate low-temperature-induced gene expression in barley. Plant Physiol. 2001, 127, 1676–1681. [Google Scholar] [CrossRef] [PubMed]
- Lejeune-Henaut, I.; Hanocq, E.; Bethencourt, L.; Fontaine, V.; Delbreil, B.; Morin, J.; Petit, A.; Devaux, R.; Boilleau, M.; Stempniak, J.J.; et al. The flowering locus Hr colocalizes with a major QTL affecting winter frost tolerance in Pisum sativum L. Theor. Appl. Genet. 2008, 116, 1105–1116. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Hwang, S.F.; Chang, K.F.; Turnbull, G.D.; Howard, R.J. Characterization of Ascochyta isolates and susceptibility of pea cultivars to the ascochyta disease complex in Alberta. Plant Pathol. 2000, 49, 540–545. [Google Scholar] [CrossRef]
- Fondevilla, S.; Rubiales, D. Powdery mildew control in pea: A review. Agron. Sustain. Dev. 2012, 32, 401–409. [Google Scholar] [CrossRef]
- Janila, P.; Sharma, B. RAPD and SCAR markers for powdery mildew resistance gene er in pea. Plant Breed. 2004, 123, 271–274. [Google Scholar] [CrossRef]
- Wilman, K.; Stepien, L.; Fabianska, I.; Kachlicki, P. Plant-pathogenic fungi in seeds of different pea cultivars in Poland. Arch. Ind. Hyg. Toxicol. 2014, 65, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Rubiales, D.; Fernandez-Aparicio, M.; Moral, A.; Barilli, E.; Sillero, J.C.; Fondevilla, S. Disease Resistance in Pea (Pisum sativum L.) Types for Autumn Sowings in Mediterranean Environments. Czech J. Genet. Plant Breed. 2009, 45, 135–142. [Google Scholar]
- Redden, R.; Leonforte, T.; Ford, R.; Croser, J.; Slattery, J. Pea (Pisum sativum L.). In Genetic Resources Chromosome Engineering and Crop Improvement Series: Grain Legumes; Jauhar, S.A., Ed.; Teylor and Francis, CRC Press: Boca Raton, FL, USA, 2005; pp. 55–70. [Google Scholar]
- Maxted, N.; Kell, S.; Ford-Lloyd, B.; Dulloo, E.; Toledo, A. Toward the Systematic Conservation of Global Crop Wild Relative Diversity. Crop Sci. 2012, 52, 774–785. [Google Scholar] [CrossRef]
- Macas, J.; Neumann, P.; Navratilova, A. Repetitive DNA in the pea (Pisum sativum L.) genome: Comprehensive characterization using 454 sequencing and comparison to soybean and Medicago truncatula. BMC Genom. 2007, 8, 427. [Google Scholar] [CrossRef] [PubMed]
- Fondevilla, S.; Fernandez-Aparicio, M.; Satovic, Z.; Emeran, A.A.; Torres, A.M.; Moreno, M.T.; Rubiales, D. Identification of quantitative trait loci for specific mechanisms of resistance to Orobanche crenata Forsk. in pea (Pisum sativum L.). Mol. Breed. 2010, 25, 259–272. [Google Scholar] [CrossRef]
- Fondevilla, S.; Martin-Sanz, A.; Satovic, Z.; Fernandez-Romero, M.D.; Rubiales, D.; Caminero, C. Identification of quantitative trait loci involved in resistance to Pseudomonas syringae pv. syringae in pea (Pisum sativum L.). Euphytica 2012, 186, 805–812. [Google Scholar] [CrossRef]
- Fondevilla, S.; Almeida, N.F.; Satovic, Z.; Rubiales, D.; Patto, M.C.V.; Cubero, J.I.; Torres, A.M. Identification of common genomic regions controlling resistance to Mycosphaerella pinodes, earliness and architectural traits in different pea genetic backgrounds. Euphytica 2011, 182, 43–52. [Google Scholar] [CrossRef]
- Srivastava, R.K.; Mishra, S.K.; Singh, A.K.; Mohapatra, T. Development of a coupling-phase SCAR marker linked to the powdery mildew resistance gene ‘er1’ in pea (Pisum sativum L.). Euphytica 2012, 186, 855–866. [Google Scholar] [CrossRef]
- Pavan, S.; Schiavulli, A.; Appiano, M.; Miacola, C.; Visser, R.G.F.; Bai, Y.; Lotti, C.; Ricciardi, L. Identification of a complete set of functional markers for the selection of er1powdery mildew resistance in Pisum sativum L. Mol. Breed. 2013, 31, 247–253. [Google Scholar] [CrossRef]
- Sun, X.; Yang, T.; Hao, J.; Zhang, X.; Ford, R.; Jiang, J.; Wang, F.; Guan, J.; Zong, X. SSR genetic linkage map construction of pea (Pisum sativum L.) based on Chinese native varieties. Crop J. 2014, 2, 170–174. [Google Scholar] [CrossRef]
- Yang, T.; Fang, L.; Zhang, X.Y.; Hu, J.G.; Bao, S.Y.; Hao, J.J.; Li, L.; He, Y.H.; Jiang, J.Y.; Wang, F.; et al. High-Throughput Development of SSR Markers from Pea (Pisum sativum L.) Based on Next Generation Sequencing of a Purified Chinese Commercial Variety. PLoS ONE 2015, 10, e0139775. [Google Scholar] [CrossRef] [PubMed]
- Sindhu, A.; Ramsay, L.; Sanderson, L.A.; Stonehouse, R.; Li, R.; Condie, J.; Shunmugam, A.S.K.; Liu, Y.; Jha, A.B.; Diapari, M.; et al. Gene-based SNP discovery and genetic mapping in pea. Theor. Appl. Genet. 2014, 127, 2225–2241. [Google Scholar] [CrossRef] [PubMed]
- Boutet, G.; Carvalho, S.A.; Falque, M.; Peterlongo, P.; Lhuillier, E.; Bouchez, O.; Lavaud, C.; Pilet-Nayel, M.L.; Riviere, N.; Baranger, A. SNP discovery and genetic mapping using genotyping by sequencing of whole genome genomic DNA from a pea RIL population. BMC Genom. 2016, 17, 121. [Google Scholar] [CrossRef] [PubMed]
| Cover Crop Desirable Traits | Cowpea | Faba Bean | Winter Pea |
|---|---|---|---|
| Source of Nitrogen | + | +++ | + |
| Reduction of N leaching | + | + | + |
| Reduction of soil erosion | +++ | + | + |
| Improvement of soil fertility | +++ | +++ | + |
| Weed management | +++ | + | +++ |
| Winter hardiness | − | +++ | + |
| Drought resistance | +++ | − | − |
| Rapid growth | +++ | + | + |
| Crops to be benefited | |||
| Cereals | + | + | +++ |
| Vegetables | + | + | + |
| Organic farming | +++ | + | + |
| Vineyard | na | +++ | na |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tani, E.; Abraham, E.; Chachalis, D.; Travlos, I. Molecular, Genetic and Agronomic Approaches to Utilizing Pulses as Cover Crops and Green Manure into Cropping Systems. Int. J. Mol. Sci. 2017, 18, 1202. https://doi.org/10.3390/ijms18061202
Tani E, Abraham E, Chachalis D, Travlos I. Molecular, Genetic and Agronomic Approaches to Utilizing Pulses as Cover Crops and Green Manure into Cropping Systems. International Journal of Molecular Sciences. 2017; 18(6):1202. https://doi.org/10.3390/ijms18061202
Chicago/Turabian StyleTani, Eleni, Eleni Abraham, Demosthenis Chachalis, and Ilias Travlos. 2017. "Molecular, Genetic and Agronomic Approaches to Utilizing Pulses as Cover Crops and Green Manure into Cropping Systems" International Journal of Molecular Sciences 18, no. 6: 1202. https://doi.org/10.3390/ijms18061202