Identification of Physiological Substrates and Binding Partners of the Plant Mitochondrial Protease FTSH4 by the Trapping Approach
Abstract
:1. Introduction
2. Results and Discussion
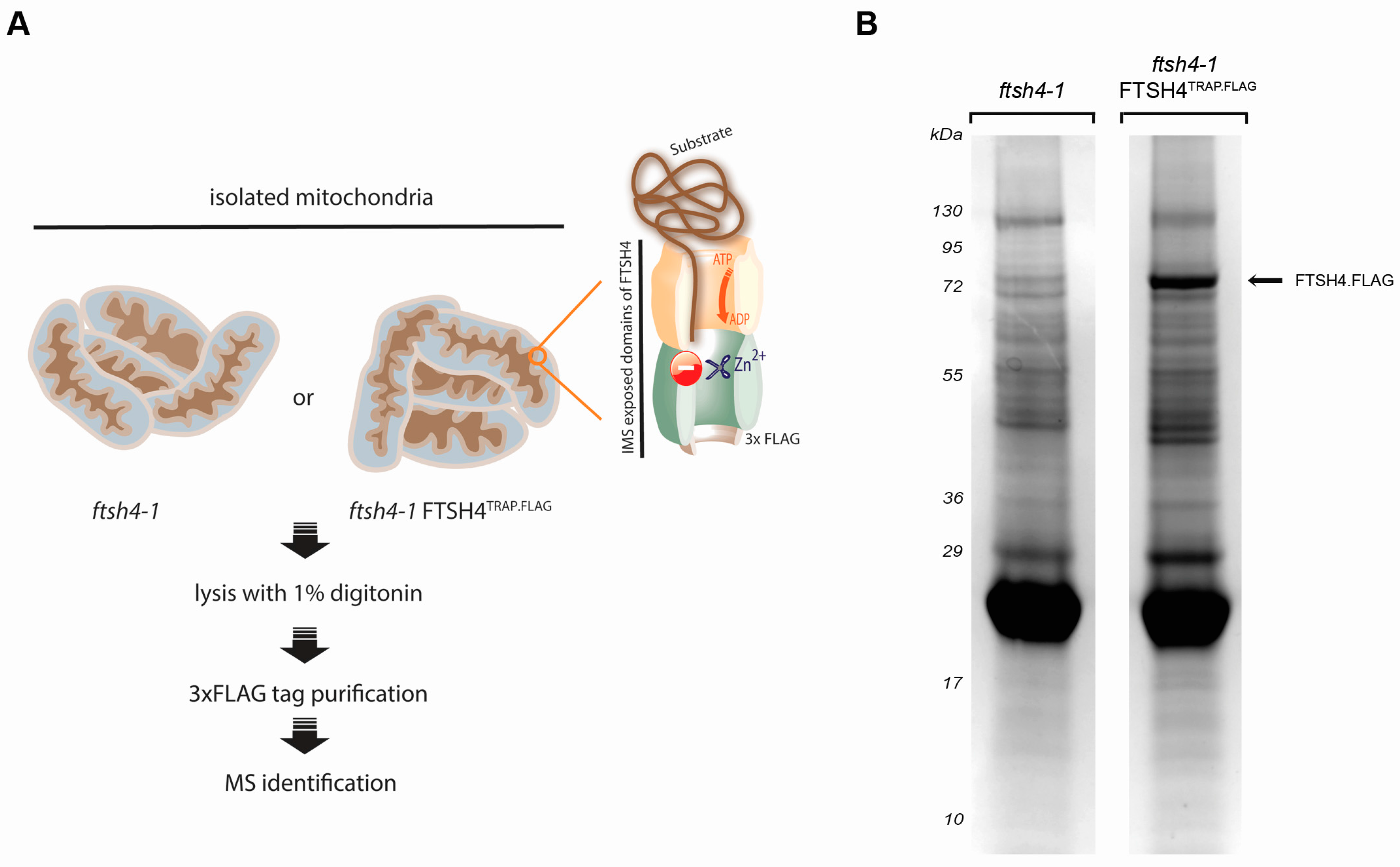
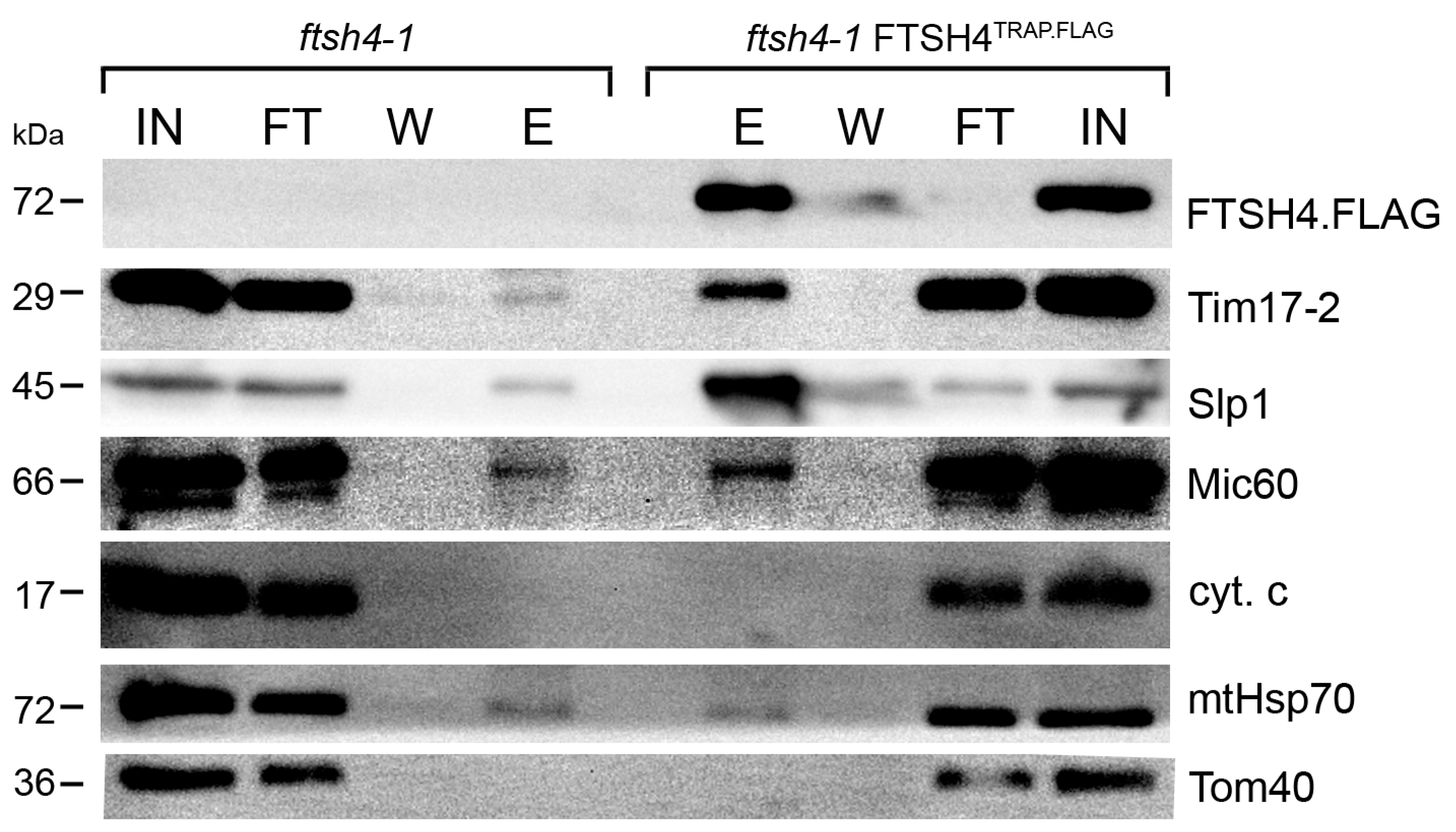
2.1. Substrate Trapping Assay Reveals a List of Potential FTSH4 Targets and Its Interacting Partners inside Mitochondria
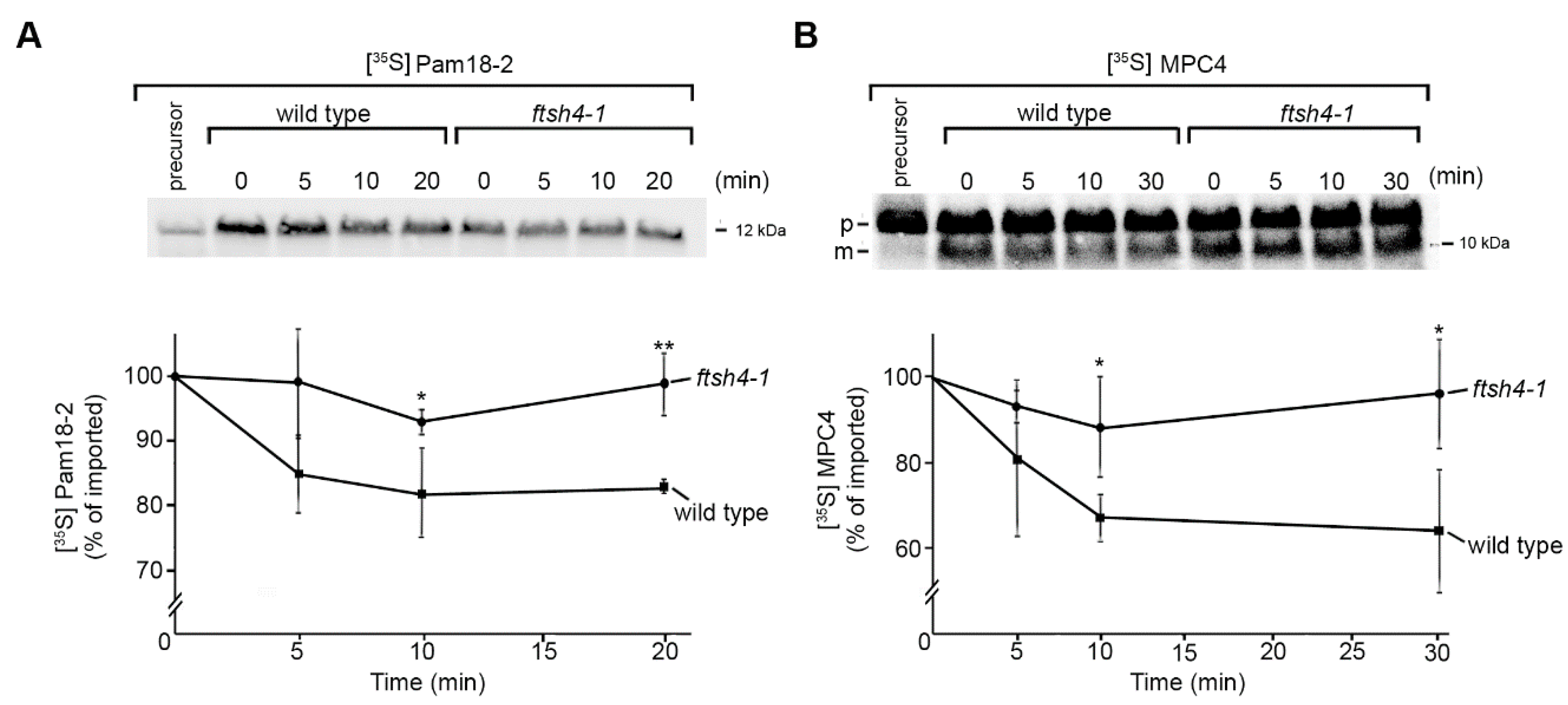
2.2. Identification of Novel Physiological Substrates of FTSH4
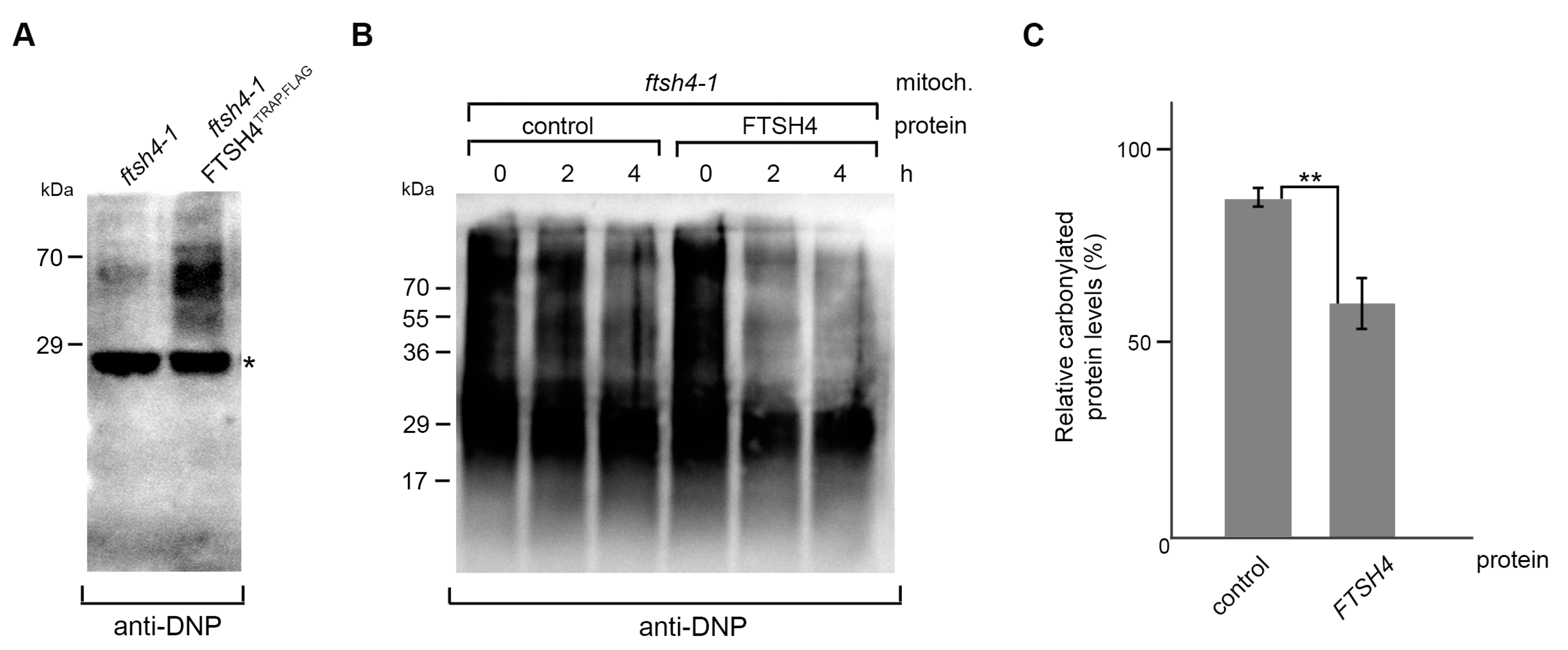
2.3. FTSH4 Degrades Mitochondrial Carbonylated Proteins
3. Materials and Methods
3.1. Plant Material and Growth Conditions
3.2. Isolation of Mitochondria from Arabidopsis thaliana
3.3. Substrate-Trapping Assay
3.4. Immunoblot Analysis
3.5. Synthesis of Radiolabeled Precursor Proteins
3.6. [35S]-Labelled Protein Uptake into the Mitochondria and in Organello Degradation Assay
3.7. Cell-Free Synthesis of FTSH4
3.8. In Vitro Degradation Assay of Carbonylated Proteins
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- McBride, H.M.; Neuspiel, M.; Wasiak, S. Mitochondria: More than just a powerhouse. Curr. Biol. 2006, 25, R551–R560. [Google Scholar] [CrossRef] [PubMed]
- Nunnari, J.; Suomalainen, A. Mitochondria: In sickness and in health. Cell 2012, 148, 1145–1159. [Google Scholar] [CrossRef] [PubMed]
- Raimundo, N. Mitochondrial pathology: Stress signals from the energy factory. Trends Mol. Med. 2014, 20, 282–292. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, R.; Nooteboom, M.; Osiewacz, H.D. Control of mitochondrial integrity in ageing and disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369. [Google Scholar] [CrossRef] [PubMed]
- Quirós, P.M.; Langer, T.; López-Otín, C. New roles for mitochondrial proteases in health, ageing and disease. Nat. Rev. Mol. Cell Biol. 2015, 16, 345–359. [Google Scholar] [CrossRef] [PubMed]
- Hamon, M.P.; Bulteau, A.L.; Friguet, B. Mitochondrial proteases and protein quality control in ageing and longevity. Ageing Res. Rev. 2015, 23, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Glynn, S.E. Multifunctional Mitochondrial AAA Proteases. Front. Mol. Biosci. 2017, 4, 34. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, B.; Wai, T.; Hu, H.; MacVicar, T.; Musante, L.; Fischer-Zirnsak, B.; Stenze, W.; Gräf, R.; van den Heuvel, L.; Ropers, H.H.; et al. Homozygous YME1L1 mutation causes mitochondriopathy with optic atrophy and mitochondrial network fragmentation. eLife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Stiburek, L.; Cesnekova, J.; Kostkova, O.; Fornuskova, D.; Vinsova, K.; Wenchich, L.; Houstek, J.; Zeman, J. YME1L controls the accumulation of respiratory chain subunits and is required for apoptotic resistance, cristae morphogenesis, and cell proliferation. Mol. Biol. Cell 2012, 23, 1010–1023. [Google Scholar] [CrossRef] [PubMed]
- Rainbolt, K.; Atanassova, N.; Genereux, J.C.; Wiseman, R.L. Stress-Regulated Translational Attenuation Adapts Mitochondrial Protein Import through Tim17A Degradation. Cell Metab. 2013, 18, 908–919. [Google Scholar] [CrossRef] [PubMed]
- Anand, R.; Wai, T.; Baker, M.J.; Kladt, N.; Schauss, A.C.; Rugarli, E.; Langer, T. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J. Cell Biol. 2014, 204, 919–929. [Google Scholar] [CrossRef] [PubMed]
- Opalińska, M.; Parys, K.; Murcha, M.W.; Jańska, H. The plant i-AAA protease controls the turnover of an essential mitochondrial protein import component. J. Cell Sci. 2017. [Google Scholar] [CrossRef] [PubMed]
- Dolzblasz, A.; Smakowska, E.; Gola, E.M.; Sokołowska, K.; Kicia, M.; Janska, H. The mitochondrial protease AtFTSH4 safeguards Arabidopsis shoot apical meristem function. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Smakowska, E.; Skibior-Blaszczyk, R.; Czarna, M.; Kolodziejczak, M.; Kwasniak-Owczarek, M.; Parys, K.; Funk, C.; Janska, H. Lack of FTSH4 Protease Affects Protein Carbonylation, Mitochondrial Morphology, and Phospholipid Content in Mitochondria of Arabidopsis: New Insights into a Complex Interplay. Plant Physiol. 2016, 171, 2516–2535. [Google Scholar] [CrossRef] [PubMed]
- Westphal, K.; Langklotz, S.; Thomanek, N.; Narberhaus, F. A trapping approach reveals novel substrates and physiological functions of the essential protease FtsH in Escherichia coli. J. Biol. Chem. 2012, 287, 42962–42971. [Google Scholar] [CrossRef] [PubMed]
- Fischer, F.; Langer, J.D.; Osiewacz, H.D. Identification of potential mitochondrial CLPXP protease interactors and substrates suggests its central role in energy metabolism. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Ben-Lulu, S.; Ziv, T.; Admon, A.; Weisman-Shomer, P.; Benhar, M. A substrate trapping approach identifies proteins regulated by reversible S-nitrosylation. Mol. Cell. Proteom. 2014, 13, 2573–2583. [Google Scholar] [CrossRef] [PubMed]
- Blanchetot, C.; Chagnon, M.; Dubé, N.; Hallé, M.; Tremblay, M.L. Substrate-trapping techniques in the identification of cellular PTP targets. Methods 2005, 35, 44–53. [Google Scholar] [CrossRef] [PubMed]
- König, T.; Tröder, S.E.; Bakka, K.; Korwitz, A.; Richter-Dennerlein, R.; Lampe, P.A.; Patron, M.; Mühlmeister, M.; Guerrero-Castillo, S.; Brandt, U.; et al. The m-AAA Protease Associated with Neurodegeneration Limits MCU Activity in Mitochondria. Mol. Cell 2016, 64, 148–162. [Google Scholar] [CrossRef] [PubMed]
- Kolodziejczak, M.; Gibala, M.; Urantówka, A.; Jańska, H. The significance of Arabidopsis AAA proteases for activity and assembly/stability of mitochondrial OXPHOS complexes. Physiol. Plant. 2006, 129, 135–142. [Google Scholar] [CrossRef]
- Palmieri, F.; Rieder, B.; Ventrella, A.; Blanco, E.; Do, P.T.; Nunes-Nesi, A.; Trauth, A.U.; Fiermonte, G.; Tjaden, J.; Agrimi, G.; et al. Molecular identification and functional characterization of Arabidopsis thaliana mitochondrial and chloroplastic NAD+ carrier proteins. J. Biol. Chem. 2009, 284, 31249–31259. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Ma, X.; Shen, J.; Li, C.; Zhang, W. The ongoing story: The mitochondria pyruvate carrier 1 in plant stress response in Arabidopsis. Plant Signal. Behav. 2014, 9, e973810. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Millar, A.H. Succinate dehydrogenase: The complex roles of a simple enzyme. Curr. Opin. Plant Biol. 2013, 16, 344–349. [Google Scholar] [CrossRef] [PubMed]
- Schertl, P.; Braun, H.P. Respiratory electron transfer pathways in plant mitochondria. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schimmeyer, J.; Bock, R.; Meyer, E.H. l-Galactono-1,4-lactone dehydrogenase is an assembly factor of the membrane arm of mitochondrial complex I in Arabidopsis. Plant Mol. Biol. 2016, 90, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Murcha, M.W.; Kmiec, B.; Kubiszewski-Jakubiak, S.; Teixeira, P.F.; Glaser, E.; Whelan, J. Protein import into plant mitochondria: Signals, machinery, processing, and regulation. J. Exp. Bot. 2014, 65, 6301–6335. [Google Scholar] [CrossRef] [PubMed]
- Huynen, M.A.; Mühlmeister, M.; Gotthardt, K.; Guerrero-Castillo, S.; Brandt, U. Evolution and structural organization of the mitochondrial contact site (MICOS) complex and the mitochondrial intermembrane space bridging (MIB) complex. Biochim. Biophys. Acta 2016, 1863, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ruan, Y.; Zhang, K.; Jian, F.; Hu, C.; Miao, L.; Gong, L.; Sun, L.; Zhang, X.; Chen, S.; et al. Mic60/Mitofilin determines MICOS assembly essential for mitochondrial dynamics and mtDNA nucleoid organization. Cell Death Differ. 2016, 23, 380–392. [Google Scholar] [CrossRef] [PubMed]
- Michaud, M.; Gros, V.; Tardif, M.; Brugière, S.; Ferro, M.; Prinz, W.A.; Toulmay, A.; Mathur, J.; Wozny, M.; Falconet, D.; et al. AtMic60 Is Involved in Plant Mitochondria Lipid Trafficking and Is Part of a Large Complex. Curr. Biol. 2016, 26, 627–639. [Google Scholar] [CrossRef] [PubMed]
- Gehl, B.; Lee, C.P.; Bota, P.; Blatt, M.R.; Sweetlove, L.J. An Arabidopsis stomatin-like protein affects mitochondrial respiratory supercomplex organization. Plant Physiol. 2014, 164, 1389–1400. [Google Scholar] [CrossRef] [PubMed]
- Tondera, D.; Grandemange, S.; Jourdain, A.; Karbowski, M.; Mattenberger, Y.; Herzig, S.; Da Cruz, S.; Clerc, P.; Raschke, I.; Merkwirth, C.; et al. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 2009, 28, 1589–1600. [Google Scholar] [CrossRef] [PubMed]
- Wai, T.; Saita, S.; Nolte, H.; Müller, S.; König, T.; Richter-Dennerlein, R.; Sprenger, H.G.; Madrenas, J.; Mühlmeister, M.; Brandt, U.; et al. The membrane scaffold SLP2 anchors a proteolytic hub in mitochondria containing PARL and the i-AAA protease YME1L. EMBO Rep. 2016, 17, 1844–1856. [Google Scholar] [CrossRef] [PubMed]
- Potting, C.; Wilmes, C.; Engmann, T.; Osman, C.; Langer, T. Regulation of mitochondrial phospholipids by Ups1/PRELI-like proteins depends on proteolysis and Mdm35. EMBO J. 2010, 29, 2888–2898. [Google Scholar] [CrossRef] [PubMed]
- Quast, R.B.; Kortt, O.; Henkel, J.; Dondapati, S.K.; Wüstenhagen, D.A.; Stech, M.; Kubick, S. Automated production of functional membrane proteins using eukaryotic cell-free translation systems. J. Biotechnol. 2015, 203, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Gibala, M.; Kicia, M.; Sakamoto, W.; Gola, E.M.; Kubrakiewicz, J.; Smakowska, E.; Janska, H. The lack of mitochondrial AtFtsH4 protease alters Arabidopsis leaf morphology at the late stage of rosette development under short-day photoperiod. Plant J. 2009, 59, 685–699. [Google Scholar] [CrossRef] [PubMed]
- Murcha, M.W.; Whelan, J. Isolation of Intact Mitochondria from the Model Plant Species Arabidopsis thaliana and Oryza sativa. Methods Mol. Biol. 2015, 1305, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ishihama, Y.; Oda, Y.; Tabata, T.; Sato, T.; Nagasu, T.; Rappsilber, J.; Mann, M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteom. 2005, 4, 1265–1272. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Chu, W.; Zhang, H.; Sun, X.; Cai, T.; Dang, L.; Wang, Q.; Yu, H.; Zhong, Y.; Chen, Z.; et al. Isolation and identification of mannose-binding proteins and estimation of their abundance in sera from hepatocellular carcinoma patients. Proteomics 2013, 13, 878–892. [Google Scholar] [CrossRef] [PubMed]
- Murcha, M.W.; Elhafez, D.; Millar, A.H.; Whelan, J. The C-terminal region of TIM17 links the outer and inner mitochondrial membranes in Arabidopsis and is essential for protein import. J. Biol. Chem. 2005, 280, 16476–16483. [Google Scholar] [CrossRef] [PubMed]
- Carrie, C.; Kühn, K.; Murcha, M.W.; Duncan, O.; Small, I.D.; O’Toole, N.; Whelan, J. Approaches to defining dual-targeted proteins in Arabidopsis. Plant J. 2009, 57, 1128–1139. [Google Scholar] [CrossRef] [PubMed]
- Mruk, D.D.; Cheng, C.Y. Enhanced chemiluminescence (ECL) for routine immunoblotting: An inexpensive alternative to commercially available kits. Spermatogenesis 2011, 1, 121–122. [Google Scholar] [CrossRef] [PubMed]
- Becker, T.; Wenz, L.S.; Krüger, V.; Lehmann, W.; Müller, J.M.; Goroncy, L.; Zufall, N.; Lithgow, T.; Guiard, B.; Chacinska, A.; et al. The mitochondrial import protein Mim1 promotes biogenesis of multispanning outer membrane proteins. J. Cell Biol. 2011, 194, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Duncan, O.; Carrie, C.; Wang, Y.; Murcha, M.W. In vitro and in vivo protein uptake studies in plant mitochondria. Methods Mol. Biol. 2015, 1305, 61–81. [Google Scholar] [CrossRef] [PubMed]
- Mossmann, D.; Vögtle, F.N.; Taskin, A.A.; Teixeira, P.F.; Ring, J.; Burkhart, J.M.; Burger, N.; Pinho, C.M.; Tadic, J.; Loreth, D.; et al. Amyloid-β peptide induces mitochondrial dysfunction by inhibition of preprotein maturation. Cell Metab. 2014, 20, 662–669. [Google Scholar] [CrossRef] [PubMed]
| Functional Category | AGI | Protein | Localization |
|---|---|---|---|
| Transmembrane transport | AT1G25380.1 | Nicotinamide adenine dinucleotide transporter 2 | inner membrane |
| AT4G22310.1 | Mitochondrial pyruvate carrier 4 (MPC4) | inner membrane | |
| Metabolism | AT4G35260.1 | Isocitrate dehydrogenase NAD regulatory subunit 1 | matrix |
| AT1G47420.1 | Succinate dehydrogenase flavoprotein 5 | inner membrane | |
| AT5G66760.1 | Succinate dehydrogenase flavoprotein 1 | inner membrane | |
| AT3G47930.1 | l-galactono-1,4-lactone dehydrogenase | intermembrane space | |
| AT5G23300.1 | Dihydroorotate dehydrogenase | inner membrane | |
| Mitochondrial protein import | AT1G53530.1 | Mitochondrial inner membrane protease subunit 1 | inner membrane |
| AT2G46470.1 | OXA1-like protein | inner membrane | |
| AT3G09700.1 | Pam18-2 | inner membrane | |
| AT5G05520.1 | Sam50-2 | outer membrane | |
| AT2G35795.1 | Pam18-1 | inner membrane | |
| AT2G37410.1 | Tim17-2 | inner membrane | |
| Membrane organization | AT5G54100.1 | Stomatin-like protein 2 (Slp2) | inner membrane |
| AT4G27585.1 | Stomatin-like protein 1 (Slp1) | inner membrane | |
| AT4G39690.1 | Mic60 | inner membrane | |
| Unknown | AT5G62270.1 | Gamete cell defective (GCD) | unknown |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Opalińska, M.; Parys, K.; Jańska, H. Identification of Physiological Substrates and Binding Partners of the Plant Mitochondrial Protease FTSH4 by the Trapping Approach. Int. J. Mol. Sci. 2017, 18, 2455. https://doi.org/10.3390/ijms18112455
Opalińska M, Parys K, Jańska H. Identification of Physiological Substrates and Binding Partners of the Plant Mitochondrial Protease FTSH4 by the Trapping Approach. International Journal of Molecular Sciences. 2017; 18(11):2455. https://doi.org/10.3390/ijms18112455
Chicago/Turabian StyleOpalińska, Magdalena, Katarzyna Parys, and Hanna Jańska. 2017. "Identification of Physiological Substrates and Binding Partners of the Plant Mitochondrial Protease FTSH4 by the Trapping Approach" International Journal of Molecular Sciences 18, no. 11: 2455. https://doi.org/10.3390/ijms18112455





