In-Depth Glyco-Peptidomics Approach Reveals Unexpected Diversity of Glycosylated Peptides and Atypical Post-Translational Modifications in Dendroaspis angusticeps Snake Venom
Abstract
:1. Introduction
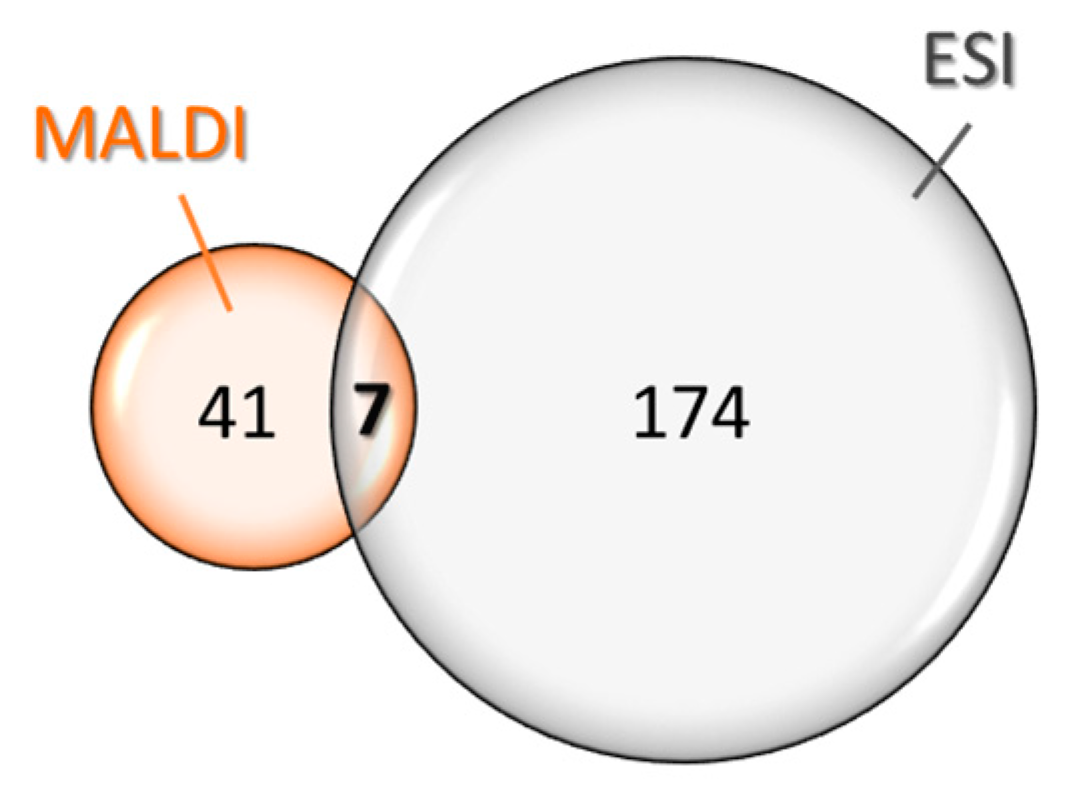
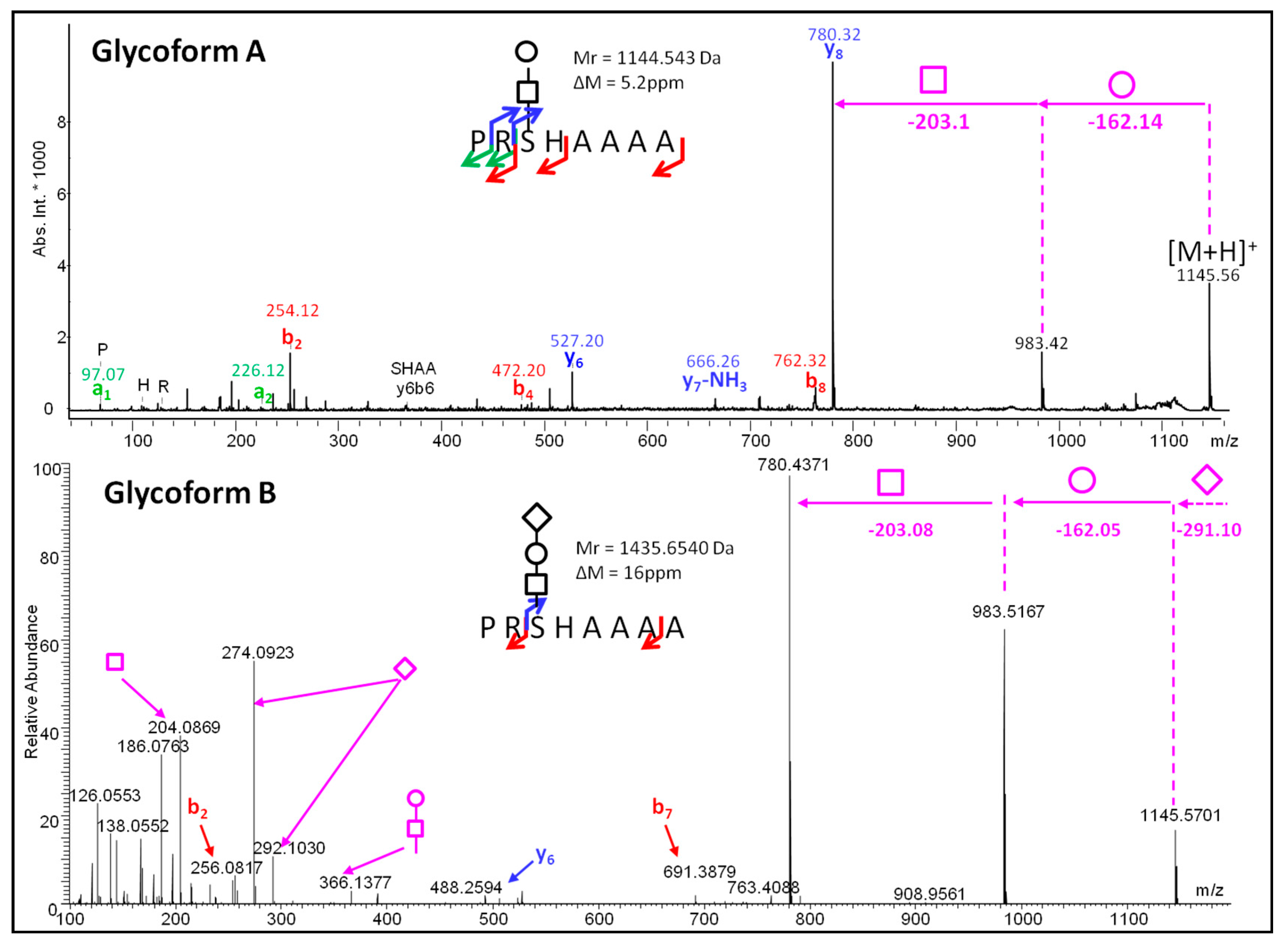
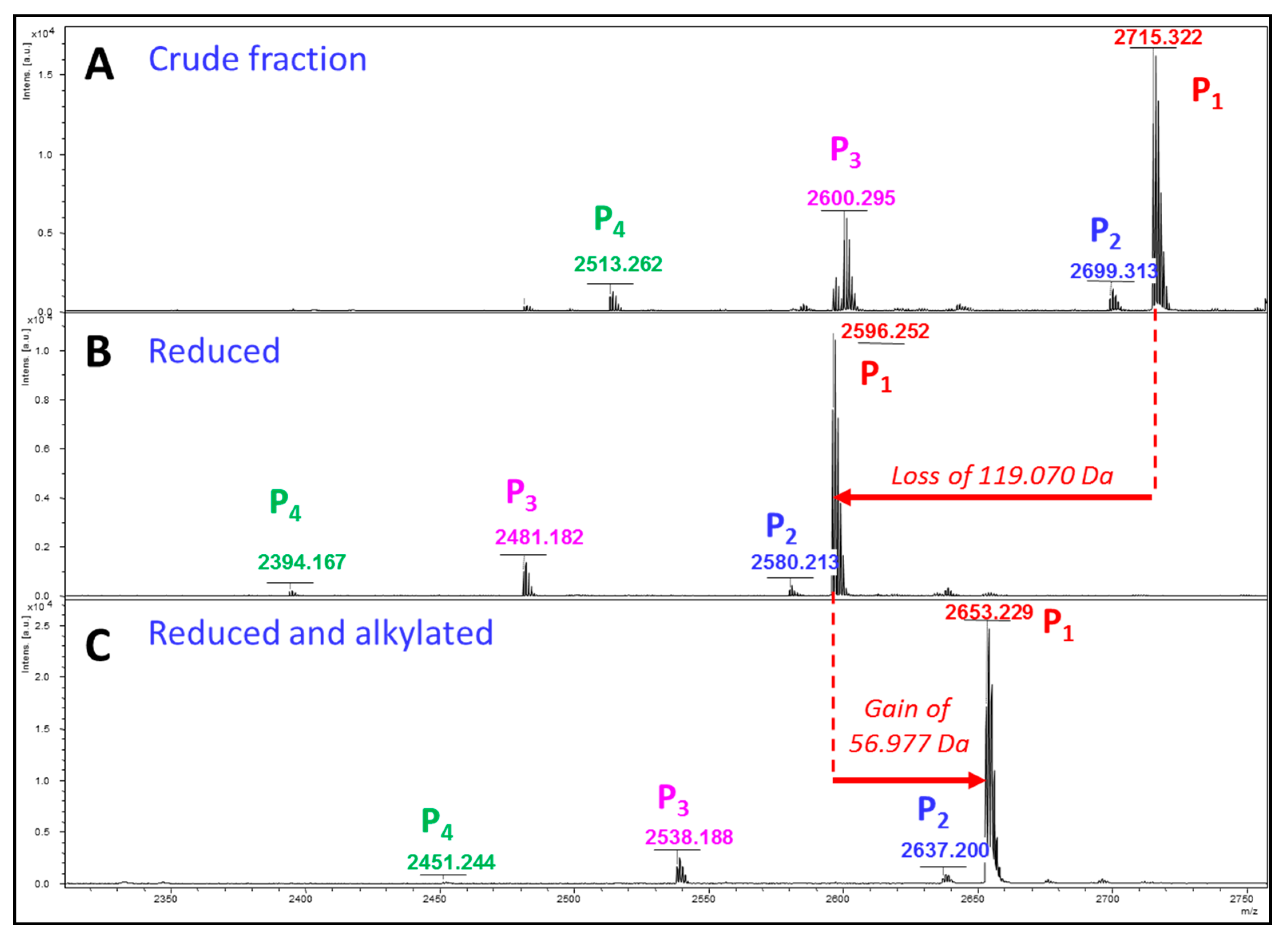
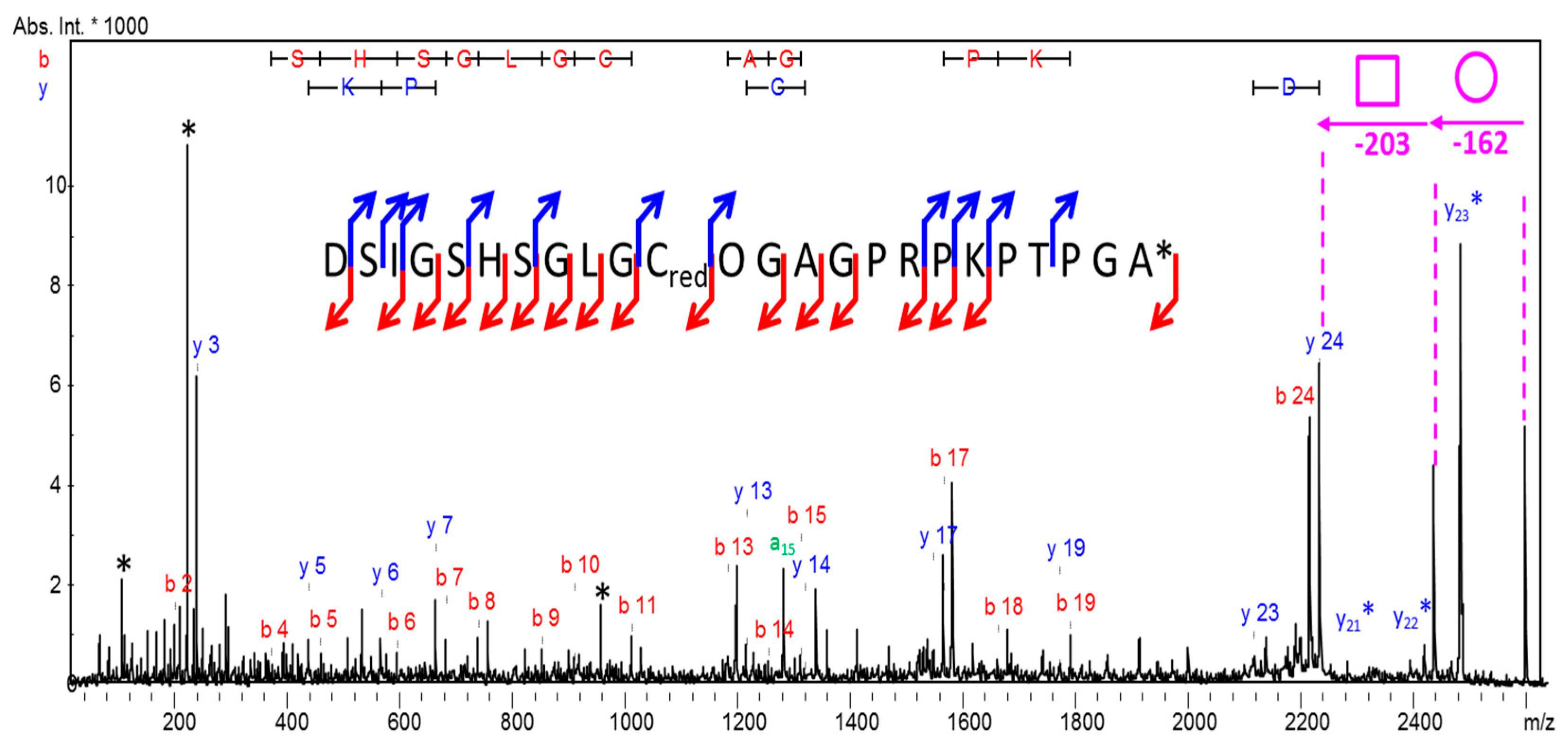
2. Results
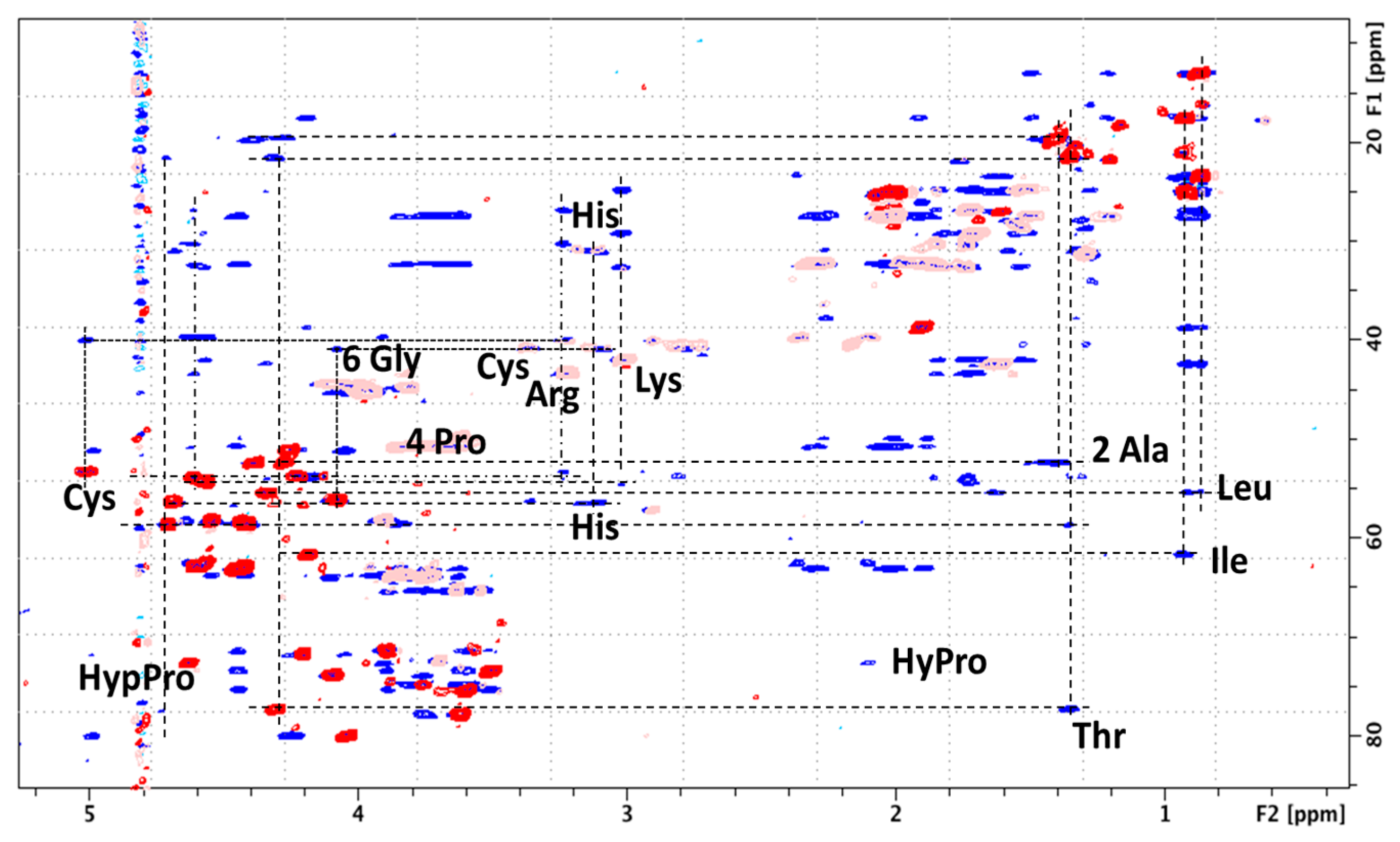
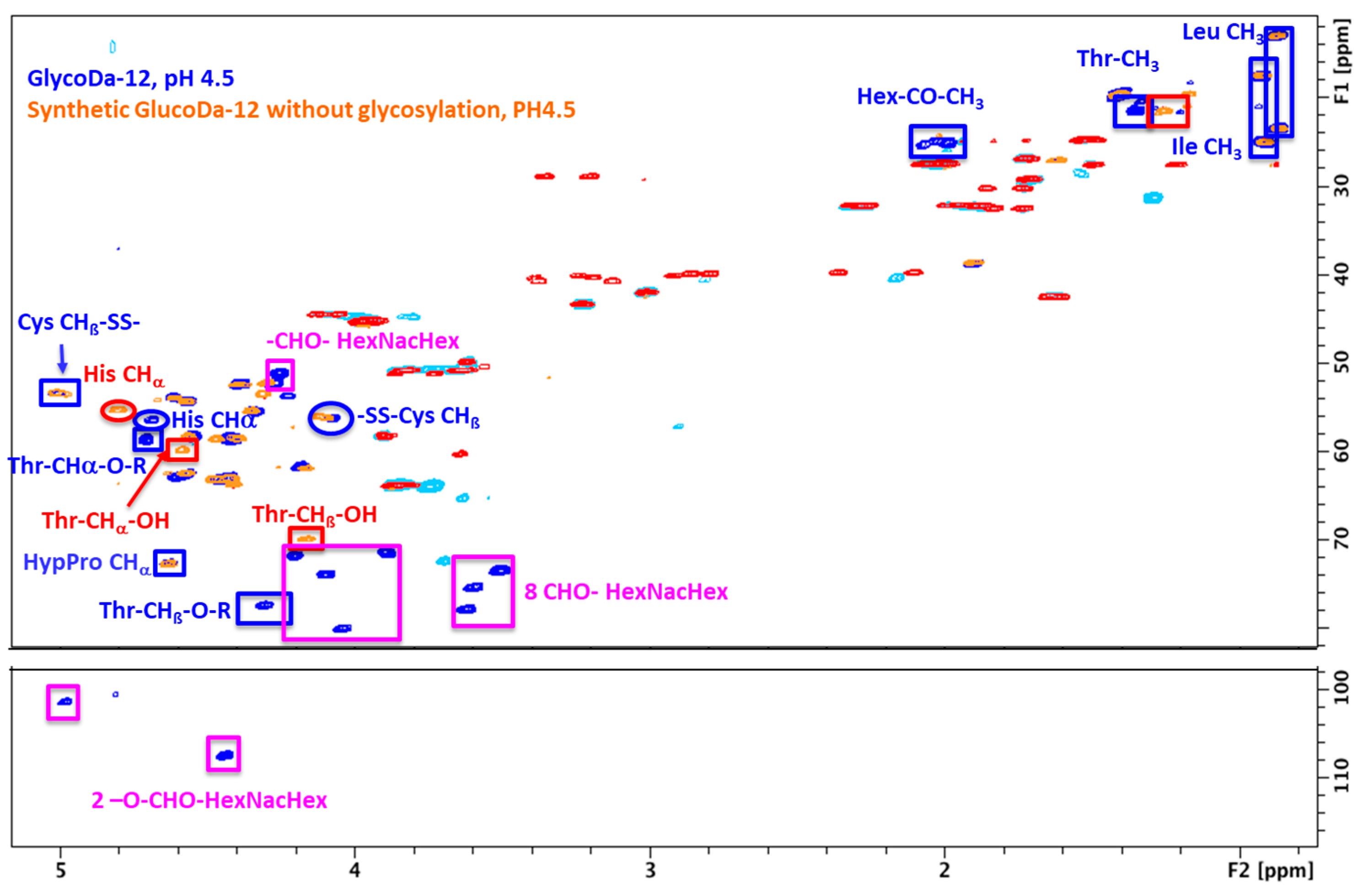
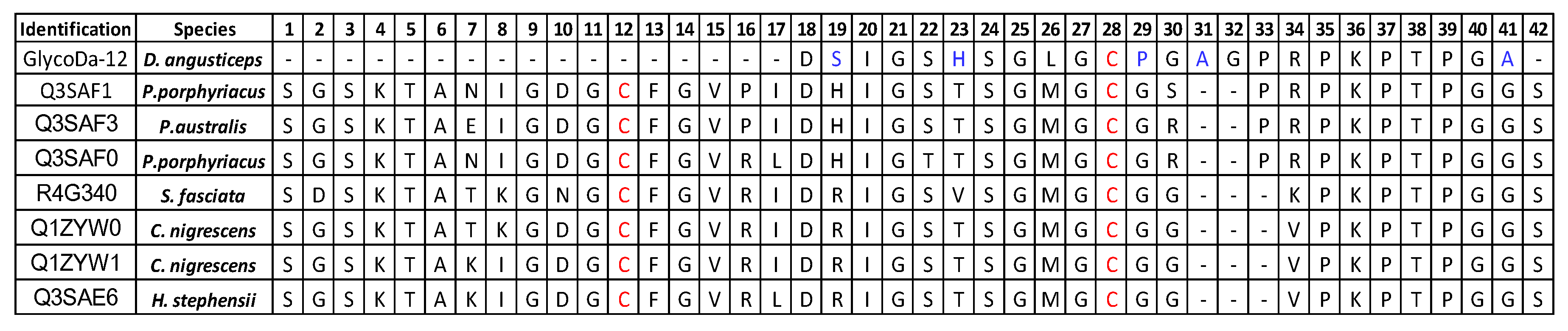
Glycopeptidome Characterization
3. Discussion
4. Material and Methods
5. Conclusions
Supplementary Materials
Author Contribution
Conflicts of Interest
References
- Quinton, L.; Girard, E.; Maiga, A.; Rekik, M.; Lluel, P.; Masuyer, G.; Larregola, M.; Marquer, C.; Ciolek, J.; Magnin, T.; et al. Isolation and pharmacological characterization of AdTx1, a natural peptide displaying specific insurmountable antagonism of the a 1A-adrenoceptor. Br. J. Pharmacol. 2010, 159, 316–325. [Google Scholar] [CrossRef] [PubMed]
- Adem, A.; Asblom, A.; Johansson, G.; Mbugua, P.M.; Karlsson, E. Toxins from the venom of the green mamba Dendroaspis angusticeps that inhibit the binding of quinuclidinyl benzilate to muscarinic acetylcholine receptors. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 1988, 968, 340–345. [Google Scholar] [CrossRef]
- Servent, D.; Blanchet, G.; Mourier, G.; Marquer, C.; Marcon, E.; Fruchart-Gaillard, C. Muscarinic toxins. Toxicon 2011, 58, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Rouget, C.; Quinton, L.; Maïga, A.; Gales, C.; Masuyer, G.; Malosse, C.; Chamot-Rooke, J.; Thai, R.; Mourier, G.; De Pauw, E.; et al. Identification of a novel snake peptide toxin displaying high affinity and antagonist behaviour for the α2-adrenoceptors. Br. J. Pharmacol. 2010, 161, 1361–1374. [Google Scholar] [CrossRef] [PubMed]
- Ciolek, J.; Reinfrank, H.; Quinton, L.; Viengchareun, S.; Stura, E.A.; Vera, L.; Sigismeau, S.; Mouillac, B.; Orcel, H.; Peigneur, S.; et al. Green mamba peptide targets type-2 vasopressin receptor against polycystic kidney disease. Proc. Natl. Acad. Sci. USA 2017, 114, 7154–7159. [Google Scholar] [CrossRef] [PubMed]
- Dutertre, S.; Jin, A.; Kaas, Q.; Jones, A.; Alewood, P.F.; Lewis, R.J. Deep venomics reveals the mechanism for expanded peptide diversity in cone snail venom. Mol. Cell. Proteom. 2013, 12, 312–329. [Google Scholar] [CrossRef] [PubMed]
- Cho, W.; Tomasselli, A.G.; Heinrikson, R.L.; Kézdy, F.J. The chemical basis for interfacial activation of monomeric phospholipase. J. Biol. Chem. 1988, 263, 11237–11241. [Google Scholar] [PubMed]
- Birrell, G.W.; Earl, S.; Masci, P.P.; Wallis, T.P.; Gorman, J.J.; Lavin, M.F. Molecular Diversity in Venom from the Australian Brown Snake, Pseudonaja textilis. Mol. Cell. Proteom. 2006, 5, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Earl, S.T.H.; Birrell, G.W.; Wallis, T.P.; Pierre, L.D.S.; Masci, P.P.; de Jersey, J.; Gorman, J.J.; Lavin, M.F. Post-translational modification accounts for the presence of varied forms of nerve growth factor in Australian elapid snake venoms. Proteomics 2006, 6, 6554–6565. [Google Scholar] [CrossRef] [PubMed]
- Quinton, L.; Gilles, N.; Smargiasso, N.; Kiehne, A.; De Pauw, E. An Unusual Family of Glycosylated Peptides Isolated from Dendroaspis angusticeps Venom and Characterized by Combination of Collision Induced and Electron Transfer Dissociation. J. Am. Soc. Mass Spectrom. 2011, 22, 1891–1897. [Google Scholar] [CrossRef] [PubMed]
- Wilm, M. Principles of Electrospray Ionization. Mol. Cell. Proteom. 2011, 10, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Knochenmuss, R.; Stortelder, A.; Breuker, K.; Zenobi, R. Secondary ion-molecule reactions in matrix assisted desorpion/ionization. J. Mass Spectrom. 2000, 35, 1237–1245. [Google Scholar] [CrossRef]
- Harvey, D.J.; Merry, A.H.; Royle, L.; Campbell, M.P.; Dwek, R.A.; Rudd, P.M. Proposal for a standard system for drawing structural diagrams of N- and O-linked carbohydrates and related compounds. Proteomics 2009, 9, 3796–3801. [Google Scholar] [CrossRef] [PubMed]
- Wuthrich, K. NMR of Proteins and Nucleic Acids; Wiley: New York, NY, USA, 1986. [Google Scholar]
- Boyer, R.D.; Johnson, R.; Krishnamurthy, K. Compensation of refocusing inefficiency with synchronized inversion sweep (CRISIS) in multiple-edited HSQC. J. Magn. Reson. 2003, 165, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Kover, K.E.; Prakash, O.; Hruby, V.J. z-Filtered heteronuclear coupled-HSQC-TOCSY experiment as a means for measuring long-range heteronuclear coupling constants. J. Magn. Reson. 1993, 103, 92–96. [Google Scholar] [CrossRef]
- Theillet, F.X.; Smet-Nocca, C.; Liokatis, S.; Thongwichian, R.; Kosten, J.; Yoon, M.K.; Kriwacki, R.W.; Landrieu, I.; Lippens, G.; Selenko, P. Cell signaling, post-translational protein modifications and NMR spectroscopy. J. Biomol. NMR 2012, 54, 217–236. [Google Scholar] [CrossRef] [PubMed]
- Dill, K.; Berman, E.; Pavia, A.A. Natural-abundance of 13C-nuclear magnetic resonance-spectral studies of carbohydrates linked to amino acids and proteins. Adv. Carbohydr. Chem. Biochem. 1985, 43, 1–49. [Google Scholar] [PubMed]
- Martin, O.A.; Villegas, M.E.; Vila, J.A.; Scheraga, H.A. Analysis of 13Ca and 13Cb chemical shifts of cysteine and cystine residues in proteins: A quantum chemical approach. J. Biomol. NMR 2010, 46, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Rajarathnam, K. 13C NMR chemcial shifts can predict disulfide bond formation. J. Biomol. NMR 2000, 18, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Harvey, D.J.; Bateman, R.H.; Bordoli, R.S.; Tyldesley, R. Ionisation and fragmentation of complex glycans with a quadrupole time-of-flight mass spectrometer fitted with a matrix-assisted laser desorption/ionisation ion source. RCMS 2000, 14, 2135–2142. [Google Scholar] [CrossRef]
- Christlet, T.H.T.; Veluraja, K. Database analysis of O-glycosylation sites in proteins. Biophys. J. 2001, 80, 952–960. [Google Scholar] [CrossRef]
- Gerwig, G.J.; Hocking, H.G.; Stöcklin, R.; Kamerling, J.P.; Boelens, R. Glycosylation of conotoxins. Mar. Drugs 2013, 11, 623–642. [Google Scholar] [CrossRef] [PubMed]
- Pierre, L.S.; Flight, S.; Masci, P.P.; Hanchard, K.J.; Lewis, R.J.; Alewood, P.F.; de Jersey, J.; Lavin, M.F. Cloning and characterisation of natriuretic peptides from the venom glands of Australian elapids. Biochimie 2006, 88, 1923–1931. [Google Scholar] [CrossRef] [PubMed]
- Rowan, E.G.; Harvey, A.L. Snake toxins from Mamba Venoms: Unique Tools dor the Physiologist. Acta Chim. Slov. 2011, 58, 689–692. [Google Scholar] [PubMed]
- Schweitz, H.; Vigne, P.; Moinier, D.; Frelin, C.; Lazdunski, M. A new member of the natriuretic peptide family is present in the venom of the green mamba (Dendroaspis angusticeps). J. Biol. Chem. 1992, 267, 13928–13932. [Google Scholar] [PubMed]
- Vink, S.; Jin, A.H.; Poth, K.J.; Head, G.A.; Alewood, P.F. Natriuretic peptide drug leads from snake venom. Toxicon 2012, 59, 434–445. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.; Chen, H.H. Natriuretic peptides as a novel target in resistant hypertension. Curr. Hypertens. Rep. 2015, 17, 530. [Google Scholar] [CrossRef] [PubMed]

| Name | RT (min) | Monoisotopic Mass (Da) | Monoisotopic Mass w/o Glycan (Da) | Sequence | Glycan |
|---|---|---|---|---|---|
| GlycoDa-01 | 7.75 | 1340.483 | 975.135 | KNPTKPEY | Hex-HexNAc |
| GlycoDa-02 | 8.50 | 1411.567 | 1046.455 | KNTPKPAEY | Hex-HexNAc |
| GlycoDa-03 | 9.00 | 1468.634 | 1103.491 | KPPTELQYE | Hex-HexNAc |
| GlycoDa-04 | 9.25 | 1469.612 | 1104.447 | KPPTELEYE | Hex-HexNAc |
| GlycoDa-05 | 9.50 | 1297.316 | 932.310 | KPTNKSGSD | Hex-HexNAc |
| GlycoDa-06 | 9.50 | 1526.674 | 1161.340 | KPEPTNGMAAF | Hex-HexNAc |
| GlycoDa-07 | 12.25 | 1073.53 | 708.196 | PRSHAAA | Hex-HexNAc |
| GlycoDa-08 | 14.25 | 1144.554 | 779.316 | PRSHAAAA | Hex-HexNAc |
| GlycoDa-09 | 17.50 | 1002.405 | 637.277 | PRSHAA | Hex-HexNAc |
| GlycoDa-10 | 24.75 | 2164.089 | 1798.645 | KSPPQALNKPLPAPSAPS | Hex-HexNAc |
| GlycoDa-11 | 26.50 | 2099.036 | 1733.885 | KPPMVMSPKRPSPEPG | Hex-HexNac |
| Monoisotopic Mass w/o Glycan (Da) | Unmodified | Hex-HexNAc | Hex2-HexNAc2 | NeuAc-Hex-HexNAc | NeuAc-Hex2-HexNAc | dHex-Hex2-HexNAc2 | NeuAc-dHex-Hex2-HexNAc2 |
|---|---|---|---|---|---|---|---|
| 637.36 | X | X | X | ||||
| 779.43 | X | X | X | ||||
| 1048.62 | X | X | X | ||||
| 1077.58 | X | X | X | X | X | ||
| 1105.58 | X | X | |||||
| 1295.61 | X | X | X | ||||
| 1311.68 | X | X | X | ||||
| 1388.71 | X | X |
| Peptide | Peptide Sequence | Mth | Mexp |
|---|---|---|---|
| GlycoDa-12 |  | 2714.228 Da | 2714.315 Da |
| GlycoDa-13 |  | 2698.232 Da | 2698.306 Da |
| GlycoDa-14 |  | 2599.200 Da | 2599.288 Da |
| GlycoDa-15 |  | 2512.169 Da | 2512.256 Da |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Degueldre, M.; Echterbille, J.; Smargiasso, N.; Damblon, C.; Gouin, C.; Mourier, G.; Gilles, N.; De Pauw, E.; Quinton, L. In-Depth Glyco-Peptidomics Approach Reveals Unexpected Diversity of Glycosylated Peptides and Atypical Post-Translational Modifications in Dendroaspis angusticeps Snake Venom. Int. J. Mol. Sci. 2017, 18, 2453. https://doi.org/10.3390/ijms18112453
Degueldre M, Echterbille J, Smargiasso N, Damblon C, Gouin C, Mourier G, Gilles N, De Pauw E, Quinton L. In-Depth Glyco-Peptidomics Approach Reveals Unexpected Diversity of Glycosylated Peptides and Atypical Post-Translational Modifications in Dendroaspis angusticeps Snake Venom. International Journal of Molecular Sciences. 2017; 18(11):2453. https://doi.org/10.3390/ijms18112453
Chicago/Turabian StyleDegueldre, Michel, Julien Echterbille, Nicolas Smargiasso, Christian Damblon, Charlotte Gouin, Gilles Mourier, Nicolas Gilles, Edwin De Pauw, and Loïc Quinton. 2017. "In-Depth Glyco-Peptidomics Approach Reveals Unexpected Diversity of Glycosylated Peptides and Atypical Post-Translational Modifications in Dendroaspis angusticeps Snake Venom" International Journal of Molecular Sciences 18, no. 11: 2453. https://doi.org/10.3390/ijms18112453





