Mutation-Structure-Function Relationship Based Integrated Strategy Reveals the Potential Impact of Deleterious Missense Mutations in Autophagy Related Proteins on Hepatocellular Carcinoma (HCC): A Comprehensive Informatics Approach
Abstract
:1. Introduction
2. Results
2.1. Missense SNP Datasets
2.2. Missense SNP Analysis
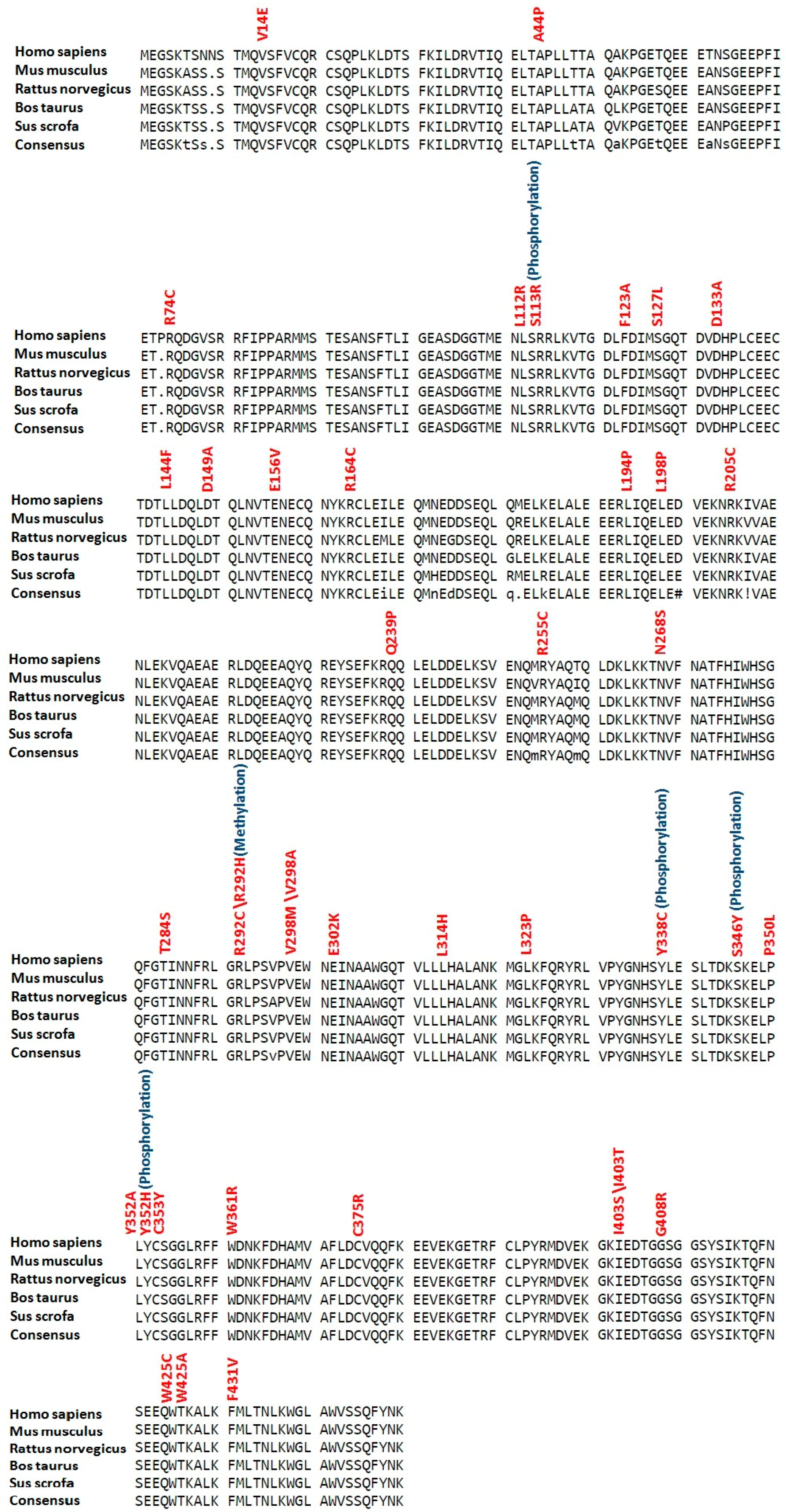
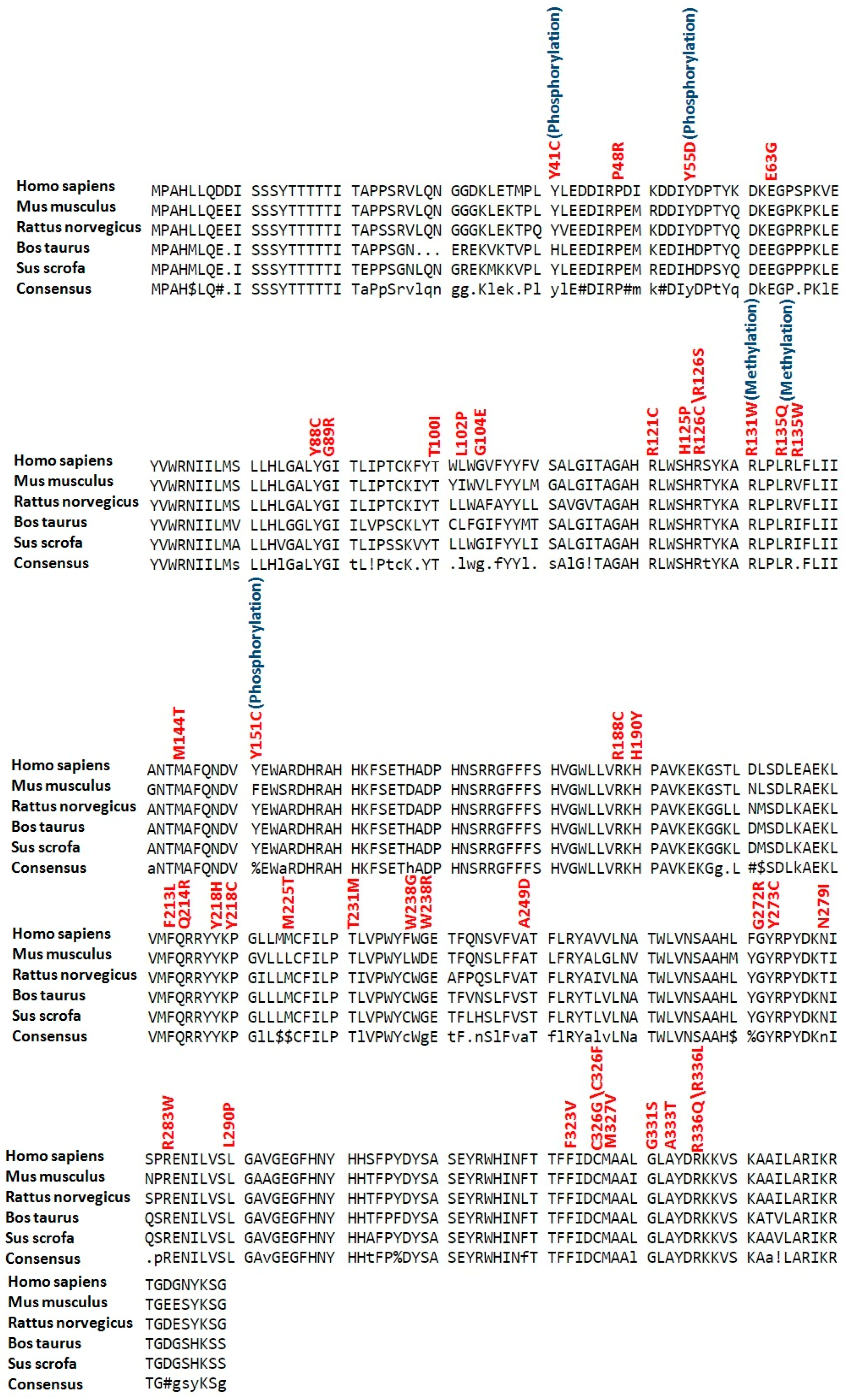
2.3. Conservation Profile of High-Risk Missense SNPs
2.4. HCC Related Mutations by PinSnps and SNPEffect4.0 Tool
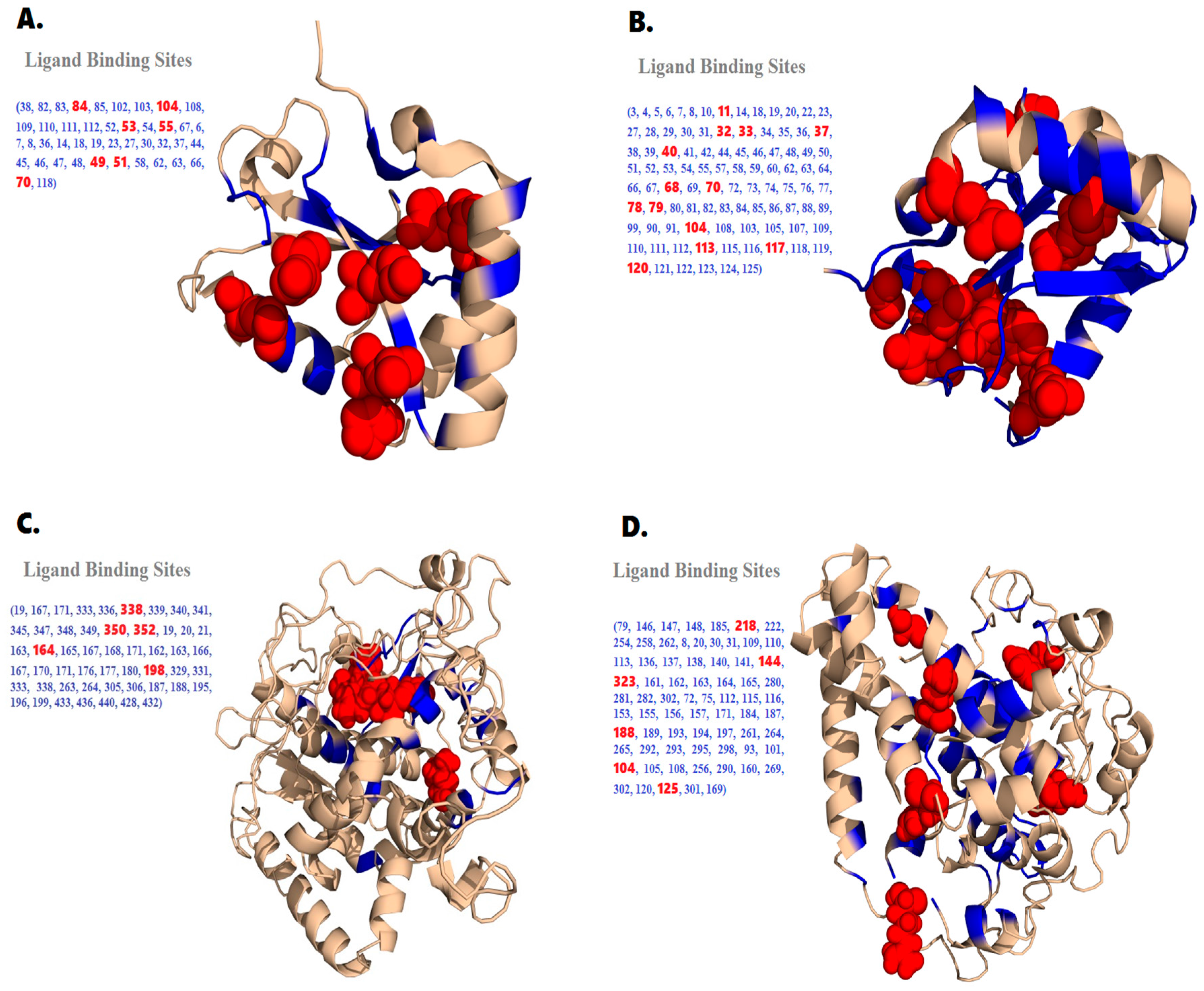
2.5. Prediction of Functional Binding Sites
2.6. Phosphorylation and Kinase-Specific Phosphorylation Sites in LC3A, LC3B, BECN1 and SCD1
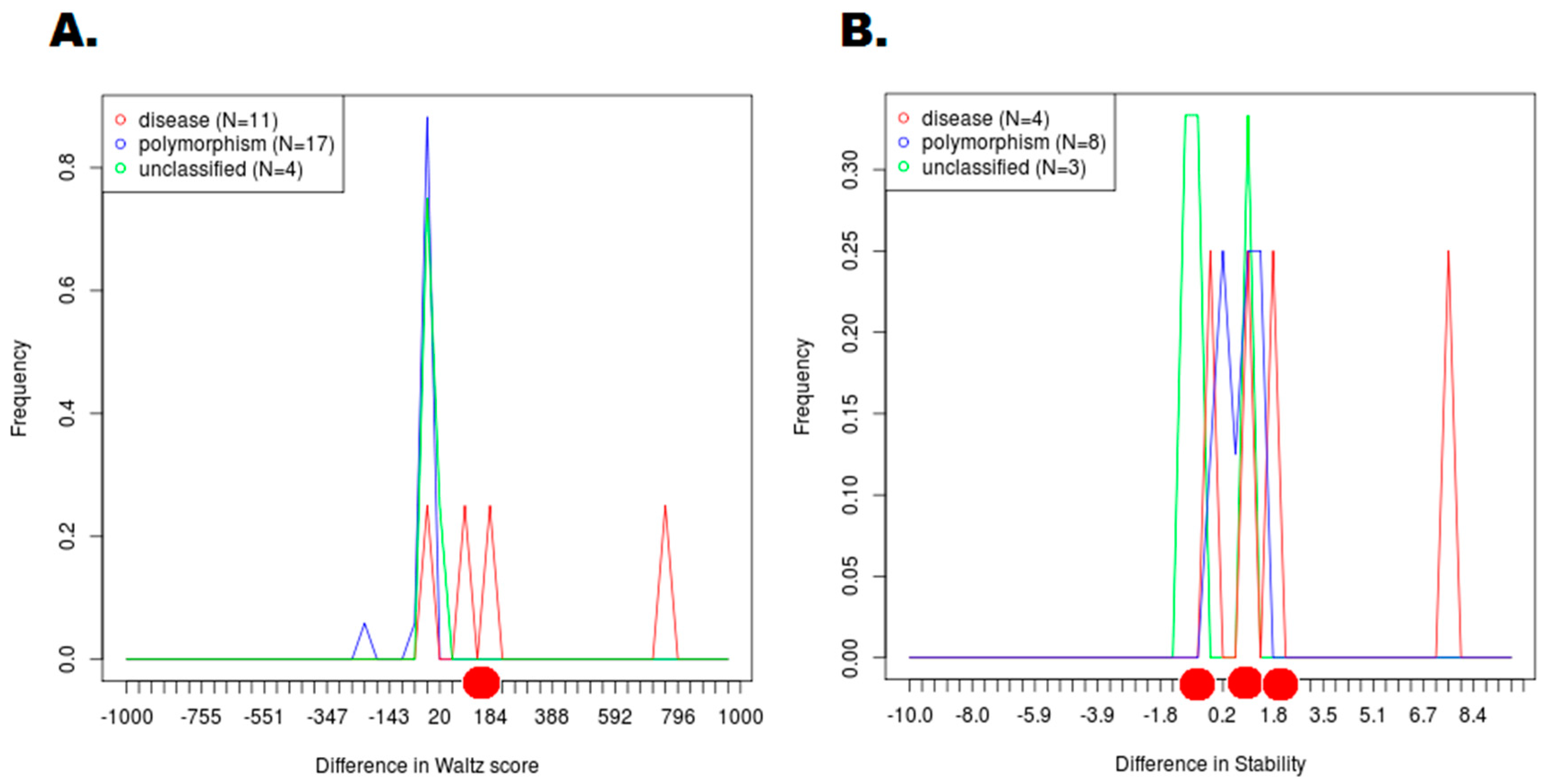
2.7. Normal Mode Analysis of Highly Deleterious HCC-Associated SNPs
2.8. Ubiquitylation Sites in LC3A, LC3B, BECN1 and SCD1
2.9. Palmitoylation and Methylation Sites in LC3A, LC3B, BECN1 and SCD1
2.10. Prediction of Acetylating Sites in LC3A, LC3B, BECN1 and SCD1
2.11. Sumoylation Sites in LC3A, LC3B, BECN1 and SCD1
2.12. Analysis of Overlap between High Risk SNPs and Potential PTM Sites
3. Discussion
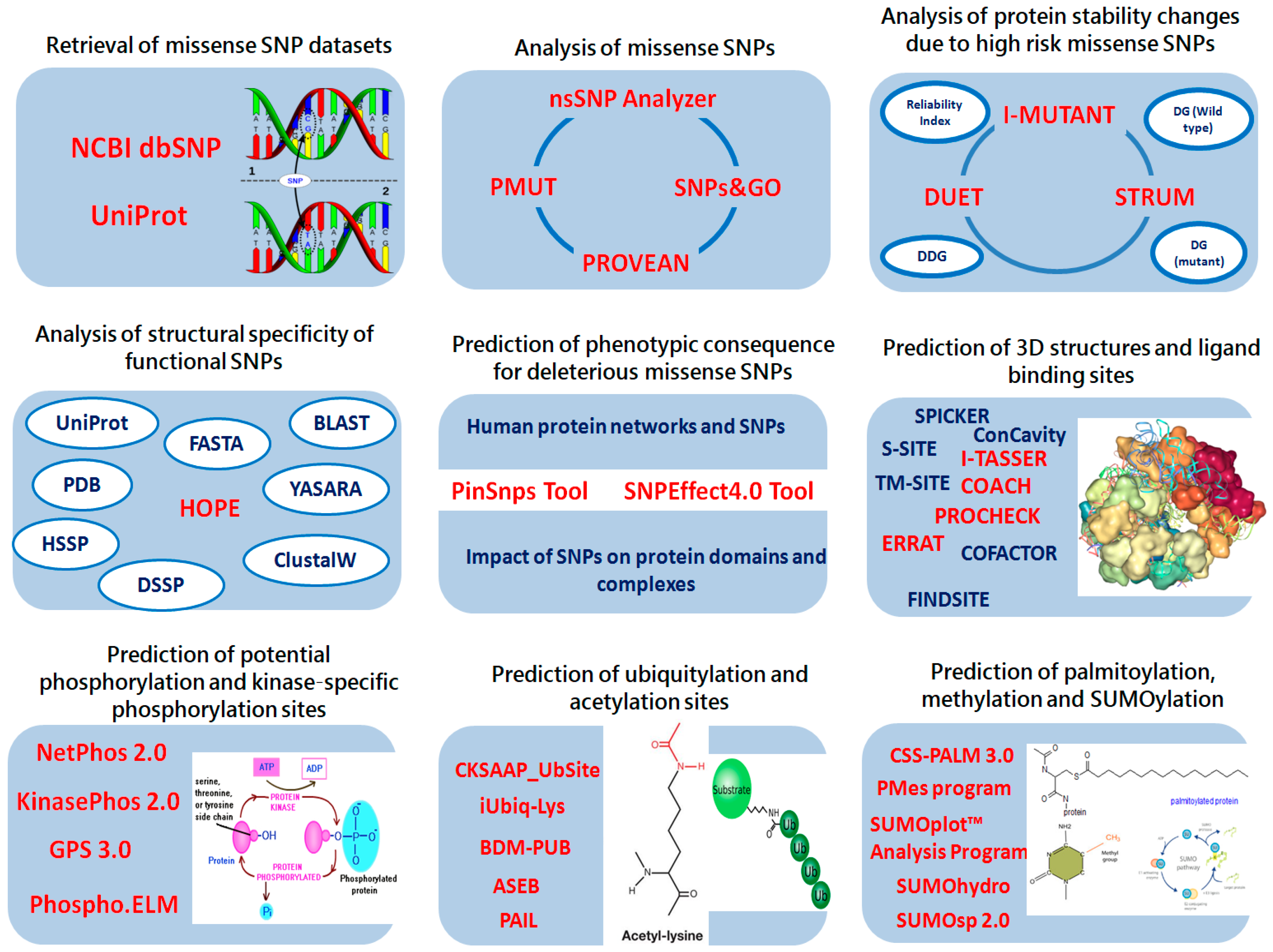
4. Materials and Methods
4.1. Protein Sequence Datasets
4.2. Retrieval of Missense SNP Datasets
4.3. Analysis of Functional Consequences of Missense SNPs
4.4. Analysis of Protein Stability Changes Due to High Risk Missense SNPs
4.5. Analysis of Structural Specificity of Functional SNPs
4.6. Prediction of Phenotypic Consequence for Deleterious Missense SNPs
4.7. Prediction of Ligand Binding Sites in LC3A, LC3B, BECN1 and SCD1 Proteins
4.8. Normal Mode Analysis
4.9. Prediction of Post-Translational Modification Sites
4.9.1. Prediction of Potential Phosphorylation and Kinase-Specific Phosphorylation Sites in LC3A, LC3B, BECN1 and SCD1
4.9.2. Prediction of Ubiquitylation Sites in LC3A, LC3B, BECN1 and SCD1
4.9.3. Prediction of Palmitoylation and Methylation Sites
4.9.4. Prediction of Acetylating Sites
4.9.5. Prediction of Sumoylation Sites in LC3A, LC3B, BECN1 and SCD1
4.10. PPI Analysis for LC3A, LC3B, BECN1 and SCD1
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kaur, J.; Debnath, J. Autophagy at the crossroads of catabolism and anabolism. Nat. Rev. Mol. Cell Biol. 2015, 16, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Gong, Z.; Shen, H.M. The role of autophagy in liver cancer: Molecular mechanisms and potential therapeutic targets. Biochim. Biophys. Acta 2013, 1836, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Levine, B. Autophagy in mammalian development and differentiation. Nat. Cell Biol. 2010, 12, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.M.; Ding, W.X.; Gao, W. Autophagy in the liver. Hepatology 2008, 47, 1773–1785. [Google Scholar] [CrossRef] [PubMed]
- Cuervo, A.M.; Knecht, E.; Terlecky, S.R.; Dice, J.F. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am. J. Physiol. 1995, 269, C1200–C1208. [Google Scholar] [PubMed]
- Mortimore, G.E.; Hutson, N.J.; Surmacz, C.A. Quantitative correlation between proteolysis and macro- and micro-autophagy in mouse hepatocytes during starvation and refeeding. Proc. Natl. Acad. Sci. USA 1983, 80, 2179–2183. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Jang, B.K. The Role of Autophagy in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2015, 16, 26629–26643. [Google Scholar] [CrossRef] [PubMed]
- Cursio, R.; Colosetti, P.; Codogno, P.; Cuervo, A.M.; Shen, H.M. The role of autophagy in liver diseases: Mechanisms and potential therapeutic targets. BioMed Res. Int. 2015. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.H.; Jia, C.C.; Chen, J.; Lin, Z.X.; Ruan, D.Y.; Li, X.; Lin, Q.; Min, D.; Ma, X.K.; Wan, X.B.; et al. Autophagic LC3B overexpression correlates with malignant progression and predicts a poor prognosis in hepatocellular carcinoma. Tumour Biol. 2014, 35, 12225–12233. [Google Scholar] [CrossRef] [PubMed]
- Xi, S.Y.; Lu, J.B.; Chen, J.W.; Cao, Y.; Luo, R.Z.; Wu, Q.L.; Cai, M.Y. The “stone-like” pattern of LC3A expression and its clinicopathologic significance in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2013, 431, 760–766. [Google Scholar] [CrossRef] [PubMed]
- Qiu, D.M.; Wang, G.L.; Chen, L.; Xu, Y.Y.; He, S.; Cao, X.L.; Qin, J.; Zhou, J.M.; Zhang, Y.X. The expression of beclin-1, an autophagic gene, in hepatocellular carcinoma associated with clinical pathological and prognostic significance. BMC Cancer 2014, 14, 327. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.M.; Jiang, Q.H.; Cai, C.; Qu, M.; Shen, W. SCD1 negatively regulates autophagy-induced cell death in human hepatocellular carcinoma through inactivation of the AMPK signaling pathway. Cancer Lett. 2015, 358, 180–190. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Klionsky, D.J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009, 43, 67–93. [Google Scholar] [CrossRef] [PubMed]
- Wani, W.Y.; Boyer-Guittaut, M.; Dodson, M.; Chatham, J.; Darley-Usmar, V.; Zhang, J. Regulation of autophagy by protein post-translational modification. Lab. Investig. 2015, 95, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Todde, V.; Veenhuis, M.; van der Klei, I.J. Autophagy: Principles and significance in health and disease. Biochim. Biophys. Acta 2009, 1792, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Datta, A.; Mazumder, M.H.; Chowdhury, A.S.; Hasan, M.A. Functional and Structural Consequences of Damaging Single Nucleotide Polymorphisms in Human Prostate Cancer Predisposition Gene RNASEL. BioMed Res. Int. 2015, 2015, 271458. [Google Scholar] [CrossRef] [PubMed]
- Thakur, R.; Shankar, J. In silico Analysis Revealed High-risk Single Nucleotide Polymorphisms in Human Pentraxin-3 Gene and their Impact on Innate Immune Response against Microbial Pathogens. Front. Microbiol. 2016, 7, 192. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Zhang, M.; Cui, P.; Li, H.; Jia, J.; Li, Y.; Xie, L. Nonsynonymous Single-Nucleotide Variations on Some Posttranslational Modifications of Human Proteins and the Association with Diseases. Comput. Math. Methods Med. 2015. [Google Scholar] [CrossRef] [PubMed]
- Benzeno, S.; Lu, F.; Guo, M.; Barbash, O.; Zhang, F.; Herman, J.G.; Klein, P.S.; Rustgi, A.; Diehl, J.A. Identification of mutations that disrupt phosphorylation-dependent nuclear export of cyclin D1. Oncogene 2006, 25, 6291–6303. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Jiang, C.; Gao, X.; Liu, Z.; Yuan, Z.; Jin, C.; Wen, L.; Zhang, Z.; Xue, Y.; Yao, X. PhosSNP for systematic analysis of genetic polymorphisms that influence protein phosphorylation. Mol. Cell Proteom. 2010, 9, 623–634. [Google Scholar] [CrossRef] [PubMed]
- Radivojac, P.; Baenziger, P.H.; Kann, M.G.; Mort, M.E.; Hahn, M.W.; Mooney, S.D. Gain and loss of phosphorylation sites in human cancer. Bioinformatics 2008, 24, i241–i247. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.Y.; Chang, C.H.; Yu, Y.L.; Lin, T.C.; Lee, S.A.; Yen, C.C.; Yang, J.M.; Lai, J.M.; Hong, Y.R.; Tseng, T.L.; et al. PhosphoPOINT: A comprehensive human kinase interactome and phospho-protein database. Bioinformatics 2008, 24, i14–i20. [Google Scholar] [CrossRef] [PubMed]
- Ryu, G.M.; Song, P.; Kim, K.W.; Oh, K.S.; Park, K.J.; Kim, J.H. Genome-wide analysis to predict protein sequence variations that change phosphorylation sites or their corresponding kinases. Nucleic Acids Res. 2009, 37, 1297–1307. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Mizushima, N. Autophagy and human diseases. Cell Res. 2014, 24, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Schmukler, E.; Kloog, Y.; Pinkas-Kramarski, R. Ras and autophagy in cancer development and therapy. Oncotarget 2014, 5, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.R.; Kim, M.S.; Oh, J.E.; Kim, Y.R.; Song, S.Y.; Kim, S.S.; Ahn, C.H.; Yoo, N.J.; Lee, S.H. Frameshift mutations of autophagy-related genes ATG2B, ATG5, ATG9B and ATG12 in gastric and colorectal cancers with microsatellite instability. J. Pathol. 2009, 217, 702–706. [Google Scholar] [CrossRef] [PubMed]
- An, C.H.; Kim, M.S.; Yoo, N.J.; Park, S.W.; Lee, S.H. Mutational and expressional analyses of ATG5, an autophagy-related gene, in gastrointestinal cancers. Pathol. Res. Pract. 2011, 207, 433–437. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; Xue, J.; He, Y.; Ma, H.; Jin, G.; Chen, J.; Hu, Z.; Liu, X.; Shen, H. Potentially functional polymorphisms in ATG10 are associated with risk of breast cancer in a Chinese population. Gene 2013, 527, 491–495. [Google Scholar] [CrossRef] [PubMed]
- Sridharan, S.; Jain, K.; Basu, A. Regulation of autophagy by kinases. Cancers (Basel) 2011, 3, 2630–2654. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; An, Z.; Zou, Z.; Sumpter, R.; Su, M.; Zang, X.; Sinha, S.; Gaestel, M.; Levine, B. The stress-responsive kinases MAPKAPK2/MAPKAPK3 activate starvation-induced autophagy through Beclin 1 phosphorylation. Elife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Menendez-Benito, V.; Neefjes, J. Autophagy in MHC class II presentation: Sampling from within. Immunity 2007, 26, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.M.; Garcia-Haro, L.; Sparks, C.A.; Guertin, D.A. mTOR-dependent cell survival mechanisms. Cold Spring Harb. Perspect. Biol. 2012, 4. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.; Grammer, T.C.; Lemon, K.P.; Kazlauskas, A.; Blenis, J. PDGF- and insulin-dependent pp70S6k activation mediated by phosphatidylinositol-3-OH kinase. Nature 1994, 370, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Scott, R.C.; Schuldiner, O.; Neufeld, T.P. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell 2004, 7, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Hardie, D.G. AMP-activated protein kinase: An energy sensor that regulates all aspects of cell function. Genes Dev. 2011, 25, 1895–1908. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, D.S.; Jariwala, J.S.; Anderson, E.; Mitra, K.; Meisenhelder, J.; Chang, J.T.; Ideker, T.; Hunter, T.; Nizet, V.; Dillin, A.; et al. Phosphorylation of LC3 by the Hippo kinases STK3/STK4 is essential for autophagy. Mol. Cell 2015, 57, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Tabata, K.; Morita, E.; Kawasaki, M.; Kato, R.; Dobson, R.C.; Yoshimori, T.; Wakatsuki, S. Structural basis of the autophagy-related LC3/ATG13 LIR complex: Recognition and interaction mechanism. Structure 2014, 22, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Narayan, S.; Bader, G.D.; Reimand, J. Frequent mutations in acetylation and ubiquitination sites suggest novel driver mechanisms of cancer. Genome Med. 2016, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Ma, H.; Wu, J.; Huang, Q.; Liu, J.O.; Yu, L. Arginine68 is an essential residue for the C-terminal cleavage of human Atg8 family proteins. BMC Cell Biol. 2013, 14, 27. [Google Scholar] [CrossRef] [PubMed]
- Birgisdottir, A.B.; Lamark, T.; Johansen, T. The LIR motif—Crucial for selective autophagy. J. Cell Sci. 2013, 126, 3237–3247. [Google Scholar] [PubMed]
- Kraft, L.J.; Nguyen, T.A.; Vogel, S.S.; Kenworthy, A.K. Size, stoichiometry, and organization of soluble LC3-associated complexes. Autophagy 2014, 10, 861–877. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.H.; Lam, H.C.; Jin, Y.; Kim, H.P.; Cao, J.; Lee, S.J.; Ifedigbo, E.; Parameswaran, H.; Ryter, S.W.; Choi, A.M. Autophagy protein microtubule-associated protein 1 light chain-3B (LC3B) activates extrinsic apoptosis during cigarette smoke-induced emphysema. Proc. Natl. Acad. Sci. USA 2010, 107, 18880–18885. [Google Scholar] [CrossRef] [PubMed]
- Cokakli, M.; Erdal, E.; Nart, D.; Yilmaz, F.; Sagol, O.; Kilic, M.; Karademir, S.; Atabey, N. Differential expression of Caveolin-1 in hepatocellular carcinoma: Correlation with differentiation state, motility and invasion. BMC Cancer 2009, 9, 65. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Smith, A.; Guo, L.; Alastalo, T.P.; Li, M.; Sawada, H.; Liu, X.; Chen, Z.H.; Ifedigbo, E.; Jin, Y.; et al. Autophagic protein LC3B confers resistance against hypoxia-induced pulmonary hypertension. Am. J. Respir. Crit. Care Med. 2011, 183, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.T.; Ji, J.; Dagda, R.K.; Jiang, J.F.; Tyurina, Y.Y.; Kapralov, A.A.; Tyurin, V.A.; Yanamala, N.; Shrivastava, I.H.; Mohammadyani, D.; et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat. Cell Biol. 2013, 15, 1197–1205. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Jeong, E.G.; Lee, S.H.; Yoo, N.J. Somatic mutations of BECN1, an autophagy-related gene, in human cancers. Acta Pathol. Microbiol. Immunol. Scand. 2007, 115, 750–756. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Zou, Z.; Becker, N.; Anderson, M.; Sumpter, R.; Xiao, G.; Kinch, L.; Koduru, P.; Christudass, C.S.; Veltri, R.W.; et al. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell 2013, 154, 1269–1284. [Google Scholar] [CrossRef] [PubMed]
- Zalckvar, E.; Berissi, H.; Mizrachy, L.; Idelchuk, Y.; Koren, I.; Eisenstein, M.; Sabanay, H.; Pinkas-Kramarski, R.; Kimchi, A. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009, 10, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; He, L.; Che, K.H.; Funderburk, S.F.; Pan, L.; Pan, N.; Zhang, M.; Yue, Z.; Zhao, Y. Imperfect interface of Beclin1 coiled-coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nat. Commun. 2012, 3, 662. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Ramanathan, A.; Glover, K.; Stanley, C.; Sanishvili, R.; Chakravarthy, S.; Yang, Z.; Colbert, C.L.; Sinha, S.C. Conformational Flexibility Enables the Function of a BECN1 Region Essential for Starvation-Mediated Autophagy. Biochemistry 2016, 55, 1945–1958. [Google Scholar] [CrossRef] [PubMed]
- Laddha, S.V.; Ganesan, S.; Chan, C.S.; White, E. Mutational landscape of the essential autophagy gene BECN1 in human cancers. Mol. Cancer Res. 2014, 12, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Jounai, N.; Kobiyama, K.; Shiina, M.; Ogata, K.; Ishii, K.J.; Takeshita, F. NLRP4 negatively regulates autophagic processes through an association with beclin1. J. Immunol. 2011, 186, 1646–1655. [Google Scholar] [CrossRef] [PubMed]
- Choubey, V.; Cagalinec, M.; Liiv, J.; Safiulina, D.; Hickey, M.A.; Kuum, M.; Liiv, M.; Anwar, T.; Eskelinen, E.L.; Kaasik, A. BECN1 is involved in the initiation of mitophagy: It facilitates PARK2 translocation to mitochondria. Autophagy 2014, 10, 1105–1119. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Choi, W.; Hu, W.; Mi, N.; Guo, Q.; Ma, M.; Liu, M.; Tian, Y.; Lu, P.; Wang, F.L.; et al. Crystal structure and biochemical analyses reveal Beclin 1 as a novel membrane binding protein. Cell Res. 2012, 22, 473–489. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Klein, M.G.; Zou, H.; Lane, W.; Snell, G.; Levin, I.; Li, K.; Sang, B.C. Crystal structure of human stearoyl-coenzyme A desaturase in complex with substrate. Nat. Struct. Mol. Biol. 2015, 22, 581–585. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.N.; Lim, Y.S.; Pham, L.V.; Shin, H.Y.; Kim, Y.S.; Hwang, S.B. Stearoyl coenzyme A desaturase 1 is associated with hepatitis C virus replication complex and regulates viral replication. J. Virol. 2014, 88, 12311–12325. [Google Scholar] [CrossRef] [PubMed]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef] [PubMed]
- Sherry, S.T.; Ward, M.H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef] [PubMed]
- Bao, L.; Zhou, M.; Cui, Y. nsSNPAnalyzer: Identifying disease-associated nonsynonymous single nucleotide polymorphisms. Nucleic Acids Res. 2005, 33, W480–W482. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Chan, A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015, 31, 2745–2747. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, R.; Capriotti, E.; Fariselli, P.; Martelli, P.L.; Casadio, R. Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum. Mutat. 2009, 30, 1237–1244. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Costa, C.; Gelpi, J.L.; Zamakola, L.; Parraga, I.; de la Cruz, X.; Orozco, M. PMUT: A web-based tool for the annotation of pathological mutations on proteins. Bioinformatics 2005, 21, 3176–3178. [Google Scholar] [CrossRef] [PubMed]
- Moreira, L.G.; Pereira, L.C.; Drummond, P.R.; de Mesquita, J.F. Structural and functional analysis of human SOD1 in amyotrophic lateral sclerosis. PLoS ONE 2013, 8, e81979. [Google Scholar] [CrossRef] [PubMed]
- Capriotti, E.; Fariselli, P.; Casadio, R. I-Mutant2.0: Predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res 2005, 33, W306–W310. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.E.; Ascher, D.B.; Blundell, T.L. DUET: A server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res. 2014, 42, W314–W319. [Google Scholar] [CrossRef] [PubMed]
- Quan, L.; Lv, Q.; Zhang, Y. STRUM: Structure-based prediction of protein stability changes upon single-point mutation. Bioinformatics 2016. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.C.; Herrera Braga, J.; Fraternali, F. PinSnps: Structural and functional analysis of SNPs in the context of protein interaction networks. Bioinformatics 2016. [Google Scholar] [CrossRef] [PubMed]
- De Baets, G.; van Durme, J.; Reumers, J.; Maurer-Stroh, S.; Vanhee, P.; Dopazo, J.; Schymkowitz, J.; Rousseau, F. SNPeffect 4.0: On-Line prediction of molecular and structural effects of protein-coding variants. Nucleic Acids Res. 2012, 40, D935–D939. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [PubMed]
- Colovos, C.; Yeates, T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Roy, A.; Zhang, Y. Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics 2013, 29, 2588–2595. [Google Scholar] [CrossRef] [PubMed]
- Ngan, C.H.; Hall, D.R.; Zerbe, B.; Grove, L.E.; Kozakov, D.; Vajda, S. FTSite: High accuracy detection of ligand binding sites on unbound protein structures. Bioinformatics 2012, 28, 286–287. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Blanco, J.R.; Aliaga, J.I.; Quintana-Orti, E.S.; Chacon, P. iMODS: Internal coordinates normal mode analysis server. Nucleic Acids Res. 2014, 42, W271–W276. [Google Scholar] [CrossRef] [PubMed]
- Blom, N.; Gammeltoft, S.; Brunak, S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 1999, 294, 1351–1362. [Google Scholar] [CrossRef] [PubMed]
- Wong, Y.H.; Lee, T.Y.; Liang, H.K.; Huang, C.M.; Wang, T.Y.; Yang, Y.H.; Chu, C.H.; Huang, H.D.; Ko, M.T.; Hwang, J.K. KinasePhos 2.0: A web server for identifying protein kinase-specific phosphorylation sites based on sequences and coupling patterns. Nucleic Acids Res. 2007, 35, W588–W594. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Ren, J.; Gao, X.; Jin, C.; Wen, L.; Yao, X. GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol. Cell. Proteom. 2008, 7, 1598–1608. [Google Scholar] [CrossRef] [PubMed]
- Diella, F.; Cameron, S.; Gemund, C.; Linding, R.; Via, A.; Kuster, B.; Sicheritz-Ponten, T.; Blom, N.; Gibson, T.J. Phospho.ELM: A database of experimentally verified phosphorylation sites in eukaryotic proteins. BMC Bioinform. 2004, 5, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, B.; Petersen, T.N.; Andersen, P.; Nielsen, M.; Lundegaard, C. A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct. Biol. 2009, 9, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Chen, Y.Z.; Wang, X.F.; Wang, C.; Yan, R.X.; Zhang, Z. Prediction of ubiquitination sites by using the composition of k-spaced amino acid pairs. PLoS ONE 2011, 6, e22930. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.R.; Xiao, X.; Lin, W.Z.; Chou, K.C. iUbiq-Lys: Prediction of lysine ubiquitination sites in proteins by extracting sequence evolution information via a gray system model. J. Biomol. Struct. Dyn. 2015, 33, 1731–1742. [Google Scholar] [CrossRef] [PubMed]
- Radivojac, P.; Vacic, V.; Haynes, C.; Cocklin, R.R.; Mohan, A.; Heyen, J.W.; Goebl, M.G.; Iakoucheva, L.M. Identification, analysis, and prediction of protein ubiquitination sites. Proteins 2010, 78, 365–380. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Wen, L.; Gao, X.; Jin, C.; Xue, Y.; Yao, X. CSS-Palm 2.0: An updated software for palmitoylation sites prediction. Protein Eng. Des. Sel. 2008, 21, 639–644. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.P.; Qiu, J.D.; Sun, X.Y.; Suo, S.B.; Huang, S.Y.; Liang, R.P. PMeS: Prediction of methylation sites based on enhanced feature encoding scheme. PLoS ONE 2012, 7, e38772. [Google Scholar] [CrossRef] [PubMed]
- Fullgrabe, J.; Kavanagh, E.; Joseph, B. Histone onco-modifications. Oncogene 2011, 30, 3391–3403. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Du, Y.; Lu, M.; Li, T. ASEB: A web server for KAT-specific acetylation site prediction. Nucleic Acids Res. 2012, 40, W376–W379. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Xue, Y.; Jin, C.; Wang, M.; Yao, X. Prediction of Nepsilon-acetylation on internal lysines implemented in Bayesian Discriminant Method. Biochem. Biophys. Res. Commun. 2006, 350, 818–824. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Zhou, F.; Fu, C.; Xu, Y.; Yao, X. SUMOsp: A web server for sumoylation site prediction. Nucleic Acids Res. 2006, 34, W254–W257. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Z.; Chen, Z.; Gong, Y.A.; Ying, G. SUMOhydro: A novel method for the prediction of sumoylation sites based on hydrophobic properties. PLoS ONE 2012, 7, e39195. [Google Scholar] [CrossRef] [PubMed]
- David, A.; Razali, R.; Wass, M.N.; Sternberg, M.J. Protein-protein interaction sites are hot spots for disease-associated nonsynonymous SNPs. Hum. Mutat. 2012, 33, 359–363. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Han, J.G.; Shyu, C.R.; Korkin, D. Determining effects of non-synonymous SNPs on protein-protein interactions using supervised and semi-supervised learning. PLoS Comput. Biol. 2014, 10, e1003592. [Google Scholar] [CrossRef] [PubMed]


| Protein | Mutation | Deleterious SNPs | Destabilizing SNPs | |||||
|---|---|---|---|---|---|---|---|---|
| nsSNP Analyzer | PROVEAN | PMUT | SNPs & GO. | I-Mutant2.0 | DUET | STRUM | ||
| LC3A | R24C | √ | √ | √ | √ | √ | √ | √ |
| S29R | √ | – | √ | √ | √ | √ | √ | |
| P55A | √ | √ | √ | √ | √ | √ | ||
| P55L | √ | √ | √ | √ | √ | √ | √ | |
| R70C | √ | √ | √ | √ | √ | √ | √ | |
| P75R | – | √ | √ | √ | √ | – | – | |
| F79V | √ | √ | √ | √ | √ | √ | √ | |
| F79S | √ | √ | √ | √ | √ | √ | √ | |
| N84K | – | √ | √ | √ | √ | – | √ | |
| A96P | – | √ | √ | √ | √ | √ | √ | |
| D104N | √ | √ | – | √ | √ | √ | √ | |
| E105K | – | √ | √ | √ | √ | – | √ | |
| K49A | √ | √ | √ | √ | √ | √ | √ | |
| K51A | √ | √ | √ | √ | √ | √ | √ | |
| L53A | √ | √ | √ | √ | √ | √ | ||
| G120A | √ | √ | √ | √ | √ | √ | √ | |
| LC3B | R11C | √ | √ | √ | √ | √ | √ | √ |
| R24Q | √ | √ | √ | √ | √ | √ | ||
| P32L | √ | √ | √ | √ | √ | √ | √ | |
| V33A | √ | √ | √ | √ | √ | √ | ||
| R37Q | √ | √ | √ | √ | √ | √ | √ | |
| G40C | √ | √ | √ | √ | √ | √ | √ | |
| R68A | √ | √ | √ | √ | √ | √ | √ | |
| R70A | √ | √ | √ | √ | √ | √ | √ | |
| R70H | √ | √ | √ | √ | √ | √ | √ | |
| A78D | – | √ | √ | √ | √ | √ | √ | |
| F79S | √ | √ | √ | √ | √ | √ | √ | |
| S92C | – | √ | √ | √ | √ | √ | √ | |
| D104N | – | √ | √ | √ | √ | √ | √ | |
| D106G | √ | √ | √ | √ | √ | √ | √ | |
| Y113S | √ | √ | √ | √ | √ | √ | √ | |
| Y113C | √ | √ | √ | √ | – | √ | √ | |
| E117V | – | √ | √ | √ | – | √ | √ | |
| G120A | √ | √ | √ | √ | – | √ | √ | |
| BECN1 | V14E | √ | √ | – | √ | √ | √ | √ |
| A44P | – | √ | √ | √ | √ | – | ||
| R74C | √ | – | √ | √ | √ | √ | √ | |
| L112R | √ | √ | √ | √ | √ | – | √ | |
| S113R | √ | √ | √ | √ | – | √ | √ | |
| S127L | √ | √ | – | √ | √ | – | √ | |
| L144F | √ | √ | √ | – | √ | √ | √ | |
| E156V | – | √ | √ | √ | √ | – | √ | |
| R164C | √ | √ | √ | √ | √ | √ | √ | |
| L194P | √ | √ | √ | √ | √ | √ | √ | |
| L198P | – | √ | √ | √ | √ | √ | √ | |
| R205C | – | √ | √ | √ | √ | √ | √ | |
| Q239P | – | √ | √ | √ | √ | – | √ | |
| R255C | √ | √ | √ | √ | √ | √ | √ | |
| N268S | √ | √ | – | √ | √ | – | √ | |
| T284S | √ | √ | – | √ | √ | √ | √ | |
| R292C | √ | √ | √ | √ | √ | √ | √ | |
| R292H | √ | √ | √ | √ | √ | √ | √ | |
| V298M | √ | √ | – | √ | √ | √ | √ | |
| V298A | √ | √ | – | √ | √ | √ | √ | |
| E302K | √ | √ | √ | √ | √ | √ | √ | |
| L314H | √ | √ | √ | √ | √ | √ | √ | |
| L323P | – | √ | √ | √ | √ | √ | √ | |
| Y338C | √ | √ | √ | √ | √ | √ | √ | |
| S346Y | √ | √ | √ | – | – | √ | √ | |
| P350L | √ | √ | √ | – | √ | – | √ | |
| Y352H | √ | √ | – | √ | √ | √ | √ | |
| C353Y | √ | √ | √ | √ | √ | √ | √ | |
| W361R | – | √ | √ | √ | √ | √ | √ | |
| C375R | √ | √ | √ | √ | √ | √ | √ | |
| I403S | √ | √ | √ | √ | √ | √ | √ | |
| I403T | √ | √ | √ | √ | √ | √ | √ | |
| G408R | √ | √ | √ | – | √ | √ | √ | |
| W425C | √ | √ | √ | √ | √ | √ | √ | |
| F431V | √ | √ | √ | √ | √ | √ | √ | |
| F123A | √ | √ | √ | √ | √ | √ | √ | |
| D133A | √ | √ | √ | √ | √ | – | √ | |
| D149A | – | √ | √ | √ | √ | – | √ | |
| Y352A | √ | √ | √ | √ | √ | √ | √ | |
| W425A | √ | √ | √ | √ | √ | √ | √ | |
| Y41C | √ | – | √ | √ | √ | √ | √ | |
| SCD1 | P48R | √ | – | √ | √ | √ | √ | √ |
| Y55D | √ | – | √ | √ | √ | √ | √ | |
| E63G | √ | – | √ | √ | √ | √ | √ | |
| Y88C | √ | √ | √ | √ | √ | √ | √ | |
| G89R | √ | √ | √ | √ | √ | √ | – | |
| G89A | √ | √ | – | √ | √ | √ | – | |
| T100I | √ | √ | √ | √ | √ | – | – | |
| L102P | √ | √ | – | √ | √ | √ | √ | |
| G104E | √ | – | √ | √ | √ | √ | √ | |
| R121C | √ | √ | √ | √ | √ | √ | √ | |
| H125P | √ | √ | √ | √ | – | √ | – | |
| R126C | √ | √ | – | √ | √ | √ | √ | |
| R126S | √ | √ | √ | √ | √ | √ | √ | |
| R131W | √ | √ | √ | √ | √ | √ | √ | |
| R135W | √ | √ | √ | √ | √ | √ | √ | |
| R135Q | – | √ | √ | √ | √ | √ | √ | |
| M144T | √ | √ | √ | √ | √ | √ | √ | |
| Y151C | √ | √ | √ | √ | – | √ | √ | |
| R188C | √ | √ | – | √ | √ | √ | √ | |
| H190Y | √ | √ | – | √ | – | – | √ | |
| F213L | √ | √ | – | √ | √ | √ | √ | |
| Q214R | √ | √ | – | √ | √ | √ | √ | |
| Y218H | √ | √ | – | √ | √ | √ | √ | |
| Y218C | √ | √ | √ | √ | √ | √ | √ | |
| M225T | √ | √ | – | √ | √ | √ | √ | |
| T231M | √ | √ | – | √ | √ | √ | √ | |
| W238G | √ | √ | √ | √ | √ | √ | √ | |
| W238R | √ | √ | √ | √ | √ | √ | √ | |
| A249D | – | √ | √ | √ | √ | √ | √ | |
| G272R | √ | √ | √ | √ | √ | √ | – | |
| Y273C | √ | – | √ | √ | √ | √ | √ | |
| N279I | – | √ | √ | √ | – | – | √ | |
| R283W | √ | √ | √ | √ | √ | √ | √ | |
| L290P | √ | √ | – | √ | √ | √ | √ | |
| F323V | √ | √ | √ | √ | √ | √ | √ | |
| C326G | √ | √ | √ | √ | √ | √ | √ | |
| C326F | √ | – | √ | √ | √ | √ | √ | |
| M327V | – | √ | √ | √ | √ | √ | √ | |
| G331S | √ | √ | √ | √ | √ | √ | √ | |
| A333T | – | √ | √ | √ | √ | √ | – | |
| R336Q | √ | – | √ | √ | √ | √ | √ | |
| R336L | √ | – | √ | √ | √ | – | – | |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Awan, F.M.; Obaid, A.; Ikram, A.; Janjua, H.A. Mutation-Structure-Function Relationship Based Integrated Strategy Reveals the Potential Impact of Deleterious Missense Mutations in Autophagy Related Proteins on Hepatocellular Carcinoma (HCC): A Comprehensive Informatics Approach. Int. J. Mol. Sci. 2017, 18, 139. https://doi.org/10.3390/ijms18010139
Awan FM, Obaid A, Ikram A, Janjua HA. Mutation-Structure-Function Relationship Based Integrated Strategy Reveals the Potential Impact of Deleterious Missense Mutations in Autophagy Related Proteins on Hepatocellular Carcinoma (HCC): A Comprehensive Informatics Approach. International Journal of Molecular Sciences. 2017; 18(1):139. https://doi.org/10.3390/ijms18010139
Chicago/Turabian StyleAwan, Faryal Mehwish, Ayesha Obaid, Aqsa Ikram, and Hussnain Ahmed Janjua. 2017. "Mutation-Structure-Function Relationship Based Integrated Strategy Reveals the Potential Impact of Deleterious Missense Mutations in Autophagy Related Proteins on Hepatocellular Carcinoma (HCC): A Comprehensive Informatics Approach" International Journal of Molecular Sciences 18, no. 1: 139. https://doi.org/10.3390/ijms18010139
APA StyleAwan, F. M., Obaid, A., Ikram, A., & Janjua, H. A. (2017). Mutation-Structure-Function Relationship Based Integrated Strategy Reveals the Potential Impact of Deleterious Missense Mutations in Autophagy Related Proteins on Hepatocellular Carcinoma (HCC): A Comprehensive Informatics Approach. International Journal of Molecular Sciences, 18(1), 139. https://doi.org/10.3390/ijms18010139






