Promotion of Ni2+ Removal by Masking Toxicity to Sulfate-Reducing Bacteria: Addition of Citrate
Abstract
:1. Introduction
2. Results
2.1. The Effect of the Addition of Citrate on Ni2+ Toxicity
| Media | % of Total Ni (1 mM) | |||||||
|---|---|---|---|---|---|---|---|---|
| Ni2+ | NiCl+ | NiSO4 (aq) | [Ni-Lactate]+ | [Ni-(Lactate)3]− | Ni-(Lactate)2 (aq) | [Ni-Citrate]− | [Ni-(Citrate)2]4− | |
| Ni/citrate (0) | 26.20 | 0.03 | 16.18 | 43.21 | 0.714 | 13.67 | – | – |
| Ni/citrate (0.5) | 13.88 | 0.01 | 8.53 | 23.10 | 0.39 | 7.38 | 45.58 | 0.84 |
| Ni/citrate (1.0) | 4.79 | – | 2.92 | 8.02 | 0.14 | 2.58 | 74.92 | 6.63 |
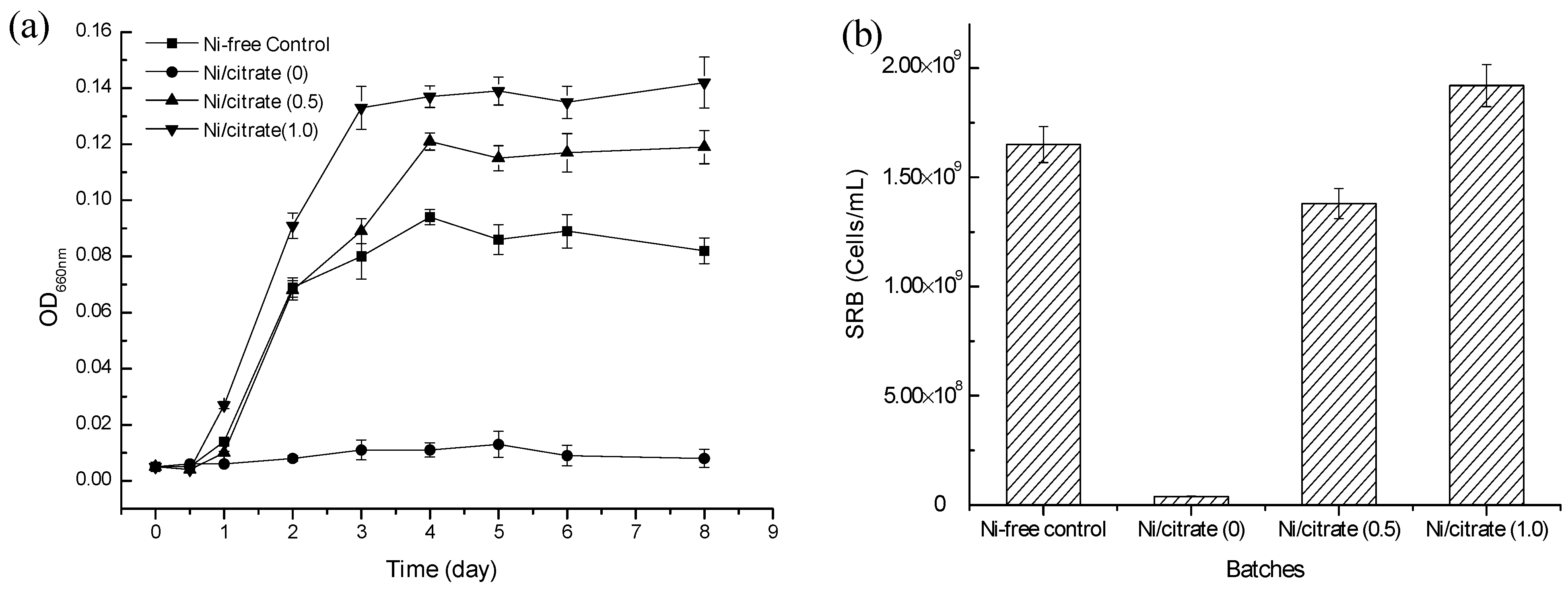
| Media | pH | Sulfate (mM) | Sulfide (mM) | Ni (mM) |
|---|---|---|---|---|
| Ni-free control | 7.09 ± 0.31 | 25.08 ± 1.57 | 1.44 ± 0.04 | – |
| Ni/citrate (0) | 6.77 ± 0.23 | 29.96 ± 1.46 | 0.08 ± 0.01 | 0.98 ± 0.04 |
| Ni/citrate (0.5) | 7.23 ± 0.18 | 24.26 ± 1.26 | 1.50 ± 0.03 | 0 |
| Ni/citrate (1.0) | 7.18 ± 0.15 | 24.00 ± 1.31 | 1.63 ± 0.05 | 0 |
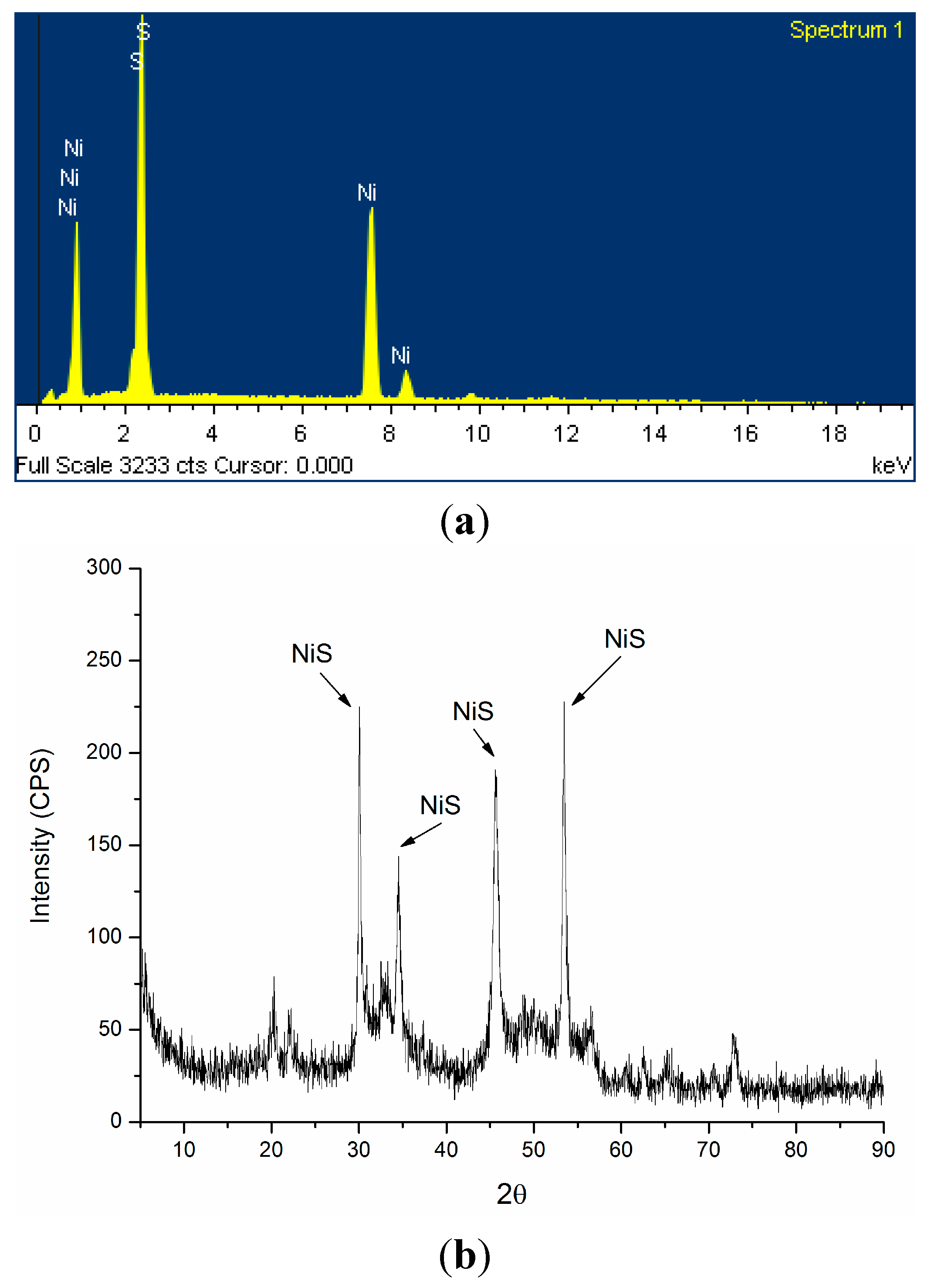
2.2. Mechanism of Ni2+ Masking and Removal
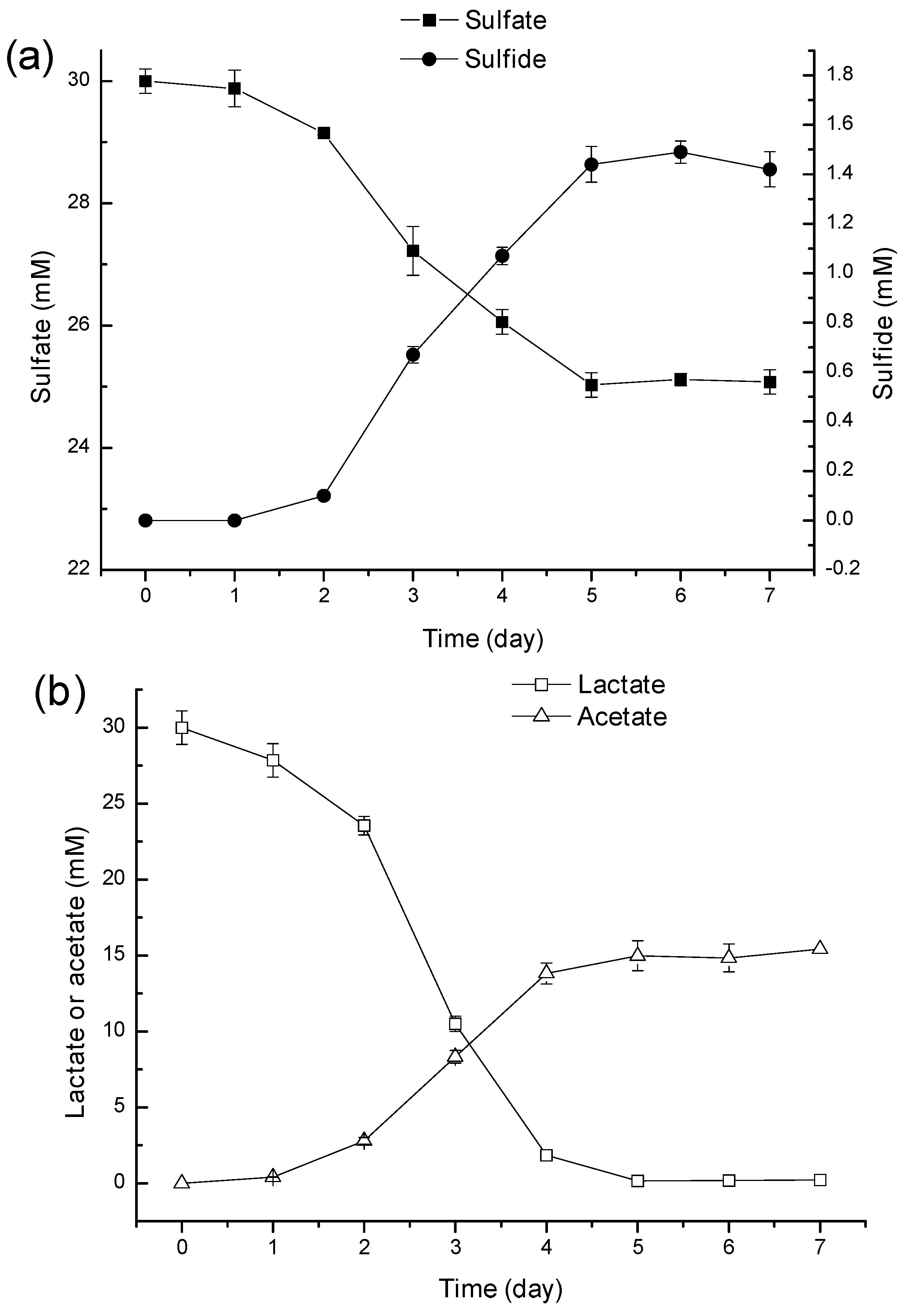
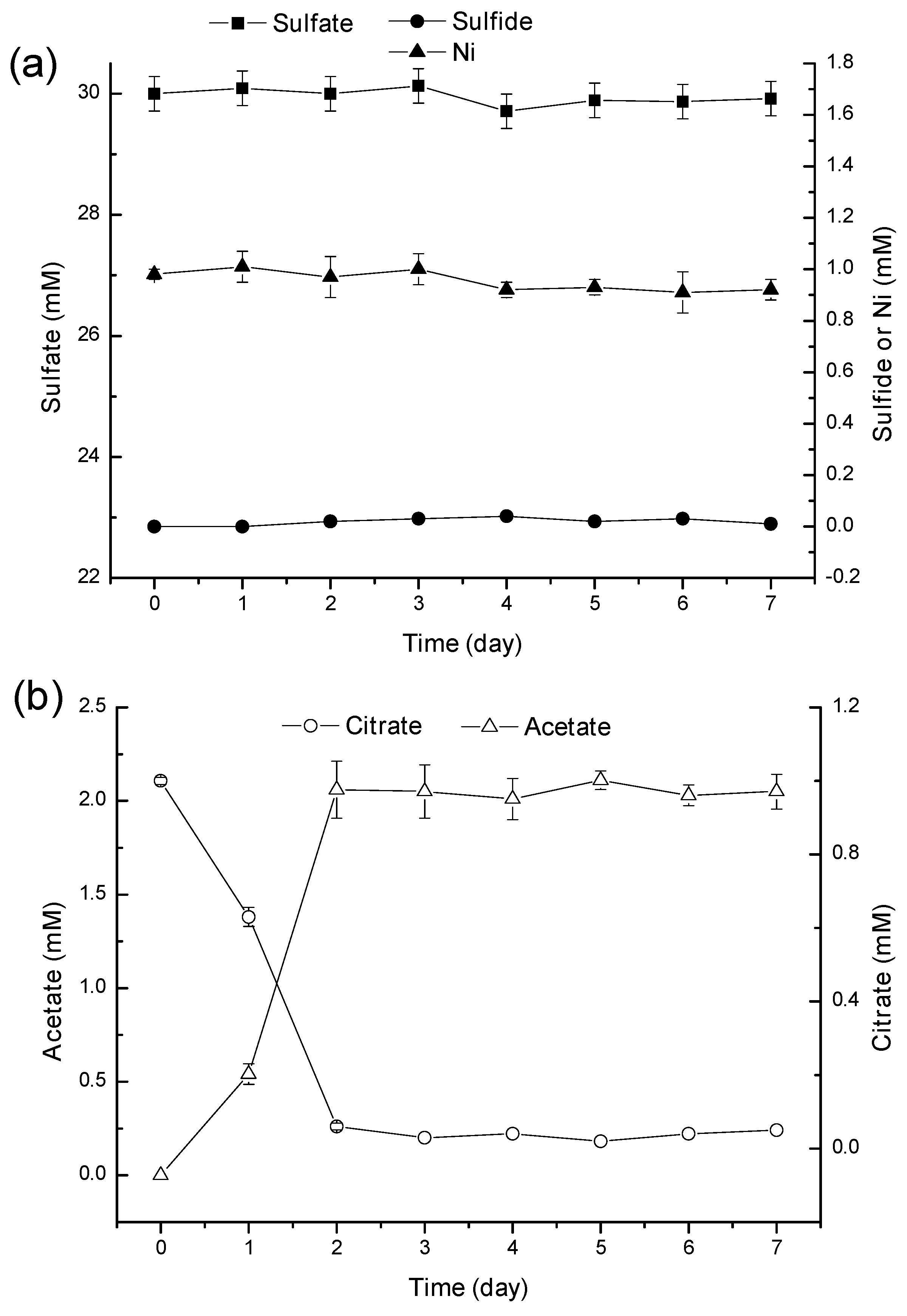
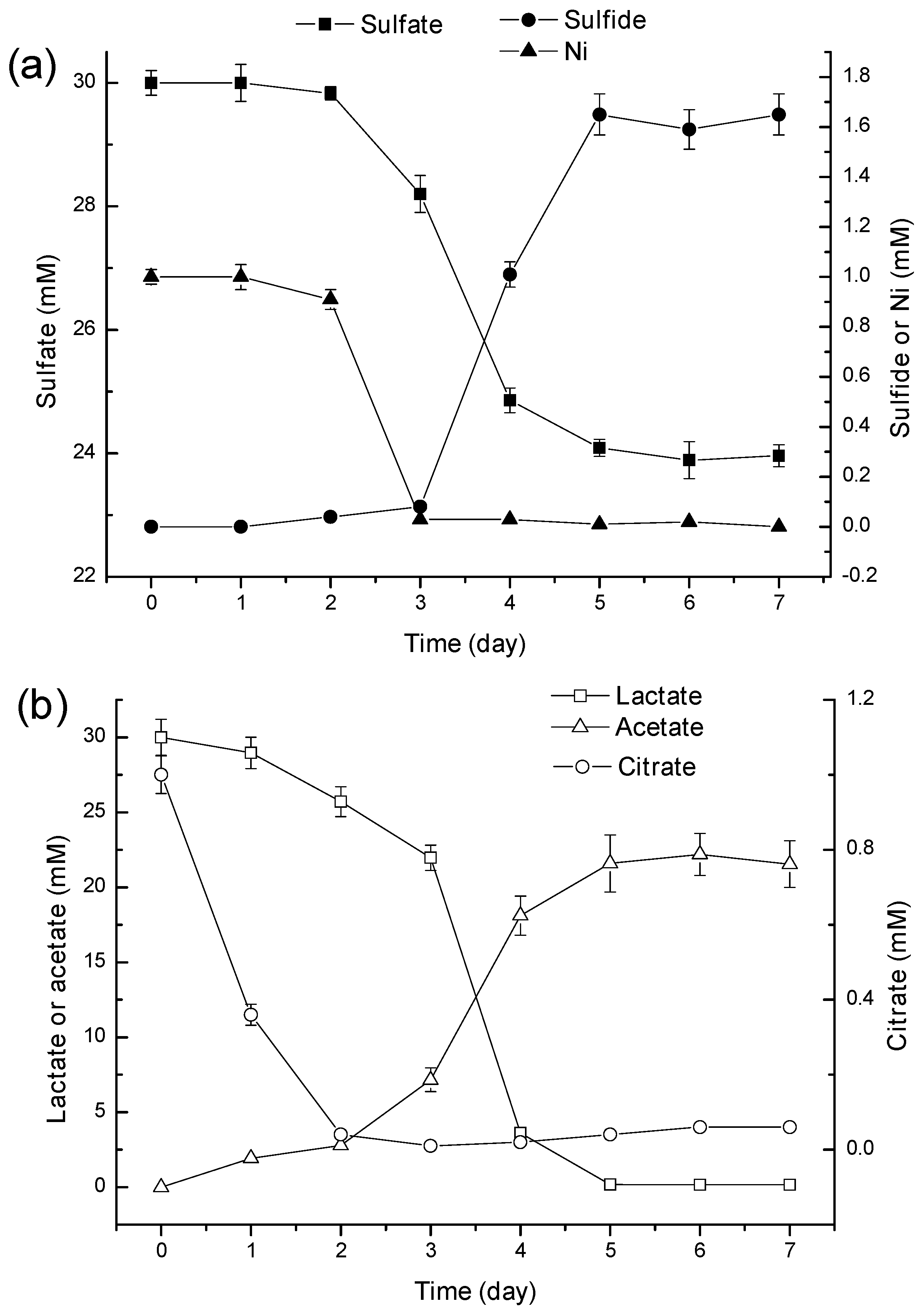
2.3. Discussion
3. Materials and Methods
3.1. Sulfate-Reducing Bacteria (SRB) Enrichment
3.2. Batch Experiments
3.3. Defined Mineral Media
3.4. Effect of Citrate on the SRB Growth and Ni2+ Removal
3.5. Biodegradation of Ni-Citrate Complex
3.6. Analytical Methods
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Albertson, O.E. Of: Changes in the biochemical oxygen demand procedure in the 21st edition of Standard Methods for the Examination of Water and Wastewater, James C. Young, Lenore S. Clesceri, sabry M. Kamhawy, 77, 404–410 (2005). Water Environ. Res. 2007, 79, 453–455; discussion 455–456. [Google Scholar]
- Liamleam, W.; Annachhatre, A.P. Electron donors for biological sulfate reduction. Biotechnol. Adv. 2007, 25, 452–463. [Google Scholar] [CrossRef] [PubMed]
- Qiu, R.; Zhao, B.; Liu, J.; Huang, X.; Li, Q.; Brewer, E.; Wang, S.; Shi, N. Sulfate reduction and copper precipitation by a Citrobacter sp. isolated from a mining area. J. Hazard. Mater. 2009, 164, 1310–1315. [Google Scholar] [CrossRef]
- Muyzer, G.; Stams, A.J.M. The ecology and biotechnology of sulphate-reducing bacteria. Nat. Rev. Microbiol. 2008, 6, 441–454. [Google Scholar] [PubMed]
- Kaksonen, A.H.; Puhakka, J.A. Sulfate reduction based bioprocesses for the treatment of acid mine drainage and the recovery of metals. Eng. Life Sci. 2007, 7, 541–564. [Google Scholar] [CrossRef]
- Dvorak, D.H.; Hedin, R.S.; Edenborn, H.M.; Mcintire, P.E. Treatment of metal-contaminated water using bacterial sulfate reduction: Results from pilot-scale reactors. Biotechnol. Bioeng. 1992, 40, 609–616. [Google Scholar] [CrossRef] [PubMed]
- Gallegos-Garcia, M.; Celis, L.B.; Rangel-Mendez, R.; Razo-Flores, E. Precipitation and recovery of metal sulfides from metal containing acidic wastewater in a sulfidogenic down-flow fluidized bed reactor. Biotechnol. Bioeng. 2009, 102, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Kieu, H.T.Q.; Muller, E.; Horn, H. Heavy metal removal in anaerobic semi-continuous stirred tank reactors by a consortium of sulfate-reducing bacteria. Water Res. 2011, 45, 3863–3870. [Google Scholar] [CrossRef] [PubMed]
- Gamez, V.M.; Sierra-Alvarez, R.; Waltz, R.J.; Field, J.A. Anaerobic degradation of citrate under sulfate reducing and methanogenic conditions. Biodegradation 2009, 20, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Stams, A.J.M.; Huisman, J.; Encina, P.A.G.; Muyzer, G. Citric acid wastewater as electron donor for biological sulfate reduction. Appl. Microbiol. Biotechnol. 2009, 83, 957–963. [Google Scholar] [CrossRef] [PubMed]
- Dodge, C.J.; Francis, A.J. Photodegradation of a ternary iron(III)–Uranium(VI)–Citric acid complex. Environ. Sci. Technol. 2002, 36, 2094–2100. [Google Scholar] [CrossRef] [PubMed]
- Francis, A.J.; JoshiTope, G.A.; Dodge, C.J. Biodegradation of nickel-citrate and modulation of nickel toxicity by iron. Environ. Sci. Technol. 1996, 30, 562–568. [Google Scholar] [CrossRef]
- Qian, J.; Li, D.; Zhan, G.; Zhang, L.; Su, W.; Gao, P. Simultaneous biodegradation of Ni-citrate complexes and removal of nickel from solutions by Pseudomonas alcaliphila. Bioresour. Technol. 2012, 116, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.W.; Tao, Y.; Zhang, W.J.; He, X.H.; Gao, P.; Li, D.P. Presence of Fe3+ and Zn2+ promoted biotransformation of Cd-citrate complex and removal of metals from solutions. J. Hazard. Mater. 2013, 263 Pt 2, 367–373. [Google Scholar] [CrossRef]
- Wakeman, K.D.; Erving, L.; Riekkola-Vanhanen, M.L.; Puhakka, J.A. Silage supports sulfate reduction in the treatment of metals- and sulfate-containing waste waters. Water Res. 2010, 44, 4932–4939. [Google Scholar] [CrossRef] [PubMed]
- Hughes, M.N.; Poole, R.K. Metal speciation and microbial-growth—The hard (and soft) facts. J. Gen. Microbiol. 1991, 137, 725–734. [Google Scholar] [CrossRef]
- Van Nostrand, J.D.; Sowder, A,G.; Bertsch, P.M.; Morris, P.J. Effect of pH on the toxicity of nickel and other divalent metals to Burkholderia cepacia PR1301. Environ. Toxicol. Chem. 2005, 24, 2742–2750. [Google Scholar]
- Hao, S.; Jiao, M.; Zhao, J.; Xing, M.; Huang, B. Reorganization and condensation of chromatin in mitotic prophase nuclei of Allium cepa. Chromosoma 1994, 103, 432–440. [Google Scholar] [CrossRef] [PubMed]
- Poulson, S.R.; Colberg, P.J.S.; Drever, J.I. Toxicity of heavy metals (Ni, Zn) to Desulfovibrio desulfuricans. Geomicrobiol. J. 1997, 14, 41–49. [Google Scholar] [CrossRef]
- Jong, T.; Parry, D.L. Removal of sulfate and heavy metals by sulfate reducing bacteria in short-term bench scale upflow anaerobic packed bed reactor runs. Water Res. 2003, 37, 3379–3389. [Google Scholar] [CrossRef] [PubMed]
- Celis-Garcia, L.B.; Razo-Flores, E.; Monroy, O. Performance of a down-flow fluidized-bed reactor under sulfate reduction conditions using volatile fatty acids as electron donors. Biotechnol. Bioeng. 2007, 97, 771–779. [Google Scholar] [CrossRef] [PubMed]
- Kaksonen, A.H.; Franzmann, P.D.; Puhakka, J.A. Effects of hydraulic retention time and sulfide toxicity on ethanol and acetate oxidation in sulfate-reducing metal-precipitating fluidized-bed reactor. Biotechnol. Bioeng. 2004, 86, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, S.; Chuichulcherm, S.; Peeva, L.; Livingston, A. Microbial sulfate reduction in a liquid-solid fluidized bed reactor. Biotechnol. Bioeng. 2000, 70, 370–380. [Google Scholar] [CrossRef] [PubMed]
- Oyekola, O.O.; van Hille, R.P.; Harrison, S.T. Study of anaerobic lactate metabolism under biosulfidogenic conditions. Water Res. 2009, 43, 3345–3354. [Google Scholar] [CrossRef] [PubMed]
- APHA. Standard Methods for the Examination of Water and Wastewater; American Public Health Association: Washington, DC, USA, 1998. [Google Scholar]
- Leloup, J.; Loy, A.; Knab, N.J.; Borowski, C.; Wagner, M.; Jorgensen, B.B. Diversity and abundance of sulfate-reducing microorganisms in the sulfate and methane zones of a marine sediment, Black Sea. Environ. Microbiol. 2007, 9, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.; Friedrich, M.; Roger, A.J.; Hugenholtz, P.; Fishbain, S.; Abicht, H.; Blackall, L.L.; Stahl, D.A.; Wagner, M. Multiple lateral transfers of dissimilatory sulfite reductase genes between major lineages of sulfate-reducing prokaryotes. J. Bacteriol. 2001, 183, 6028–6035. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qian, J.; Zhu, X.; Tao, Y.; Zhou, Y.; He, X.; Li, D. Promotion of Ni2+ Removal by Masking Toxicity to Sulfate-Reducing Bacteria: Addition of Citrate. Int. J. Mol. Sci. 2015, 16, 7932-7943. https://doi.org/10.3390/ijms16047932
Qian J, Zhu X, Tao Y, Zhou Y, He X, Li D. Promotion of Ni2+ Removal by Masking Toxicity to Sulfate-Reducing Bacteria: Addition of Citrate. International Journal of Molecular Sciences. 2015; 16(4):7932-7943. https://doi.org/10.3390/ijms16047932
Chicago/Turabian StyleQian, Junwei, Xiaoyu Zhu, Yong Tao, Yan Zhou, Xiaohong He, and Daping Li. 2015. "Promotion of Ni2+ Removal by Masking Toxicity to Sulfate-Reducing Bacteria: Addition of Citrate" International Journal of Molecular Sciences 16, no. 4: 7932-7943. https://doi.org/10.3390/ijms16047932





