High-Throughput Screening for a Moderately Halophilic Phenol-Degrading Strain and Its Salt Tolerance Response
Abstract
:1. Introduction
2. Results and Discussion
2.1. High-Throughput Cultivation of Halophilic Bacterial Communities
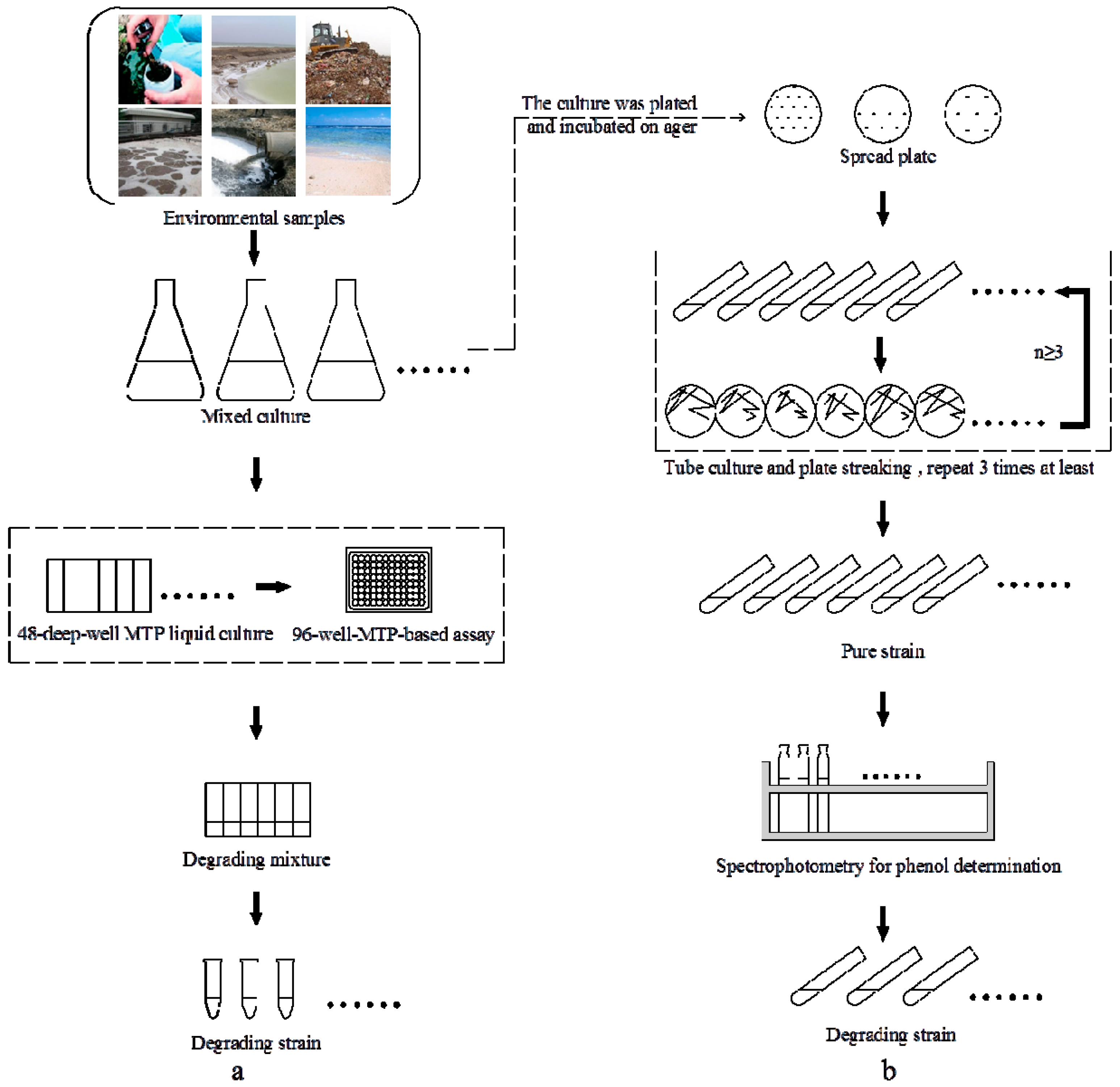
2.2. High-Throughput Phenol Measurement Performed in Microplates
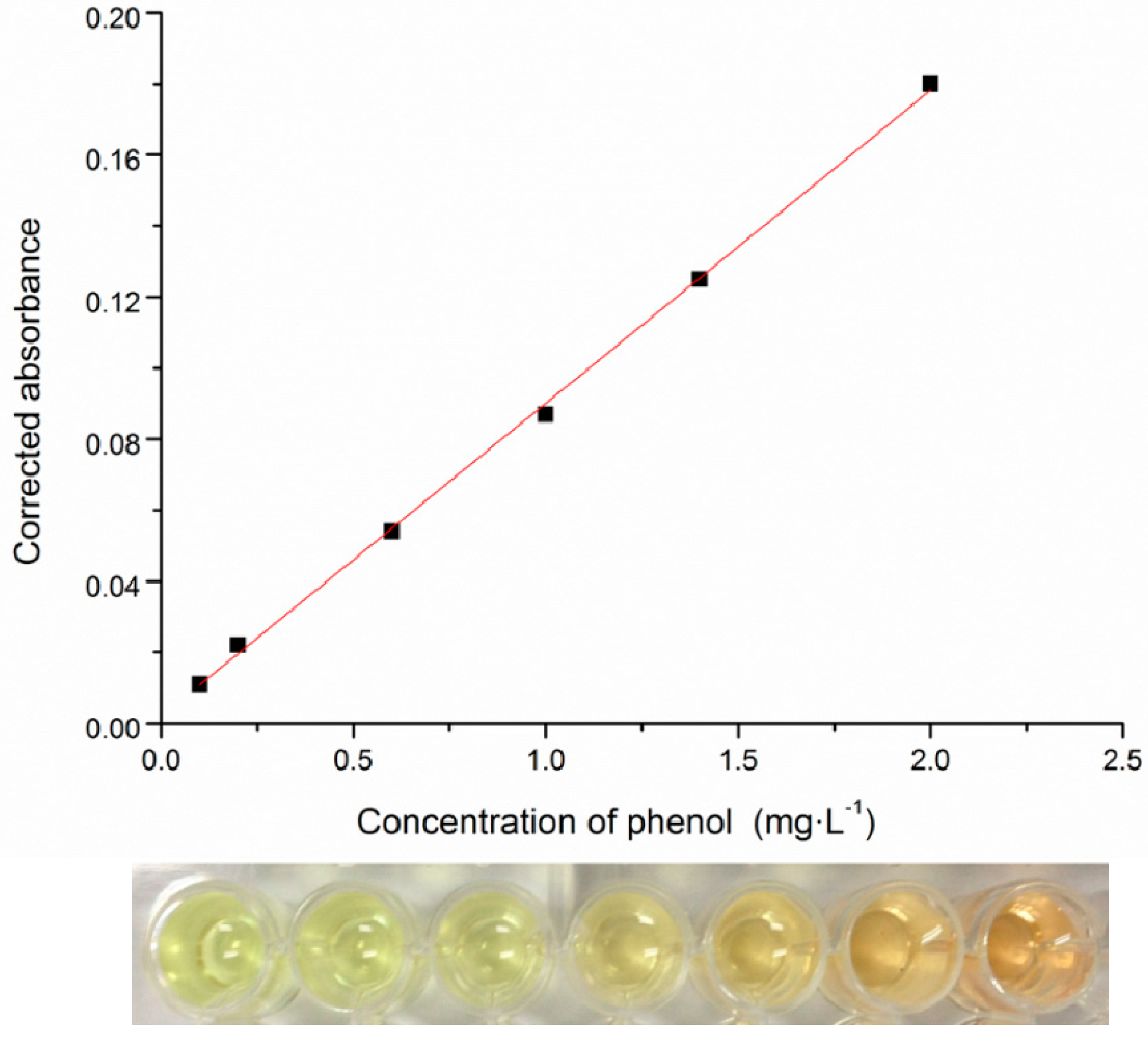
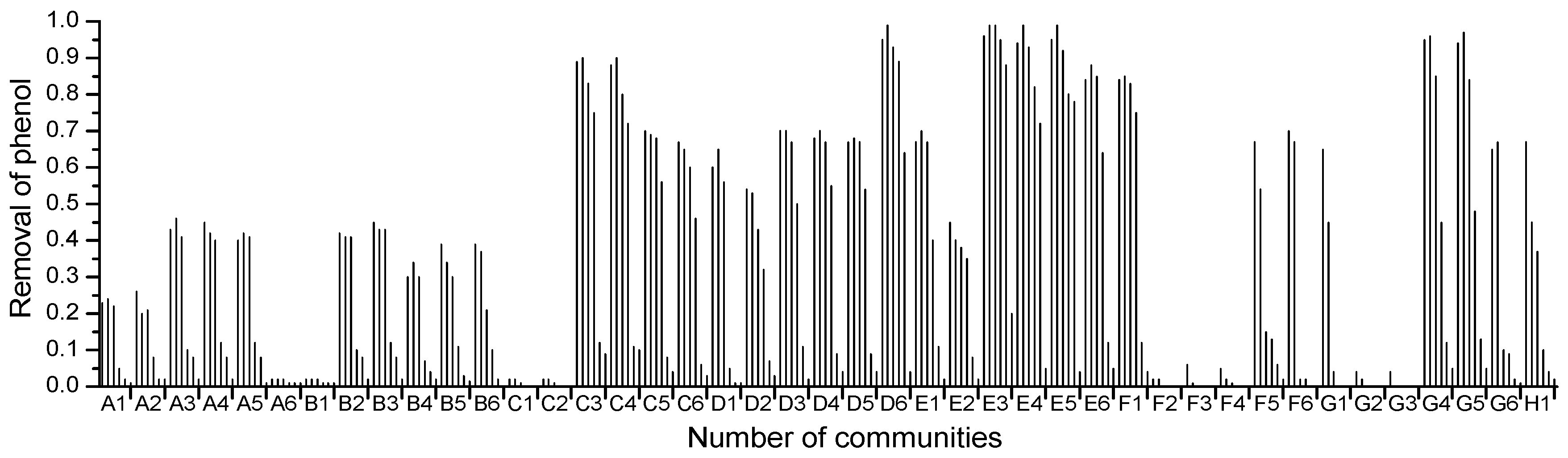

2.3. 16S rRNA Gene Sequence Analysis and Identification of the Isolated Strain

2.4. Phenol Degradation by Halomonas sp. Strain 4-5 under Various Salinities
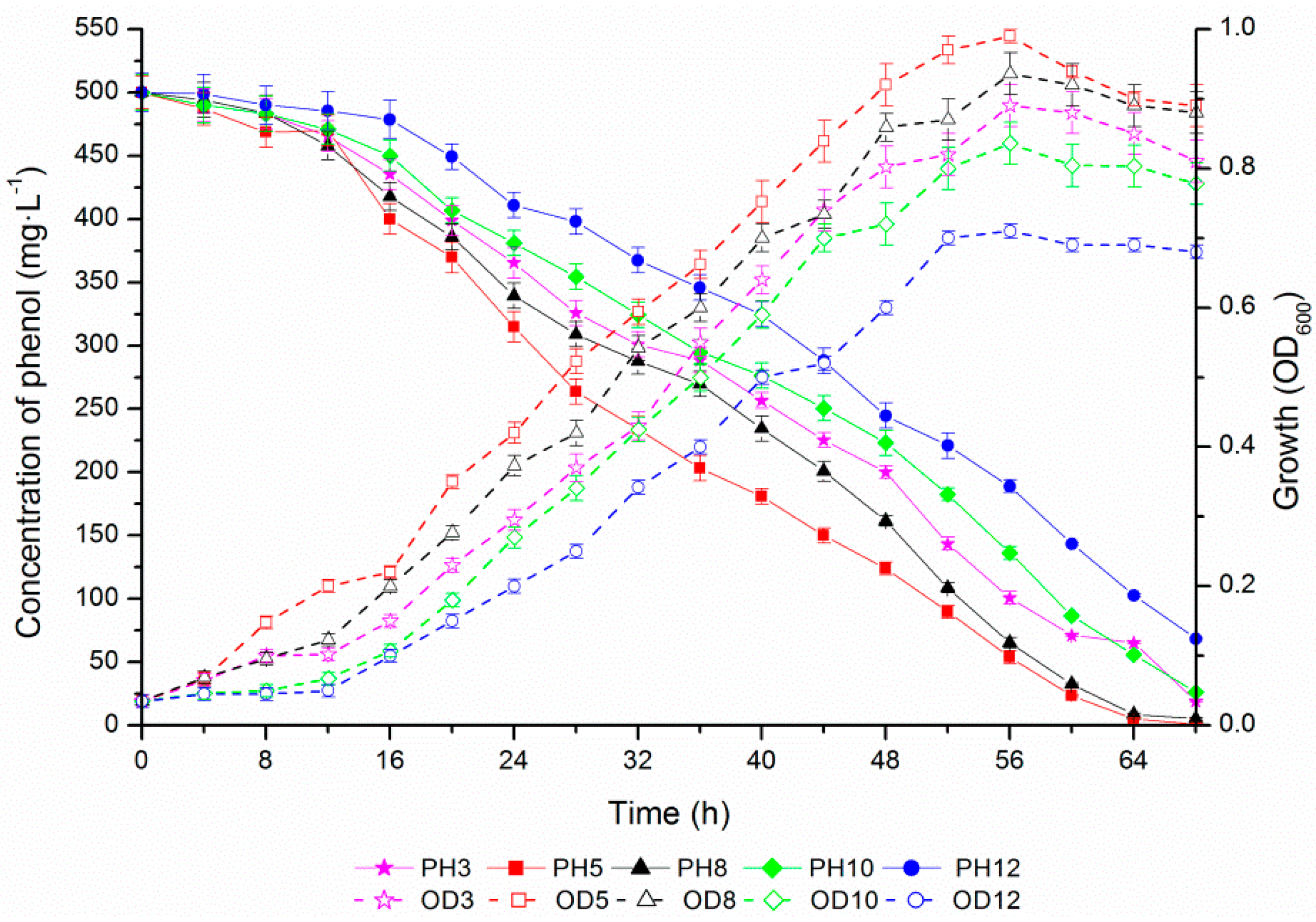
2.5. Osmoprotection Response of Halomonas sp. Strain 4-5
| NaCl (%, w/v) | Ectoine (mg·g−1) |
|---|---|
| 3 | 0.92 |
| 5 | 1.04 |
| 8 | 5.41 |
| 10 | 13.53 |
| 12 | 15.29 |
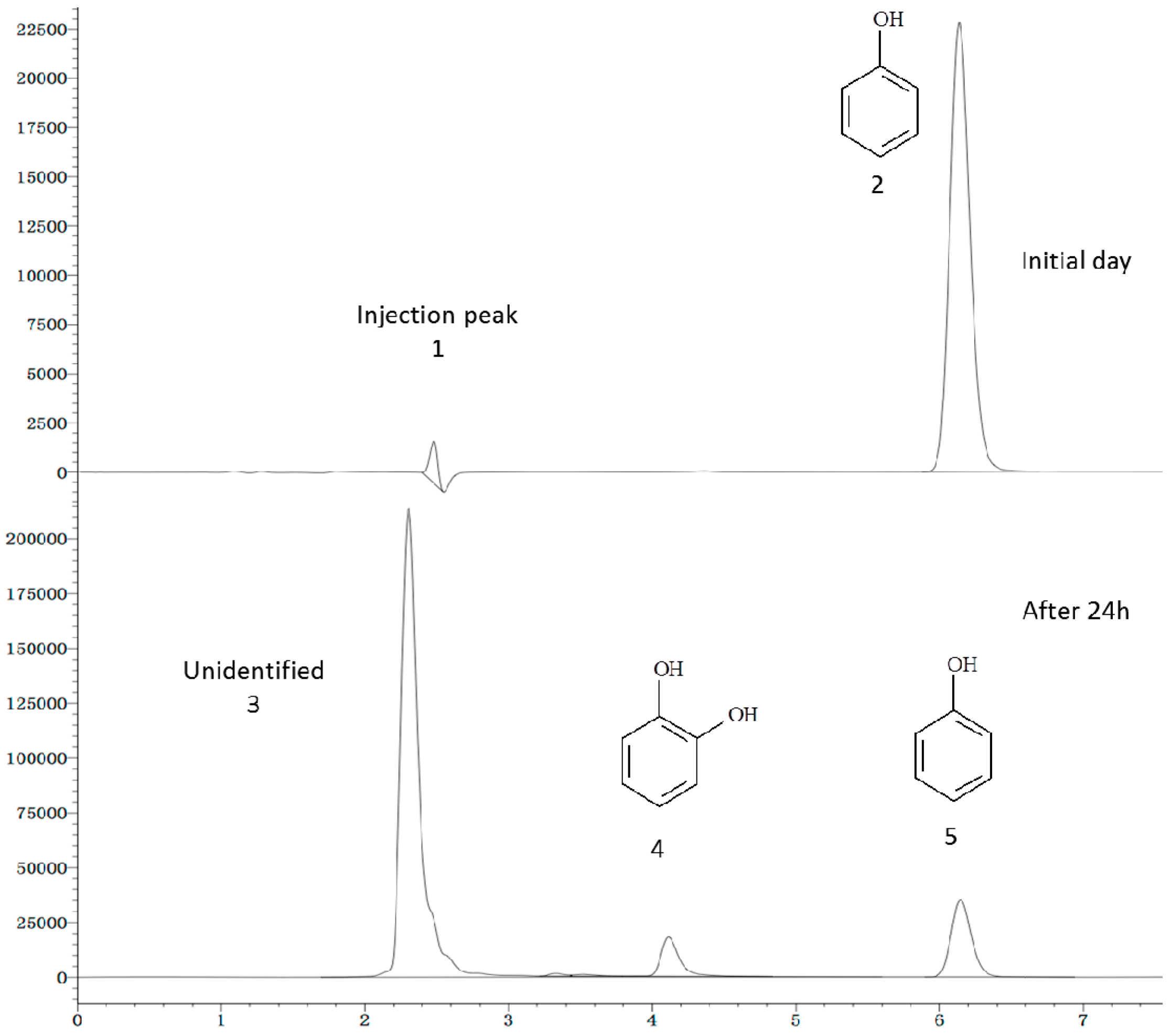
2.6. Phenol Degradation Pathway
3. Experimental Section
3.1. Sampling and Enrichment of Halophilic Bacterial Communities
| Sampling Site | Description | Sample Numbers |
|---|---|---|
| Qarhan Salt Lake | Surface soil | A1–A2 |
| Upper sediment | A3–A5 | |
| Water | A6, B1 | |
| Xin Jiang Salt Lake | Surface soil | B2–B3 |
| Upper sediment | B4–B6 | |
| Water | C1–C2 | |
| Saline-alkaline soil in Daqing, Heilongjiang | Surface soil | C3–C5 |
| Deep soil | C6, D1–D2 | |
| Shanghai Old Port Landfill Factory waste | Mineralized waste, landfill established in1990 | D3–D5 |
| Mineralized waste, landfill established in 1991 | D6, E1–E2 | |
| Mineralized waste, landfill established in 1994 | E3–E5 | |
| Leachate-contaminated soil under the landfill pit | E6, F1 | |
| Surface soil near the landfill site | F2 | |
| Seaside of the East China Sea | Sea water | F3–F4 |
| Upper sediment | F5–F6, G1 | |
| Beach silt | G2–G3 | |
| Shanghai Sinopec Gaoqiao petrochemical factory | Biochemical reaction basin | G4–G5 |
| Excess sludge | G6, H1 |
3.2. High-Throughput Cultivation of Halophilic Bacterial Communities
3.3. Phenol Measurement Performed in 96-Well Microplates
3.4. Identification of the Isolated Strain
3.5. Phenol Degradation Assay
3.6. Ectoine Determination with HPLC Method
3.7. Detection of Genotypes Involved in Aerobic Phenol Biodegradation
3.8. Nucleotide Sequence Accession Numbers
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Maszenan, A.M.; Liu, Y.; Jern Ng, W. High-performance anaerobic granulation processes for treatment of wastewater-containing recalcitrant compounds. Crit. Rev. Environ. Sci. Technol. 2011, 41, 1271–1308. [Google Scholar] [CrossRef]
- Bajaj, M.; Gallert, C.; Winter, J. Biodegradation of high phenol containing synthetic wastewater by an aerobic fixed bed reactor. Bioresour. Technol. 2008, 99, 8376–8381. [Google Scholar] [CrossRef] [PubMed]
- Moussavi, G.; Barikbin, B.; Mahmoudi, M. The removal of high concentrations of phenol from saline wastewater using aerobic granular SBR. Chem. Eng. J. 2010, 158, 498–504. [Google Scholar] [CrossRef]
- Rosenkranz, F.; Cabrol, L.; Carballa, M.; Donoso-Bravo, A.; Cruz, L.; Ruiz-Filippi, G.; Chamy, R.; Lema, J.M. Relationship between phenol degradation efficiency and microbial community structure in an anaerobic SBR. Water Res. 2013, 47, 6739–6749. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Han, Z.; Bai, Z.; Zhuang, G.; Shim, H. Progress in decontamination by halophilic microorganisms in saline wastewater and soil. Environ. Pollut. 2010, 158, 1119–1126. [Google Scholar] [CrossRef] [PubMed]
- Mϕller, M.F.; Kjeldsen, K.U.; Ingvorsen, K. Marinimicrobium haloxylanilyticum sp. nov., a new moderately halophilic, polysaccharide-degrading bacterium isolated from Great Salt Lake, Utah. Anton Leeuw 2010, 98, 553–565. [Google Scholar] [CrossRef]
- Sorokin, D.Y.; Tourova, T.P.; Abbas, B.; Suhacheva, M.V.; Muyzer, G. Desulfonatronovibrio halophilus sp. nov., a novel moderately halophilic sulfate-reducing bacterium from hypersaline chloride-sulfate lakes in Central Asia. Extremophiles 2012, 16, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Zhou, Y.; Ja, M.; Shi, R.; Chun-Yu, W.X.; Yang, L.L.; Tang, S.K.; Li, W.J. Virgibacillus albus sp. nov., a novel moderately halophilic bacterium isolated from Lop Nur salt lake in Xinjiang province, China. Anton Leeuw 2012, 102, 553–560. [Google Scholar] [CrossRef]
- Qi, X.; Zhang, Y.; Tu, R.; Lin, Y.; Li, X.; Wang, Q. High-throughput screening and characterization of xylose-utilizing, ethanol-tolerant thermophilic bacteria for bioethanol production. J. Appl. Microbiol. 2011, 110, 1584–1591. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Chu, J.; Hao, Y.; Guo, Y.; Zhuang, Y.; Zhang, S. High-throughput system for screening of Cephalosporin C high-yield strain by 48-deep-well microtiter plates. Appl. Biochem. Biotechnol. 2013, 169, 1683–1695. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Jin, J.; Xue, N.; Song, X.; Chen, X. Development and validation of high-throughput screening assays for poly(ADP-ribose) polymerase-2 inhibitors. Anal. Biochem. 2014, 449, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.Z.; Wang, P.; Li, H.; Lin, K.F.; Lu, Z.Y.; Guo, X.J.; Liu, Y.D. Community analysis and metabolic pathway of halophilic bacteria for phenol degradation in saline environment. Int. Biodeterior. Biodegrad. 2014, 94, 115–120. [Google Scholar] [CrossRef]
- Ventosa, A.; Nieto, J.J.; Oren, A. Biology of moderately halophilic aerobic bacteria. Microbiol. Mol. Biol. Rev. 1998, 62, 504–544. [Google Scholar] [PubMed]
- Leitão, A.L.; Duarte, M.P.; Oliveira, J.S. Degradation of phenol by a halotolerant strain of Penicillium chrysogenum. Int. Biodeterior. Biodegrad. 2007, 59, 220–225. [Google Scholar] [CrossRef]
- Arulazhagan, P.; Vasudevan, N.; Yeom, I.T. Biodegradation of polycyclic aromatic hydrocarbon by a halotolerant bacterial consortium isolated from marine environment. Int. J. Environ. Sci. Technol. 2010, 7, 639–652. [Google Scholar] [CrossRef]
- Haddadi, A.; Shavandi, M. Biodegradation of phenol in hypersaline conditions by Halomonas sp. strain PH2–2 isolated from saline soil. Int. Biodeterior. Biodegrad. 2013, 85, 29–34. [Google Scholar] [CrossRef]
- Wang, P.; Qu, Y.; Zhou, J. Changes of microbial community structures and functional genes during biodegradation of phenolic compounds under high salt condition. J. Environ. Sci. 2009, 21, 821–826. [Google Scholar] [CrossRef]
- Kim, M.S.; Roh, S.W.; Bae, J.W. Halomonas jeotgali sp. nov., a new moderate halophilic bacterium isolated from a traditional fermented seafood. J. Microbiol. 2010, 48, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.X.; Xiao, W.; Dong, M.H.; Zhao, Q.; Li, Z.Y.; Lai, Y.H.; Cui, X.L. Halomonas qiaohouensis sp. nov., isolated from salt mine soil in southwest China. Anton Leeuw 2014, 106, 253–260. [Google Scholar] [CrossRef]
- Zhao, B.; Wang, H.; Mao, X.; Li, R.; Zhang, Y.J.; Tang, S.; Li, W.J. Halomonas xianhensis sp. nov., a moderately halophilic bacterium isolated from a saline soil contaminated with crude oil. Int. J. Syst. Evol. Microbiol. 2012, 62, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Peyton, B.M.; Wilson, T.; Yonge, D.R. Kinetics of phenol biodegradation in high salt solutions. Water Res. 2002, 36, 4811–4820. [Google Scholar] [CrossRef] [PubMed]
- Bonfá, M.R.; Grossman, M.J.; Piubeli, F.; Mellado, E.; Durrant, L.R. Phenol degradation by halophilic bacteria isolated from hypersaline environments. Biodegradation 2013, 24, 699–709. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, F.; Maki, T.; Nakamura, Y. Biodegradation of phenol in seawater using bacteria isolated from the intestinal contents of marine creatures. Int. Biodeterior. Biodegrad. 2012, 69, 113–118. [Google Scholar] [CrossRef]
- Gayathri, K.V.; Vasudevan, N. Enrichment of phenol degrading moderately halophilic bacterial consortium from saline environment. J. Bioremediat. Biodegrad. 2010, 1, 104–110. [Google Scholar] [CrossRef]
- Bursy, J.; Kuhlmann, A.U.; Pittelkow, M.; Hartmann, H.; Jebbar, M.; Pierik, A.J.; Bremer, E. Synthesis and uptake of the compatible solutes ectoine and 5-hydroxyectoine by streptomyces coelicolor A3(2) in response to salt and heat stresses. Appl. Environ. Microbiol. 2008, 74, 7286–7296. [Google Scholar] [CrossRef] [PubMed]
- Pastor, J.M.; Salvador, M.; Argandoña, M.; Bernal, V.; Reina-Bueno, M.; Csonka, L.N.; Iborra, J.L.; Vargas, C.; Nieto, J.J.; Cánovas, M. Ectoines in cell stress protection: uses and biotechnological production. Biotechnol. Adv. 2010, 28, 782–801. [Google Scholar] [CrossRef] [PubMed]
- Vargas, C.; Jebbar, M.; Carrasco, R.; Blanco, C.; Calderón, M.I.; Iglesias-Guerra, F.; Nieto, J.J. Ectoines as compatible solutes and carbon and energy sources for the halophilic bacterium Chromohalobacter salexigens. J. Appl. Microbiol. 2006, 100, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Veenagayathri, K.; Vasudevan, N. Ortho and meta cleavage dioxygenases detected during the degradation of phenolic compounds by a moderately halophilic bacterial consortium. Int. Res. J. Microbiol. 2011, 2, 406–414. [Google Scholar]
- García, M.T.; Ventosa, A.; Mellado, E. Catabolic versatility of aromatic compound-degrading halophilic bacteria. FEMS Microbiol. Ecol. 2005, 54, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Kuhlmann, A.U.; Bremer, E. Osmotically regulated synthesis of the compatible solute ectoine in Bacillus pasteurii and related Bacillus spp. Appl. Environ. Microbiol. 2002, 68, 772–783. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Chen, M.; Zhang, W.; Lin, M. Genetic organization of genes encoding phenol hydroxylase, benzoate 1,2-dioxygenase alpha subunit and its regulatory proteins in Acinetobacter calcoaceticus PHEA-2. Curr. Microbiol. 2003, 46, 235–240. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Z.-Y.; Guo, X.-J.; Li, H.; Huang, Z.-Z.; Lin, K.-F.; Liu, Y.-D. High-Throughput Screening for a Moderately Halophilic Phenol-Degrading Strain and Its Salt Tolerance Response. Int. J. Mol. Sci. 2015, 16, 11834-11848. https://doi.org/10.3390/ijms160611834
Lu Z-Y, Guo X-J, Li H, Huang Z-Z, Lin K-F, Liu Y-D. High-Throughput Screening for a Moderately Halophilic Phenol-Degrading Strain and Its Salt Tolerance Response. International Journal of Molecular Sciences. 2015; 16(6):11834-11848. https://doi.org/10.3390/ijms160611834
Chicago/Turabian StyleLu, Zhi-Yan, Xiao-Jue Guo, Hui Li, Zhong-Zi Huang, Kuang-Fei Lin, and Yong-Di Liu. 2015. "High-Throughput Screening for a Moderately Halophilic Phenol-Degrading Strain and Its Salt Tolerance Response" International Journal of Molecular Sciences 16, no. 6: 11834-11848. https://doi.org/10.3390/ijms160611834





