Casein Kinase 1 Epsilon Expression Predicts Poorer Prognosis in Low T-Stage Oral Cancer Patients
Abstract
:1. Introduction
2. Results and Discussion
2.1. Patient Characteristics
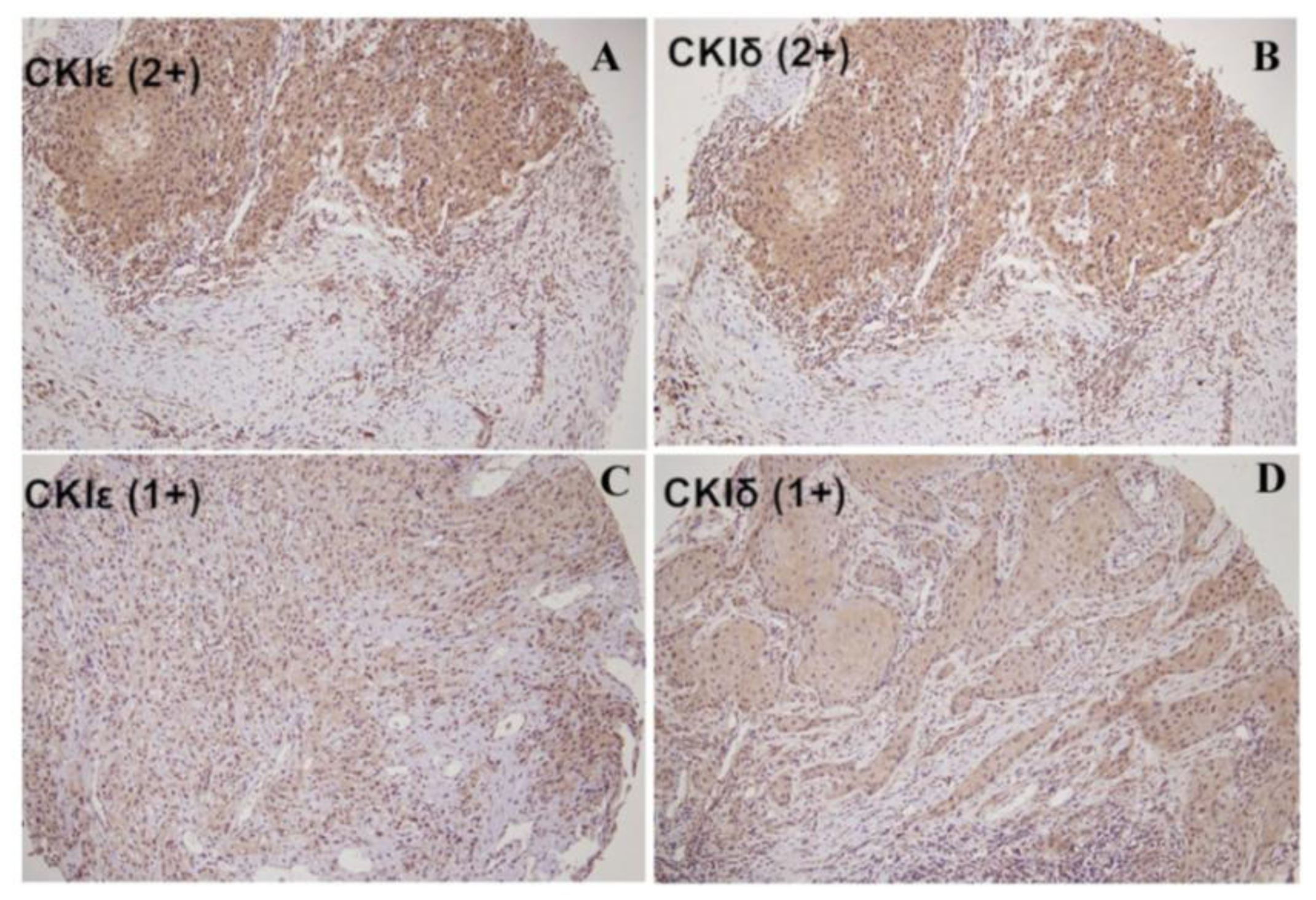
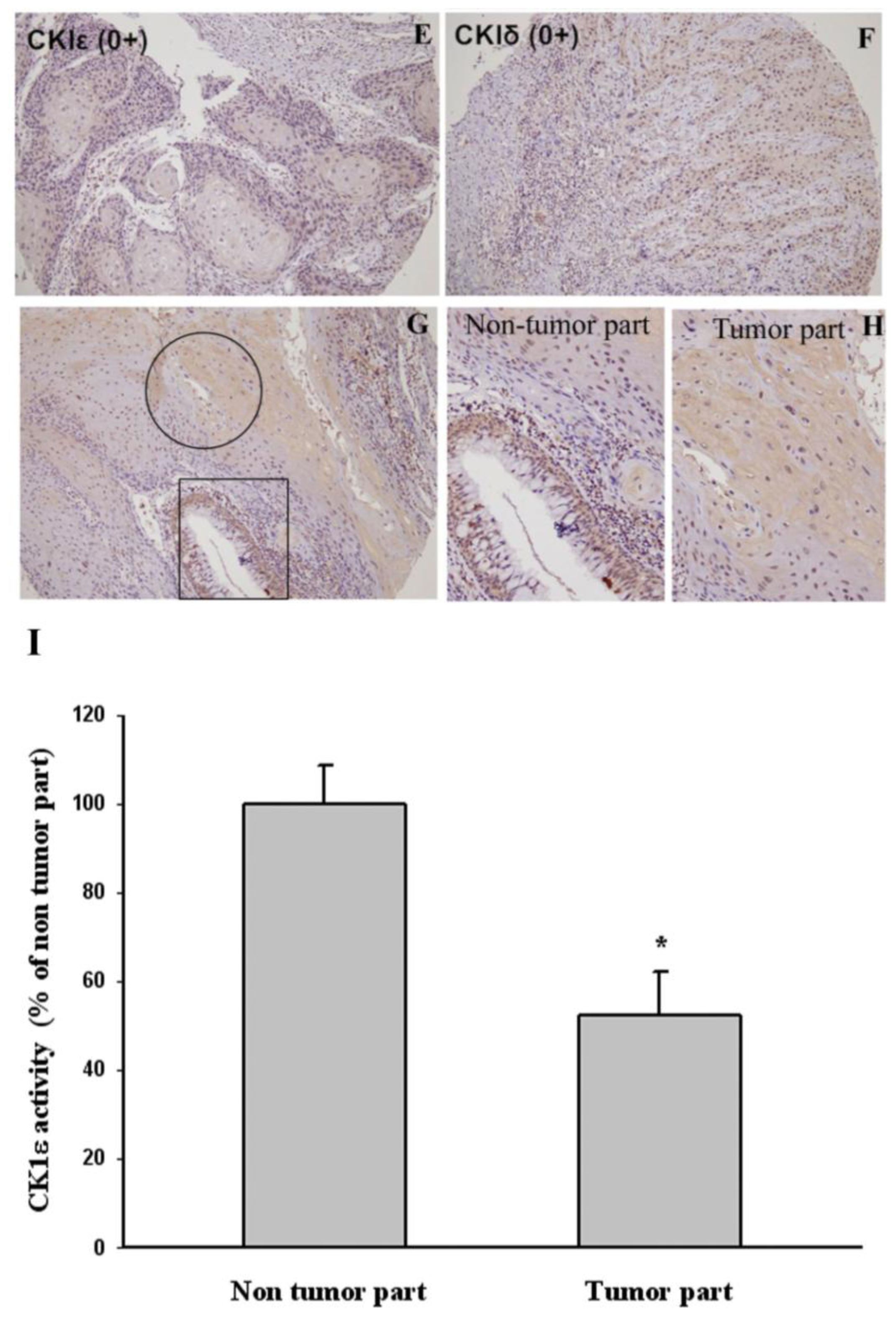
2.2. The Correlation between Cytoplasmic CK1ɛ and CK1δ Expressions in Oral Cancers and Various Clinicopathologic Characteristics
2.3. Correlations between Cytoplasmic CK1ɛ Expression and Patient Survival
3. Experimental Section
3.1. Patients and Samples
3.2. Tissue Microarrays (TMAs)
3.3. Immunohistochemistry
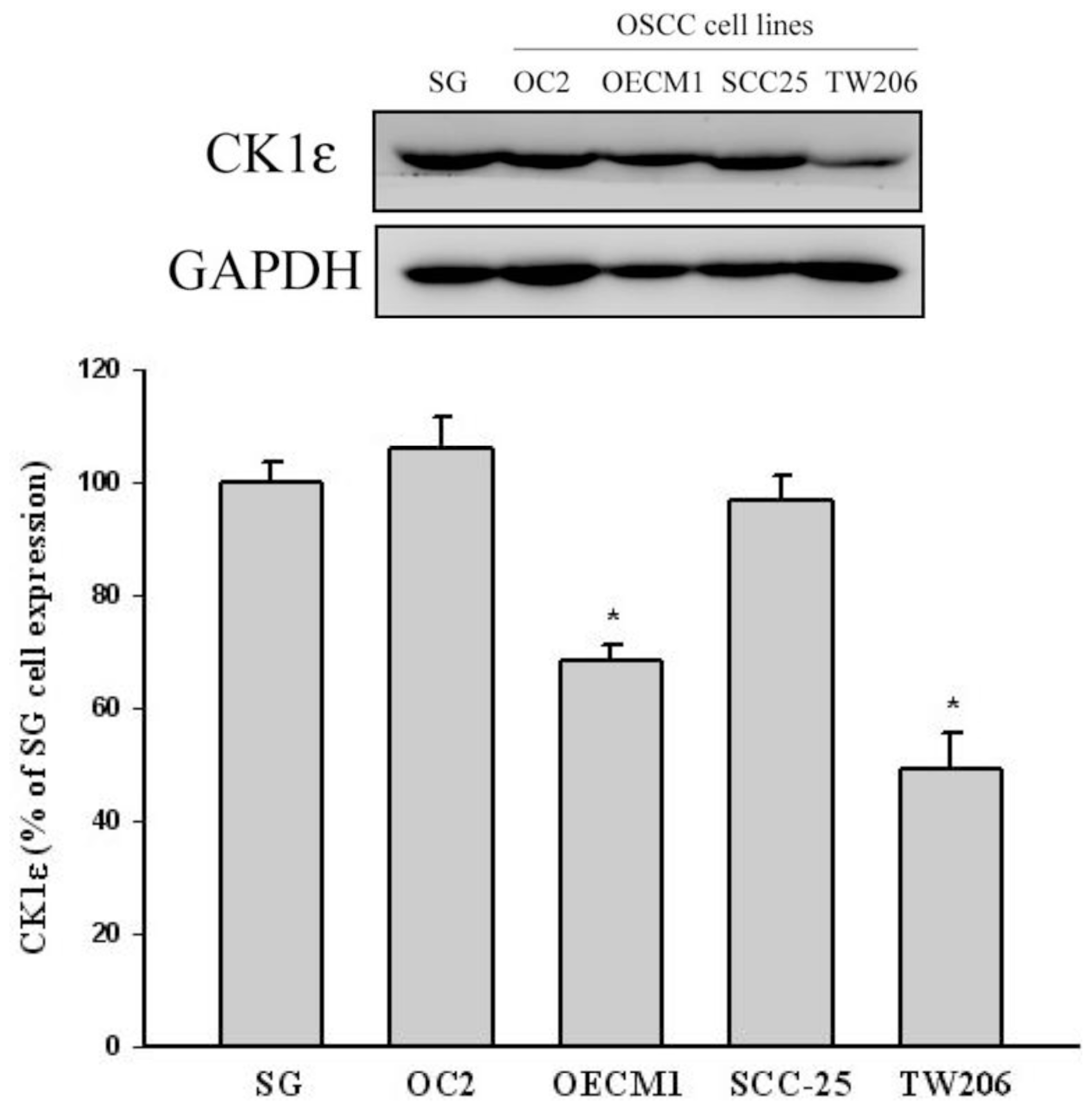
3.4. Cell and Cell Culture
3.5. Western Blot Analysis
3.6. CK1ɛ Kinase Activity
3.7. Statistical Analysis
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Cancer Registration System Annual Report; Bureau of Health Promotion, Department of Health: New Taipei City, Taiwan, 2010.
- Leite, I.C.; Koifman, S. Survival analysis in a sample of oral cancer patients at a reference hospital in Rio de Janeiro, Brazil. Oral Oncol 1998, 34, 347–352. [Google Scholar]
- Lo, W.L.; Kao, S.Y.; Chi, L.Y.; Wong, Y.K.; Chang, R.C. Outcomes of oral squamous cell carcinoma in Taiwan after surgical therapy: Factors affecting survival. J. Oral Maxillofac. Surg 2003, 61, 751–758. [Google Scholar]
- Argiris, A.; Karamouzis, M.V.; Raben, D.; Ferris, R.L. Head and neck cancer. Lancet 2008, 371, 1695–1709. [Google Scholar]
- Chiesa, F.; Mauri, S.; Tradati, N.; Calabrese, L.; Giugliano, G.; Ansarin, M.; Andrle, J.; Zurrida, S.; Orecchia, R.; Scully, C. Surfing prognostic factors in head and neck cancer at the millennium. Oral Oncol 1999, 35, 590–596. [Google Scholar]
- Helliwell, T.R. Molecular markers of metastasis in squamous carcinomas. J. Pathol 2001, 194, 289–293. [Google Scholar]
- Lopes, M.A.; Nikitakis, N.G.; Reynolds, M.A.; Ord, R.A.; Sauk, J., Jr. Biomarkers predictive of lymph node metastases in oral squamous cell carcinoma. J. Oral Maxillofac. Surg 2002, 60, 142–147. [Google Scholar]
- Rodrigo, J.P.; Suarez, C.; Ferlito, A.; Devaney, K.O.; Petruzzelli, G.J.; Rinaldo, A. Potential molecular prognostic markers for lymph node metastasis in head and neck squamous cell carcinoma. Acta Otolaryngol 2003, 123, 100–105. [Google Scholar]
- Lallemant, B.; Evrard, A.; Combescure, C.; Chapuis, H.; Chambon, G.; Raynal, C.; Reynaud, C.; Sabra, O.; Joubert, D.; Hollande, F.; et al. Clinical relevance of nine transcriptional molecular markers for the diagnosis of head and neck squamous cell carcinoma in tissue and saliva rinse. BMC Cancer 2009, 9, 370:1–370:10. [Google Scholar]
- Shah, N.G.; Trivedi, T.I.; Tankshali, R.A.; Goswami, J.V.; Jetly, D.H.; Shukla, S.N.; Shah, P.M.; Verma, R.J. Prognostic significance of molecular markers in oral squamous cell carcinoma: A multivariate analysis. Head Neck 2009, 31, 1544–1556. [Google Scholar]
- Gross, S.D.; Anderson, R.A. Casein kinase I: Spatial organization and positioning of a multifunctional protein kinase family. Cell Signal 1998, 10, 699–711. [Google Scholar]
- Knippschild, U.; Gocht, A.; Wolff, S.; Huber, N.; Löhler, J.; Stöter, M. The casein kinase I family: Participation in multiple cellular processes in eukaryotes. Cell Signal 2005, 17, 675–689. [Google Scholar]
- Price, M.A. CKI: There’s more than one: Casein kinase I family members in Wnt and Hedgehog signaling. Genes Dev 2006, 20, 399–410. [Google Scholar]
- Brockschmidt, C.; Hirner, H.; Huber, N.; Eismann, T.; Hillenbrand, A.; Giamas, G.; Radunsky, B.; Ammerpohl, O.; Bohm, B.; Henne-Bruns, D.; et al. Anti-apoptotic and growth-stimulatory functions of CKI delta and epsilon in ductal adenocarcinoma of the pancreas are inhibited by IC261 in vitro and in vivo. Gut 2008, 57, 799–806. [Google Scholar]
- Frierson, H.F., Jr.; El-Naggar, A.K.; Welsh, J.B.; Sapinoso, L.M.; Su, A.I.; Cheng, J.; Saku, T.; Moskaluk, C.A.; Hampton, G.M. Large scale molecular analysis identifies genes with altered expression in salivary adenoid cystic carcinoma. Am. J. Pathol 2002, 161, 1315–1323. [Google Scholar]
- Peters, J.M.; McKay, R.M.; McKay, J.P.; Graff, J.M. Casein kinase I transduces Wnt signals. Nature 1999, 401, 345–350. [Google Scholar]
- Polakis, P. The many ways of Wnt in cancer. Curr. Opin. Genet. Dev 2007, 17, 45–51. [Google Scholar]
- Rodriguez, N.; Yang, J.; Hasselblatt, K.; Liu, S.; Zhou, Y.; Rauh-Hain, J.A.; Ng, S.K.; Choi, P.W.; Fong, W.P.; Agar, N.Y.; et al. Casein kinase I epsilon interacts with mitochondrial proteins for the growth and survival of human ovarian cancer cells. EMBO Mol. Med 2012, 4, 952–963. [Google Scholar]
- Modak, C.; Chai, J. Potential of casein kinase I in digestive cancer screening. World J. Gastrointest. Oncol 2009, 1, 26–33. [Google Scholar]
- Yang, Y.H.; Chen, C.H.; Chang, J.S.; Lin, C.C.; Cheng, T.C.; Shieh, T.Y. Incidence rates of oral cancer and oral pre-cancerous lesions in a 6-year follow-up study of a Taiwanese aboriginal community. J. Oral Pathol. Med 2005, 34, 596–601. [Google Scholar]
- Fedele, S. Diagnostic aids in the screening of oral cancer. Head Neck Oncol 2009, 1, 5:1–5:6. [Google Scholar]
- Fuja, T.J.; Lin, F.; Osann, K.E.; Bryant, P.J. Somatic mutations and altered expression of the candidate tumor suppressors CSNK1; DLG1; and EDD/hHYDin mammary ductal carcinoma. Cancer Res 2004, 64, 942–951. [Google Scholar]
- Hsu, C.M.; Lin, S.F.; Lu, C.T.; Lin, P.M.; Yang, M.Y. Altered expression of circadian clock genes in head and neck squamous cell carcinoma. Tumour Biol 2012, 33, 149–155. [Google Scholar]
- Flajolet, M.; He, G.; Heiman, M.; Lin, A.; Nairn, A.C.; Greengard, P. Regulation of Alzheimer’s disease amyloid-beta formation by casein kinase I. Proc. Natl. Acad. Sci. USA 2007, 104, 4159–4164. [Google Scholar]
- Umar, S.; Wang, Y.; Morris, A.P.; Sellin, J.H. Dual alterations in casein kinase I-epsilon and GSK-3beta modulate beta-catenin stability in hyperproliferating colonic epithelia. Am. J. Physiol. Gastrointest. Liver Physiol 2007, 292, G599–G607. [Google Scholar]
- Tsai, I.C.; Amack, J.D.; Gao, Z.H.; Band, V.; Yost, H.J.; Virshup, D.M. A Wnt-CKIvarepsilon-Rap1 pathway regulates gastrulation by modulating SIPA1L1: A Rap GTPase activating protein. Dev. Cell 2007, 12, 335–347. [Google Scholar]
- Cegielska, A.; Gietzen, K.F.; Rivers, A.; Virshup, D.M. Autoinhibition of casein kinase I epsilon (CKI epsilon) is relieved by protein phosphatases and limited proteolysis. J. Biol. Chem 1998, 273, 1357–1364. [Google Scholar]
- Michl, P.; Downward, J. Mechanisms of disease: PI3K/AKT signaling in gastrointestinal cancers. Z. Gastroenterol 2005, 43, 1133–1139. [Google Scholar]
- Foldynová-Trantírková, S.; Sekyrová, P.; Tmejová, K.; Brumovská, E.; Bernatík, O.; Blankenfeldt, W. Breast cancer-specific mutations in CK1ɛ inhibit Wnt/β-catenin and activate the Wnt/Rac1/JNK and NFAT pathways to decrease cell adhesion and promote cell migration. Breast Cancer Res 2010, 12, R30:1–R30:14. [Google Scholar]
- Kim, S.Y.; Dunn, I.F.; Firestein, R.; Gupta, P.; Wardwell, L.; Repich, K.; Schinzel, A.C.; Wittner, B.; Silver, S.J.; Root, D.E.; et al. CK1ɛ is required for breast cancers dependent on β-catenin activity. PLoS One 2010, 5, e8979. [Google Scholar]
- Lin, C.W.; Chen, P.N.; Chen, M.K.; Yang, W.E.; Tang, C.H.; Yang, S.F.; Hsieh, Y.S. Kaempferol reduces matrix metalloproteinase-2 expression by down-regulating ERK1/2 and the activator protein-1 signaling pathways in oral cancer cells. PLoS One 2013, 8, e80883. [Google Scholar]


| Variable | Oral squamous cell carcinoma | ||
|---|---|---|---|
| n = 195 | % | ||
| Gender | Female | 36 | 18.5 |
| Male | 159 | 81.5 | |
| CK1ɛ expression | 0 | 62 | 31.8 |
| 1+ | 98 | 50.3 | |
| 2+ | 35 | 17.9 | |
| Smoke or Betel nuts | No | 71 | 36.4 |
| Yes | 89 | 45.6 | |
| Unknown | 35 | 17.9 | |
| Stage | I | 60 | 30.8 |
| II | 56 | 28.7 | |
| III | 38 | 19.5 | |
| IV | 41 | 21.0 | |
| Lymph Node Metastasis | No | 133 | 68.2 |
| Yes | 62 | 31.8 | |
| Grade | Well | 27 | 13.8 |
| Moderate | 161 | 82.6 | |
| Poor | 1 | 3.6 | |
| Tumor status | T1 | 80 | 41.0 |
| T2 | 93 | 47.7 | |
| T3 | 22 | 11.3 | |
| Age | Mean | 55.9 years | |
| Median | 55.0 years | ||
| Range | 31−88 years | ||
| Overall-survival | Mean | 4.1 years | |
| Median | 3.9 years | ||
| Range | 0.1−9.6 years | ||
| Follow up | Mean | 5.4 years | |
| Median | 6.0 years | ||
| Range | 0.1−13.2 years | ||
| Characteristics | Cytoplasmic staining of CK1ɛ (n = 195) | p value | Cytoplasmic staining of CK1δ (n = 173) | p value | ||
|---|---|---|---|---|---|---|
| Negative (%) | Positive (%) | Negative (%) | Positive (%) | |||
| Gender | ||||||
| F | 11 (17.7) | 25 (18.8) | 0.860 | 8 (6.8) | 3 (5.4) | 1.000 a |
| M | 51 (82.3) | 108 (81.2) | 109 (93.2) | 53 (94.6) | ||
| Age | ||||||
| ≤55 years | 33 (53.2) | 69 (51.9) | 0.861 | 58 (49.6) | 30 (53.6) | 0.623 |
| >55 years | 29 (46.8) | 64 (48.1) | 59 (50.4) | 26 (46.4) | ||
| Grade | ||||||
| well | 10 (16.1) | 17 (12.8) | 0.529 | 17 (14.5) | 8 (14.3) | 0.966 |
| moderate, poor | 52 (83.9) | 116 (87.2) | 100 (85.5) | 48 (85.7) | ||
| T status | ||||||
| T1 | 29 (46.8) | 51 (38.3) | 0.211 | 46 (39.3) | 21 (37.5) | 0.586 |
| T2 | 24 (38.7) | 69 (51.9) | 55 (47.0) | 30 (53.6) | ||
| T3 | 9 (14.5) | 13 (9.8) | 16 (13.7) | 5 (8.9) | ||
| Lymph Node Metastasis | ||||||
| no | 38 (61.3) | 95 (71.4) | 0.157 | 75 (64.1) | 41 (73.2) | 0.233 |
| yes | 24 (38.7) | 38 (28.6) | 42 (35.9) | 15 (26.8) | ||
| Distance Metastasis | ||||||
| no | 61 (98.4) | 133 (100) | 0.413 a | 116 (99.1) | 56 (100) | 1.000 a |
| yes | 1 (1.6) | 0 (0) | 1 (0.9) | 0 (0) | ||
| Stage | ||||||
| I | 19 (30.6) | 41 (30.8) | 0.079 | 33 (28.2) | 17 (30.4) | 0.365 |
| II | 12 (19.4) | 44 (33.1) | 36 (30.8) | 20 (35.7) | ||
| III | 12 (19.4) | 26 (19.6) | 20 (17.1) | 12 (21.4) | ||
| IV | 19 (30.6) | 22 (16.5) | 28 (23.9) | 7 (12.5) | ||
| Survival | ||||||
| ≤4 years | 42 (67.7) | 58 (43.6) | 0.002 | 45 (38.5) | 17 (30.4) | 0.298 |
| >4 years | 20 (32.3) | 75 (56.4) | 72 (61.5) | 39 (69.6) | ||
| Smoking or Betel Nuts | ||||||
| no | 22 (44) | 49 (44.5) | 0.949 | 36 (40.4) | 18 (46.2) | 0.548 |
| yes | 28 (56) | 61 (55.5) | 53 (59.6) | 21 (53.8) | ||
| Variable | Hazard ratio | 95% CI | p value |
|---|---|---|---|
| Expression of CK1ɛ (n = 195) | |||
| negative | 1.7 | 0.361–0.930 | 0.024 |
| positive | 1.0 | ||
| Grade | |||
| well | 1.0 | 1.601–26.628 | 0.009 |
| moderate/poor | 6.5 | ||
| Stage | |||
| I + II | 1.0 | 1.470–3.717 | <0.001 |
| III + IV | 2.3 | ||
| Lymph Node Metastasis | |||
| no | 1.0 | 1.740–4.379 | <0.001 |
| yes | 2.8 | ||
| Expression of CK1δ (n = 173) | |||
| negative | 1.2 | 0.462–1.432 | 0.475 |
| positive | 1.0 | ||
| Grade | |||
| well | 1.0 | 1.328–13.458 | 0.015 |
| moderate/poor | 4.2 | ||
| Stage | |||
| I + II | 1.0 | 1.560–4.101 | <0.001 |
| III + IV | 2.5 | ||
| Lymph Node Metastasis | |||
| no | 1.0 | 1.549–4.060 | <0.001 |
| yes | 2.5 | ||
| Variable | Hazard ratio | 95% CI | p value |
|---|---|---|---|
| Expression of CK1ɛ (n = 195) | |||
| negative | 1.6 | 0.382–1.004 | 0.052 |
| positive | 1.0 | ||
| Grade | |||
| well | 1.0 | 1.490–25.080 | 0.012 |
| moderate/poor | 6.1 | ||
| Stage | |||
| I + II | 1.1 | 0.364–2.424 | 0.897 |
| III + IV | 1.0 | ||
| Lymph Node Metastasis | |||
| no | 1.0 | 1.390–4.665 | 0.002 |
| yes | 2.5 | ||
| Expression of CK1δ (n = 173) | |||
| negative | 1.1 | 0.495–1.540 | 0.638 |
| positive | 1.0 | ||
| Grade | |||
| well | 1.0 | 1.180–19.992 | 0.029 |
| moderate/poor | 4.9 | ||
| Stage | |||
| I + II | 1.0 | 0.828–4.106 | 0.134 |
| III + IV | 1.8 | ||
| Lymph Node Metastasis | |||
| no | 1.0 | 0.771–3.771 | 0.188 |
| yes | 1.7 | ||
© 2014 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Lin, S.-H.; Lin, Y.-M.; Yeh, C.-M.; Chen, C.-J.; Chen, M.-W.; Hung, H.-F.; Yeh, K.-T.; Yang, S.-F. Casein Kinase 1 Epsilon Expression Predicts Poorer Prognosis in Low T-Stage Oral Cancer Patients. Int. J. Mol. Sci. 2014, 15, 2876-2891. https://doi.org/10.3390/ijms15022876
Lin S-H, Lin Y-M, Yeh C-M, Chen C-J, Chen M-W, Hung H-F, Yeh K-T, Yang S-F. Casein Kinase 1 Epsilon Expression Predicts Poorer Prognosis in Low T-Stage Oral Cancer Patients. International Journal of Molecular Sciences. 2014; 15(2):2876-2891. https://doi.org/10.3390/ijms15022876
Chicago/Turabian StyleLin, Shu-Hui, Yueh-Min Lin, Chung-Min Yeh, Chih-Jung Chen, Mei-Wen Chen, Hsiao-Fang Hung, Kun-Tu Yeh, and Shun-Fa Yang. 2014. "Casein Kinase 1 Epsilon Expression Predicts Poorer Prognosis in Low T-Stage Oral Cancer Patients" International Journal of Molecular Sciences 15, no. 2: 2876-2891. https://doi.org/10.3390/ijms15022876






