PhosphoTyrosyl Phosphatase Activator of Plasmodium falciparum: Identification of Its Residues Involved in Binding to and Activation of PP2A
Abstract
:1. Introduction
2. Results and Discussion
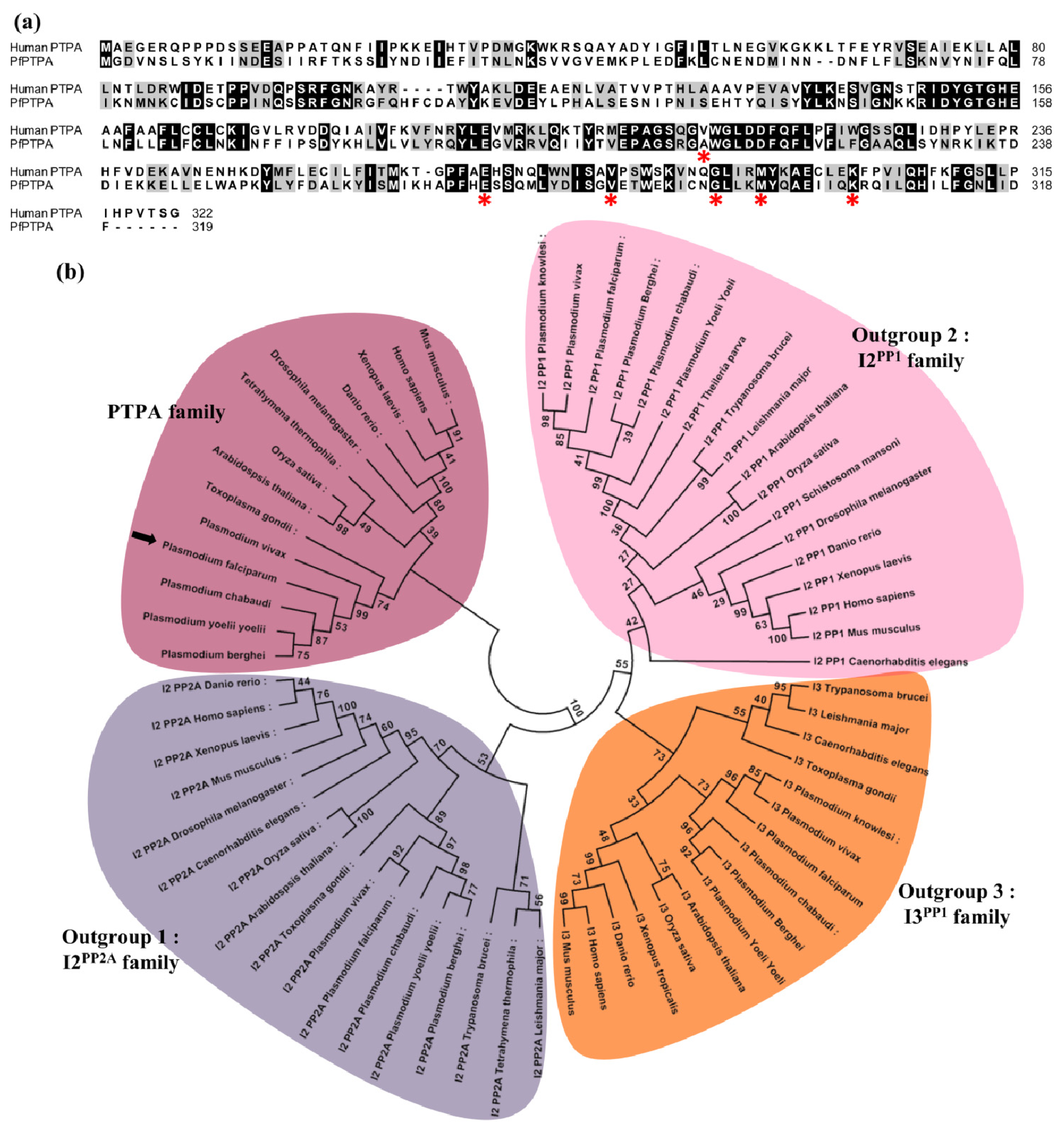
2.1. Molecular Cloning and Sequence Analysis of PfPTPA
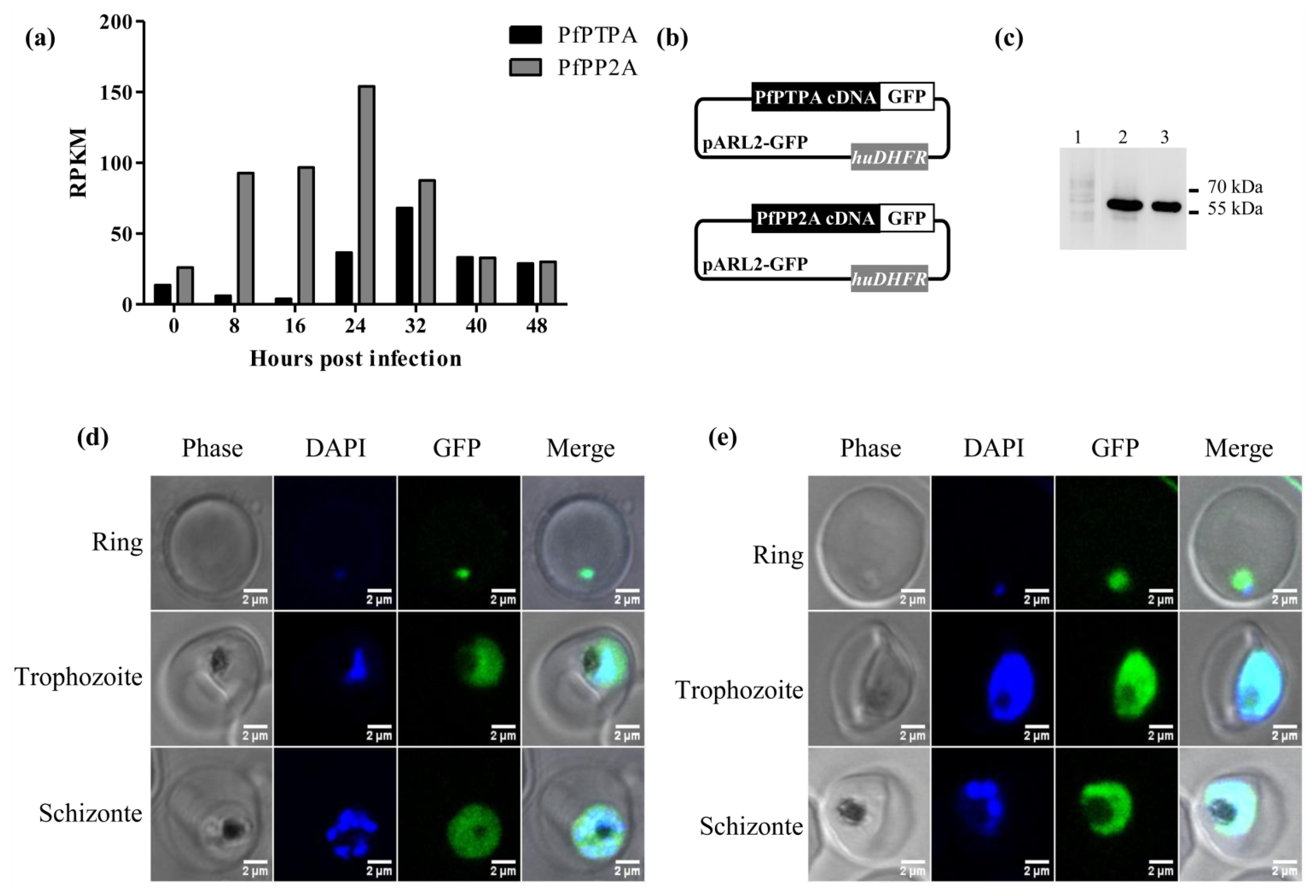
2.2. Expression of PfPTPA and PfPP2A by Plasmodium falciparum and Localization
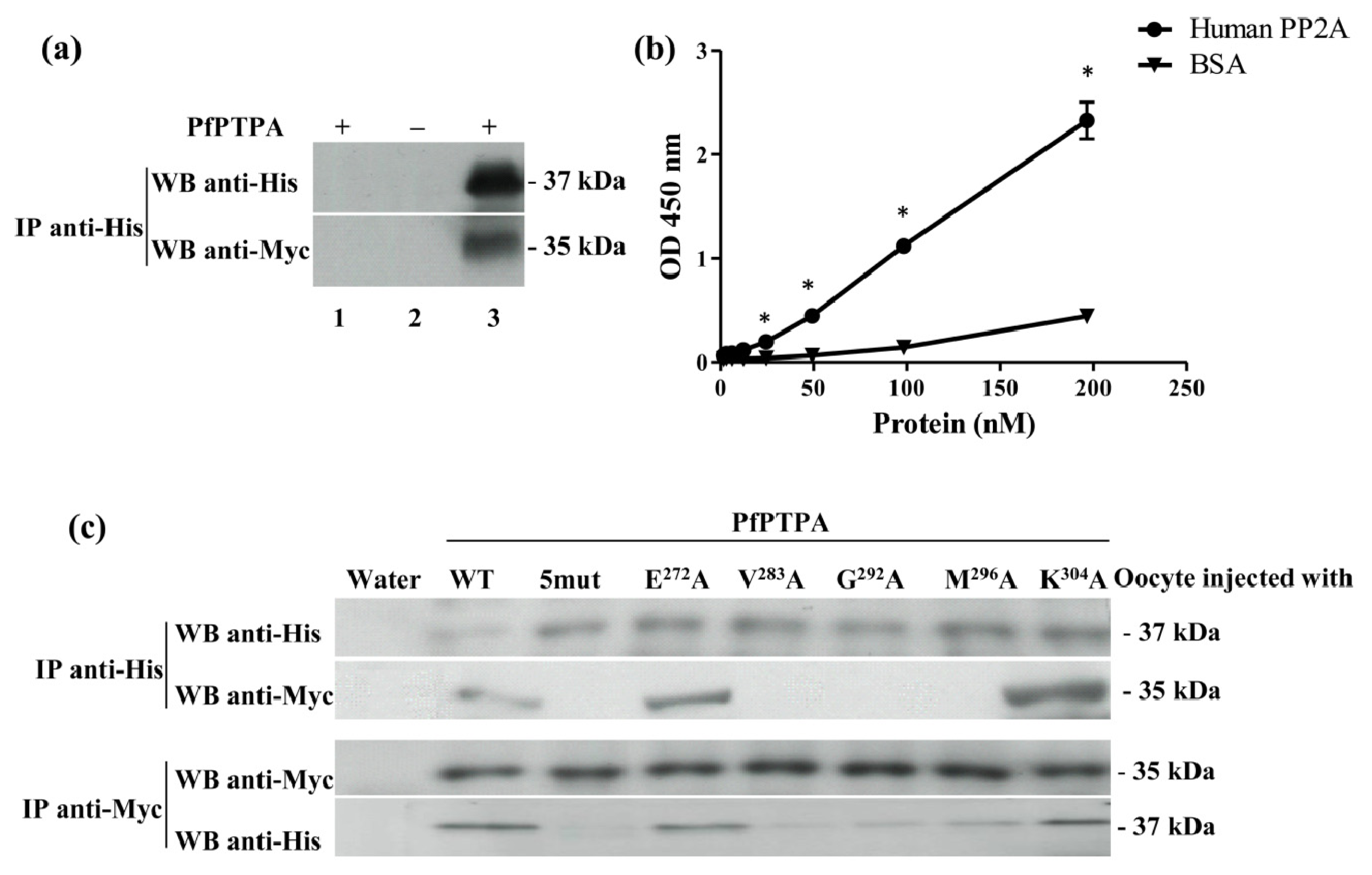
2.3. Binding of PfPTPA to PP2A
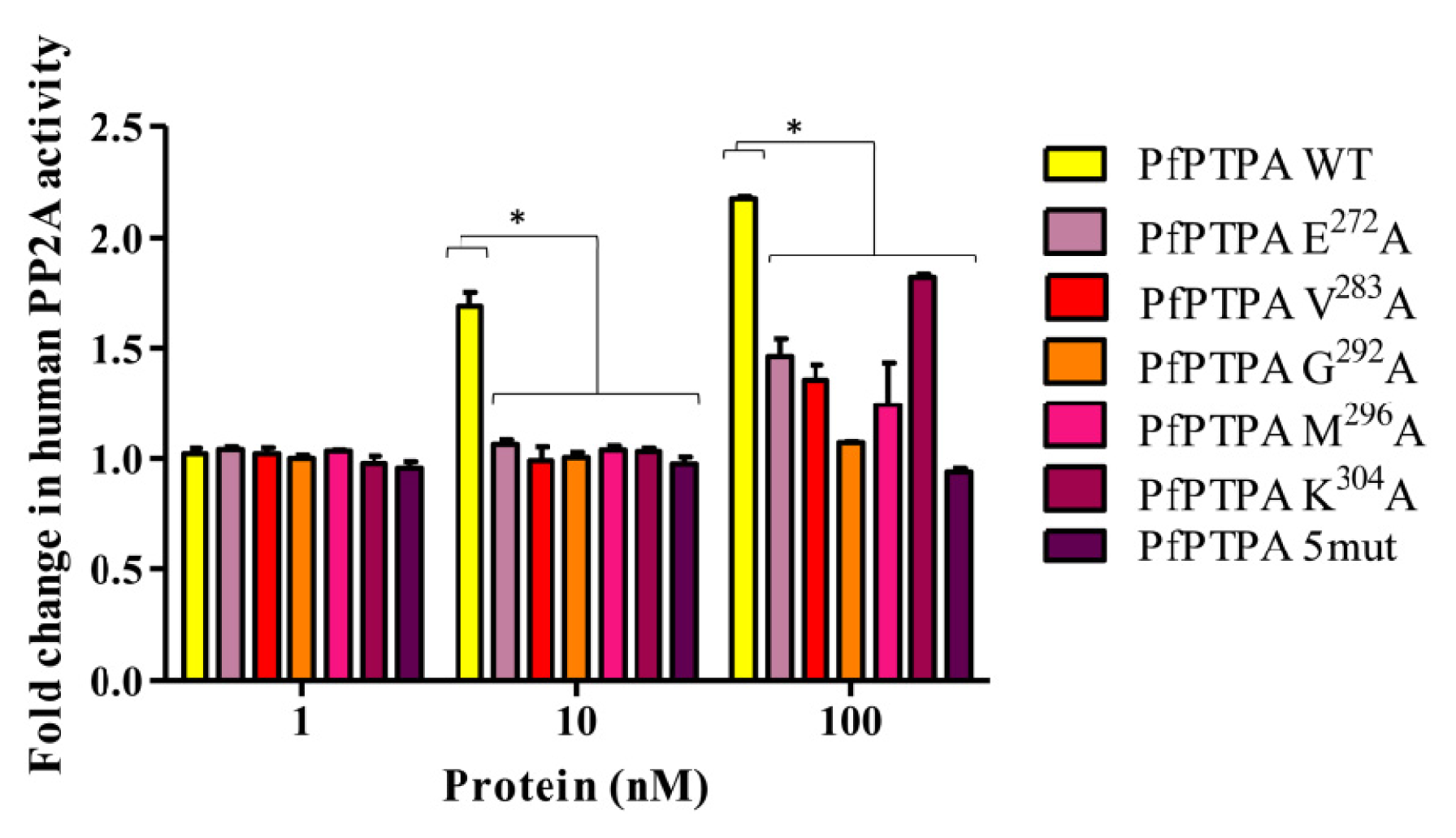
2.4. Effect of PfPTPA on the Activity of PP2A
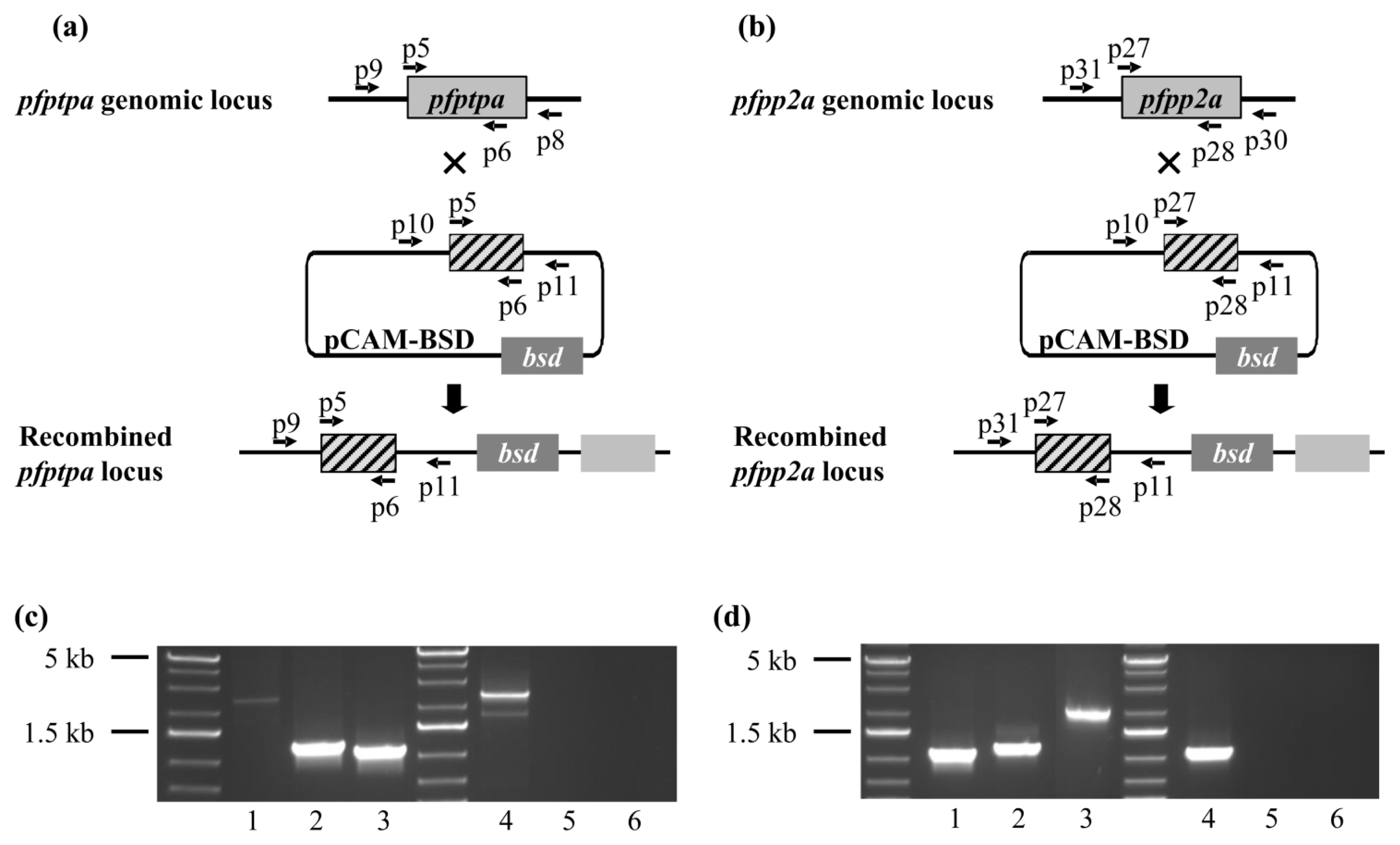
2.5. Genetic Manipulations of pfptpa and pfpp2a in Plasmodium falciparum
2.6. Inhibition of G2/M Transition of Xenopus oocytes by PfPTPA
3. Experimental Section
3.1. Materials
3.2. Phylogenetic Analyses and Secondary Structure Prediction
3.3. Preparation of Parasites
3.4. Localization of PfPTPA and PfPP2A
3.5. Generation of P. falciparum Transgenic Parasites
3.6. Genotype and Phenotype Analysis of P. falciparum Transfectants
3.7. Recombinant Proteins Expression and Purification
3.8. RNA Synthesis of PfPP2A
3.9. Induction of Xenopus Oocytes Germinal Vesicle Breakdown and Co-Immunoprecipitation
3.10. Assays for PP2A and Effect of PfPTPA
3.11. Measurement of Binding of PfPTPA to PP2A
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [Google Scholar]
- Sacco, F.; Perfetto, L.; Castagnoli, L.; Cesareni, G. The human phosphatase interactome: An intricate family portrait. FEBS Lett 2012, 586, 2732–2739. [Google Scholar]
- Bollen, M.; Peti, W.; Ragusa, M.J.; Beullens, M. The extended PP1 toolkit: Designed to create specificity. Trends Biochem. Sci 2010, 35, 450–458. [Google Scholar]
- Fardilha, M.; Esteves, S.L.; Korrodi-Gregorio, L.; da Cruz e Silva, O.A.; da Cruz e Silva, F.F. The physiological relevance of protein phosphatase 1 and its interacting proteins to health and disease. Curr. Med. Chem 2010, 17, 3996–4017. [Google Scholar]
- Hendrickx, A.; Beullens, M.; Ceulemans, H.; Den Abt, T.; van Eynde, A.; Nicolaescu, E.; Lesage, B.; Bollen, M. Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem. Biol 2009, 16, 365–371. [Google Scholar]
- Cayla, X.; Goris, J.; Hermann, J.; Hendrix, P.; Ozon, R.; Merlevede, W. Isolation and characterization of a tyrosyl phosphatase activator from rabbit skeletal muscle and Xenopus laevis oocytes. Biochemistry 1990, 29, 658–667. [Google Scholar]
- Junttila, M.R.; Puustinen, P.; Niemela, M.; Ahola, R.; Arnold, H.; Bottzauw, T.; Ala-aho, R.; Nielsen, C.; Ivaska, J.; Taya, Y.; et al. CIP2A inhibits PP2A in human malignancies. Cell 2007, 130, 51–62. [Google Scholar]
- Kong, M.; Ditsworth, D.; Lindsten, T.; Thompson, C.B. Alpha4 is an essential regulator of PP2A phosphatase activity. Mol. Cell 2009, 36, 51–60. [Google Scholar]
- Li, M.; Guo, H.; Damuni, Z. Purification and characterization of two potent heat-stable protein inhibitors of protein phosphatase 2A from bovine kidney. Biochemistry 1995, 34, 1988–1996. [Google Scholar]
- Mochida, S.; Maslen, S.L.; Skehel, M.; Hunt, T. Greatwall phosphorylates an inhibitor of protein phosphatase 2A that is essential for mitosis. Science 2010, 330, 1670–1673. [Google Scholar]
- Smetana, J.H.; Zanchin, N.I. Interaction analysis of the heterotrimer formed by the phosphatase 2A catalytic subunit, alpha 4 and the mammalian ortholog of yeast Tip41 (TIPRL). FEBS J 2007, 274, 5891–5904. [Google Scholar]
- Bernal, M.; Sanchez-Romero, M.A.; Salas-Pino, S.; Daga, R.R. Regulation of fission yeast morphogenesis by PP2A activator pta2. PLoS One 2012, 7, e32823. [Google Scholar]
- Mitchell, D.A.; Sprague, G.F., Jr. The PhosphoTyrosyl Phosphatase Activator, Ncs1p (Rrd1p), functions with Cla4p to regulate the G(2)/M transition in Saccharomyces cerevisiae. Mol. Cell. Biol 2001, 21, 488–500. [Google Scholar]
- Rempola, B.; Kaniak, A.; Migdalski, A.; Rytka, J.; Slonimski, P.P.; di Rago, J.P. Functional analysis of RRD1 (YIL153w) and RRD2 (YPL152w), which encode two putative activators of the PhosphoTyrosyl Phosphatase Activity of PP2A in Saccharomyces cerevisiae. Mol. Gen. Genet.: MGG 2000, 262, 1081–1092. [Google Scholar]
- Van Hoof, C.; Janssens, V.; de Baere, I.; de Winde, J.H.; Winderickx, J.; Dumortier, F.; Thevelein, J.M.; Merlevede, W.; Goris, J. The Saccharomyces cerevisiae homologue YPA1 of the mammalian PhosphoTyrosyl Phosphatase Activator of protein phosphatase 2A controls progression through the G1 phase of the yeast cell cycle. J. Mol. Biol 2000, 302, 103–120. [Google Scholar]
- Van Hoof, C.; Janssens, V.; de Baere, I.; Stark, M.J.; de Winde, J.H.; Winderickx, J.; Thevelein, J.M.; Merlevede, W.; Goris, J. The Saccharomyces cerevisiae PhosphoTyrosyl Phosphatase Activator proteins are required for a subset of the functions disrupted by protein phosphatase 2A mutations. Exp. Cell Res 2001, 264, 372–387. [Google Scholar]
- Cayla, X.; van Hoof, C.; Bosch, M.; Waelkens, E.; Vandekerckhove, J.; Peeters, B.; Merlevede, W.; Goris, J. Molecular cloning, expression, and characterization of PTPA, a protein that activates the tyrosyl phosphatase activity of protein phosphatase 2A. J. Biol. Chem 1994, 269, 15668–15675. [Google Scholar]
- Van Hoof, C.; Janssens, V.; Dinishiotu, A.; Merlevede, W.; Goris, J. Functional analysis of conserved domains in the PhosphoTyrosyl Phosphatase Activator. Molecular cloning of the homologues from Drosophila melanogaster and Saccharomyces cerevisiae. Biochemistry 1998, 37, 12899–12908. [Google Scholar]
- Jordens, J.; Janssens, V.; Longin, S.; Stevens, I.; Martens, E.; Bultynck, G.; Engelborghs, Y.; Lescrinier, E.; Waelkens, E.; Goris, J.; et al. The protein phosphatase 2A phosphatase activator is a novel peptidyl-prolyl cis/trans-isomerase. J. Biol. Chem 2006, 281, 6349–6357. [Google Scholar]
- Chao, Y.; Xing, Y.; Chen, Y.; Xu, Y.; Lin, Z.; Li, Z.; Jeffrey, P.D.; Stock, J.B.; Shi, Y. Structure and mechanism of the PhosphoTyrosyl Phosphatase Activator. Mol. Cell 2006, 23, 535–546. [Google Scholar]
- Leulliot, N.; Vicentini, G.; Jordens, J.; Quevillon-Cheruel, S.; Schiltz, M.; Barford, D.; van Tilbeurgh, H.; Goris, J. Crystal structure of the PP2A phosphatase activator: Implications for its PP2A-specific PPIase activity. Mol. Cell 2006, 23, 413–424. [Google Scholar]
- Magnusdottir, A.; Stenmark, P.; Flodin, S.; Nyman, T.; Hammarstrom, M.; Ehn, M.; Bakali, H.M.; Berglund, H.; Nordlund, P. The crystal structure of a human PP2A phosphatase activator reveals a novel fold and highly conserved cleft implicated in protein-protein interactions. J. Biol. Chem 2006, 281, 22434–22438. [Google Scholar]
- Bhattacharyya, M.K.; Hong, Z.; Kongkasuriyachai, D.; Kumar, N. Plasmodium falciparum protein phosphatase type 1 functionally complements a glc7 mutant in Saccharomyces cerevisiae. Int. J. Parasitol 2002, 32, 739–747. [Google Scholar]
- Blisnick, T.; Vincensini, L.; Fall, G.; Braun-Breton, C. Protein phosphatase 1, a Plasmodium falciparum essential enzyme, is exported to the host cell and implicated in the release of infectious merozoites. Cell. Microbiol 2006, 8, 591–601. [Google Scholar]
- Dobson, S.; Kar, B.; Kumar, R.; Adams, B.; Barik, S. A novel tetratricopeptide repeat (TPR) containing PP5 serine/threonine protein phosphatase in the malaria parasite Plasmodium falciparum. BMC Microbiol 2001, 1, 31. [Google Scholar]
- Dobson, S.; May, T.; Berriman, M.; del Vecchio, C.; Fairlamb, A.H.; Chakrabarti, D.; Barik, S. Characterization of protein Ser/Thr phosphatases of the malaria parasite, Plasmodium falciparum: Inhibition of the parasitic calcineurin by cyclophilin-cyclosporin complex. Mol. Biochem. Parasitol 1999, 99, 167–181. [Google Scholar]
- Fernandez-Pol, S.; Slouka, Z.; Bhattacharjee, S.; Fedotova, Y.; Freed, S.; An, X.; Holder, A.A.; Campanella, E.; Low, P.S.; Mohandas, N.; et al. A bacterial phosphatase-like enzyme of the malaria parasite Plasmodium falciparum possesses tyrosine phosphatase activity and is implicated in the regulation of band 3 dynamics during parasite invasion. Eukaryot. Cell 2013, 12, 1179–1191. [Google Scholar]
- Guttery, D.S.; Poulin, B.; Ferguson, D.J.; Szoor, B.; Wickstead, B.; Carroll, P.L.; Ramakrishnan, C.; Brady, D.; Patzewitz, E.M.; Straschil, U.; et al. A unique protein phosphatase with kelch-like domains (PPKL) in Plasmodium modulates ookinete differentiation, motility and invasion. PLoS Pathog 2012, 8, e1002948. [Google Scholar]
- Kumar, R.; Adams, B.; Oldenburg, A.; Musiyenko, A.; Barik, S. Characterisation and expression of a PP1 serine/threonine protein phosphatase (PfPP1) from the malaria parasite, Plasmodium falciparum: Demonstration of its essential role using RNA interference. Malar. J 2002, 1, 5. [Google Scholar]
- Kumar, R.; Musiyenko, A.; Barik, S. Plasmodium falciparum calcineurin and its association with heat shock protein 90: Mechanisms for the antimalarial activity of cyclosporin A and synergism with geldanamycin. Mol. Biochem. Parasitol 2005, 141, 29–37. [Google Scholar]
- Kumar, R.; Musiyenko, A.; Oldenburg, A.; Adams, B.; Barik, S. Post-translational generation of constitutively active cores from larger phosphatases in the malaria parasite, Plasmodium falciparum: Implications for proteomics. BMC Mol. Biol 2004, 5, 6. [Google Scholar]
- Li, J.L.; Baker, D.A. Protein phosphatase beta, a putative type-2A protein phosphatase from the human malaria parasite Plasmodium falciparum. Eur. J. Biochem./FEBS 1997, 249, 98–106. [Google Scholar]
- Li, J.L.; Baker, D.A. A putative protein serine/threonine phosphatase from Plasmodium falciparum contains a large N-terminal extension and five unique inserts in the catalytic domain. Mol. Biochem. Parasitol 1998, 95, 287–295. [Google Scholar]
- Lindenthal, C.; Klinkert, M.Q. Identification and biochemical characterisation of a protein phosphatase 5 homologue from Plasmodium falciparum. Mol. Biochem. Parasitol 2002, 120, 257–268. [Google Scholar]
- Mamoun, C.B.; Goldberg, D.E. Plasmodium protein phosphatase 2C dephosphorylates translation elongation factor 1beta and inhibits its PKC-mediated nucleotide exchange activity in vitro. Mol. Microbiol. 2001, 39, 973–981. [Google Scholar]
- Mamoun, C.B.; Sullivan, D.J., Jr.; Banerjee, R.; Goldberg, D.E. Identification and characterization of an unusual double serine/threonine protein phosphatase 2C in the malaria parasite Plasmodium falciparum. J. Biol. Chem 1998, 273, 11241–11247. [Google Scholar]
- Patzewitz, E.M.; Guttery, D.S.; Poulin, B.; Ramakrishnan, C.; Ferguson, D.J.; Wall, R.J.; Brady, D.; Holder, A.A.; Szoor, B.; Tewari, R. An ancient protein phosphatase, SHLP1, is critical to microneme development in Plasmodium ookinetes and parasite transmission. Cell Rep 2013, 3, 622–629. [Google Scholar]
- Philip, N.; Vaikkinen, H.J.; Tetley, L.; Waters, A.P. A unique Kelch domain phosphatase in Plasmodium regulates ookinete morphology, motility and invasion. PLoS One 2012, 7, e44617. [Google Scholar]
- Singh, S.; More, K.R.; Chitnis, C.E. Role of calcineurin and actin dynamics in regulated secretion of microneme proteins in Plasmodium falciparum merozoites during erythrocyte invasion. Cell. Microbiol 2013. [Google Scholar] [CrossRef]
- Daher, W.; Browaeys, E.; Pierrot, C.; Jouin, H.; Dive, D.; Meurice, E.; Dissous, C.; Capron, M.; Tomavo, S.; Doerig, C.; et al. Regulation of protein phosphatase type 1 and cell cycle progression by PfLRR1, a novel leucine-rich repeat protein of the human malaria parasite Plasmodium falciparum. Mol. Microbiol 2006, 60, 578–590. [Google Scholar]
- Freville, A.; Cailliau-Maggio, K.; Pierrot, C.; Tellier, G.; Kalamou, H.; Lafitte, S.; Martoriati, A.; Pierce, R.J.; Bodart, J.F.; Khalife, J. Plasmodium falciparum encodes a conserved active inhibitor-2 for Protein Phosphatase type 1: Perspectives for novel anti-plasmodial therapy. BMC Biol 2013, 11, 80. [Google Scholar]
- Freville, A.; Landrieu, I.; Garcia-Gimeno, M.A.; Vicogne, J.; Montbarbon, M.; Bertin, B.; Verger, A.; Kalamou, H.; Sanz, P.; Werkmeister, E.; et al. Plasmodium falciparum inhibitor-3 homolog increases protein phosphatase type 1 activity and is essential for parasitic survival. J. Biol. Chem 2012, 287, 1306–1321. [Google Scholar]
- Dobson, S.; Kumar, R.; Bracchi-Ricard, V.; Freeman, S.; Al-Murrani, S.W.; Johnson, C.; Damuni, Z.; Chakrabarti, D.; Barik, S. Characterization of a unique aspartate-rich protein of the SET/TAF-family in the human malaria parasite, Plasmodium falciparum, which inhibits protein phosphatase 2A. Mol. Biochem. Parasitol 2003, 126, 239–250. [Google Scholar]
- Aurrecoechea, C.; Brestelli, J.; Brunk, B.P.; Dommer, J.; Fischer, S.; Gajria, B.; Gao, X.; Gingle, A.; Grant, G.; Harb, O.S.; et al. PlasmoDB: A functional genomic database for malaria parasites. Nucleic Acids Res 2009, 37, D539–D543. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol 2011, 28, 2731–2739. [Google Scholar]
- Pieper, U.; Webb, B.M.; Barkan, D.T.; Schneidman-Duhovny, D.; Schlessinger, A.; Braberg, H.; Yang, Z.; Meng, E.C.; Pettersen, E.F.; Huang, C.C.; et al. ModBase, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res 2011, 39, D465–D474. [Google Scholar]
- Bowyer, P.W.; Simon, G.M.; Cravatt, B.F.; Bogyo, M. Global profiling of proteolysis during rupture of Plasmodium falciparum from the host erythrocyte. Mol. Cell. Proteomics 2011, 10. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Barragan, M.J.; Lemieux, J.; Quinones, M.; Williamson, K.C.; Molina-Cruz, A.; Cui, K.; Barillas-Mury, C.; Zhao, K.; Su, X.Z. Directional gene expression and antisense transcripts in sexual and asexual stages of Plasmodium falciparum. BMC Genomics 2011, 12, 587. [Google Scholar]
- Treeck, M.; Sanders, J.L.; Elias, J.E.; Boothroyd, J.C. The phosphoproteomes of Plasmodium falciparum and Toxoplasma gondii reveal unusual adaptations within and beyond the parasites’ boundaries. Cell Host Microbe 2011, 10, 410–419. [Google Scholar]
- Longin, S.; Jordens, J.; Martens, E.; Stevens, I.; Janssens, V.; Rondelez, E.; de Baere, I.; Derua, R.; Waelkens, E.; Goris, J.; et al. An inactive protein phosphatase 2A population is associated with methylesterase and can be re-activated by the PhosphoTyrosyl Phosphatase Activator. Biochem. J 2004, 380, 111–119. [Google Scholar]
- Kuhn, Y.; Sanchez, C.P.; Ayoub, D.; Saridaki, T.; van Dorsselaer, A.; Lanzer, M. Trafficking of the phosphoprotein PfCRT to the digestive vacuolar membrane in Plasmodium falciparum. Traffic 2010, 11, 236–249. [Google Scholar]
- Lee, T.Y.; Lai, T.Y.; Lin, S.C.; Wu, C.W.; Ni, I.F.; Yang, Y.S.; Hung, L.Y.; Law, B.K.; Chiang, C.W. The B56gamma3 regulatory subunit of protein phosphatase 2A (PP2A) regulates S phase-specific nuclear accumulation of PP2A and the G1 to S transition. J. Biol. Chem 2010, 285, 21567–21580. [Google Scholar]
- Van Hoof, C.; Martens, E.; Longin, S.; Jordens, J.; Stevens, I.; Janssens, V.; Goris, J. Specific interactions of PP2A and PP2A-like phosphatases with the yeast PTPA homologues, Ypa1 and Ypa2. Biochem. J 2005, 386, 93–102. [Google Scholar]
- Kinoshita, N.; Ohkura, H.; Yanagida, M. Distinct, essential roles of type 1 and 2A protein phosphatases in the control of the fission yeast cell division cycle. Cell 1990, 63, 405–415. [Google Scholar]
- Gotz, J.; Probst, A.; Ehler, E.; Hemmings, B.; Kues, W. Delayed embryonic lethality in mice lacking protein phosphatase 2A catalytic subunit Calpha. Proc. Natl. Acad. Sci. USA 1998, 95, 12370–12375. [Google Scholar]
- Yokoyama, D.; Saito-Ito, A.; Asao, N.; Tanabe, K.; Yamamoto, M.; Matsumura, T. Modulation of the growth of Plasmodium falciparum in vitro by protein serine/threonine phosphatase inhibitors. Biochem. Biophys. Res. Commun 1998, 247, 18–23. [Google Scholar]
- Jeong, A.L.; Yang, Y. PP2A function toward mitotic kinases and substrates during the cell cycle. BMB Rep 2013, 46, 289–294. [Google Scholar]
- Jiang, Y. Regulation of the cell cycle by protein phosphatase 2A in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev 2006, 70, 440–449. [Google Scholar]
- Krasinska, L.; Domingo-Sananes, M.R.; Kapuy, O.; Parisis, N.; Harker, B.; Moorhead, G.; Rossignol, M.; Novak, B.; Fisher, D. Protein phosphatase 2A controls the order and dynamics of cell-cycle transitions. Mol. Cell 2011, 44, 437–450. [Google Scholar]
- Lorca, T.; Bernis, C.; Vigneron, S.; Burgess, A.; Brioudes, E.; Labbe, J.C.; Castro, A. Constant regulation of both the MPF amplification loop and the Greatwall-PP2A pathway is required for metaphase II arrest and correct entry into the first embryonic cell cycle. J. Cell Sci 2010, 123, 2281–2291. [Google Scholar]
- Ferrell, J.E., Jr. Xenopus oocyte maturation: New lessons from a good egg. BioEssays 1999, 21, 833–842. [Google Scholar]
- Trager, W.; Jensen, J.B. Human malaria parasites in continuous culture. Science 1976, 193, 673–675. [Google Scholar]
- Vernes, A.; Haynes, J.D.; Tapchaisri, P.; Williams, J.L.; Dutoit, E.; Diggs, C.L. Plasmodium falciparum strain-specific human antibody inhibits merozoite invasion of erythrocytes. Am. J. Trop. Med. Hyg 1984, 33, 197–203. [Google Scholar]
- Umlas, J.; Fallon, J.N. New thick-film technique for malaria diagnosis. Use of saponin stromatolytic solution for lysis. Am. J. Trop. Med. Hyg 1971, 20, 527–529. [Google Scholar]
- Sidhu, A.B.; Valderramos, S.G.; Fidock, D.A. pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol. Microbiol 2005, 57, 913–926. [Google Scholar]
- Vicogne, J.; Cailliau, K.; Tulasne, D.; Browaeys, E.; Yan, Y.T.; Fafeur, V.; Vilain, J.P.; Legrand, D.; Trolet, J.; Dissous, C. Conservation of epidermal growth factor receptor function in the human parasitic helminth Schistosoma mansoni. J. Biol. Chem 2004, 279, 37407–37414. [Google Scholar]
- Azzi, L.; Meijer, L.; Ostvold, A.C.; Lew, J.; Wang, J.H. Purification of a 15-kDa cdk4- and cdk5-binding protein. J. Biol. Chem 1994, 269, 13279–13288. [Google Scholar]


© 2014 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Vandomme, A.; Fréville, A.; Cailliau, K.; Kalamou, H.; Bodart, J.-F.; Khalife, J.; Pierrot, C. PhosphoTyrosyl Phosphatase Activator of Plasmodium falciparum: Identification of Its Residues Involved in Binding to and Activation of PP2A. Int. J. Mol. Sci. 2014, 15, 2431-2453. https://doi.org/10.3390/ijms15022431
Vandomme A, Fréville A, Cailliau K, Kalamou H, Bodart J-F, Khalife J, Pierrot C. PhosphoTyrosyl Phosphatase Activator of Plasmodium falciparum: Identification of Its Residues Involved in Binding to and Activation of PP2A. International Journal of Molecular Sciences. 2014; 15(2):2431-2453. https://doi.org/10.3390/ijms15022431
Chicago/Turabian StyleVandomme, Audrey, Aline Fréville, Katia Cailliau, Hadidjatou Kalamou, Jean-François Bodart, Jamal Khalife, and Christine Pierrot. 2014. "PhosphoTyrosyl Phosphatase Activator of Plasmodium falciparum: Identification of Its Residues Involved in Binding to and Activation of PP2A" International Journal of Molecular Sciences 15, no. 2: 2431-2453. https://doi.org/10.3390/ijms15022431




