Folding and Biogenesis of Mitochondrial Small Tim Proteins
Abstract
:1. Introduction
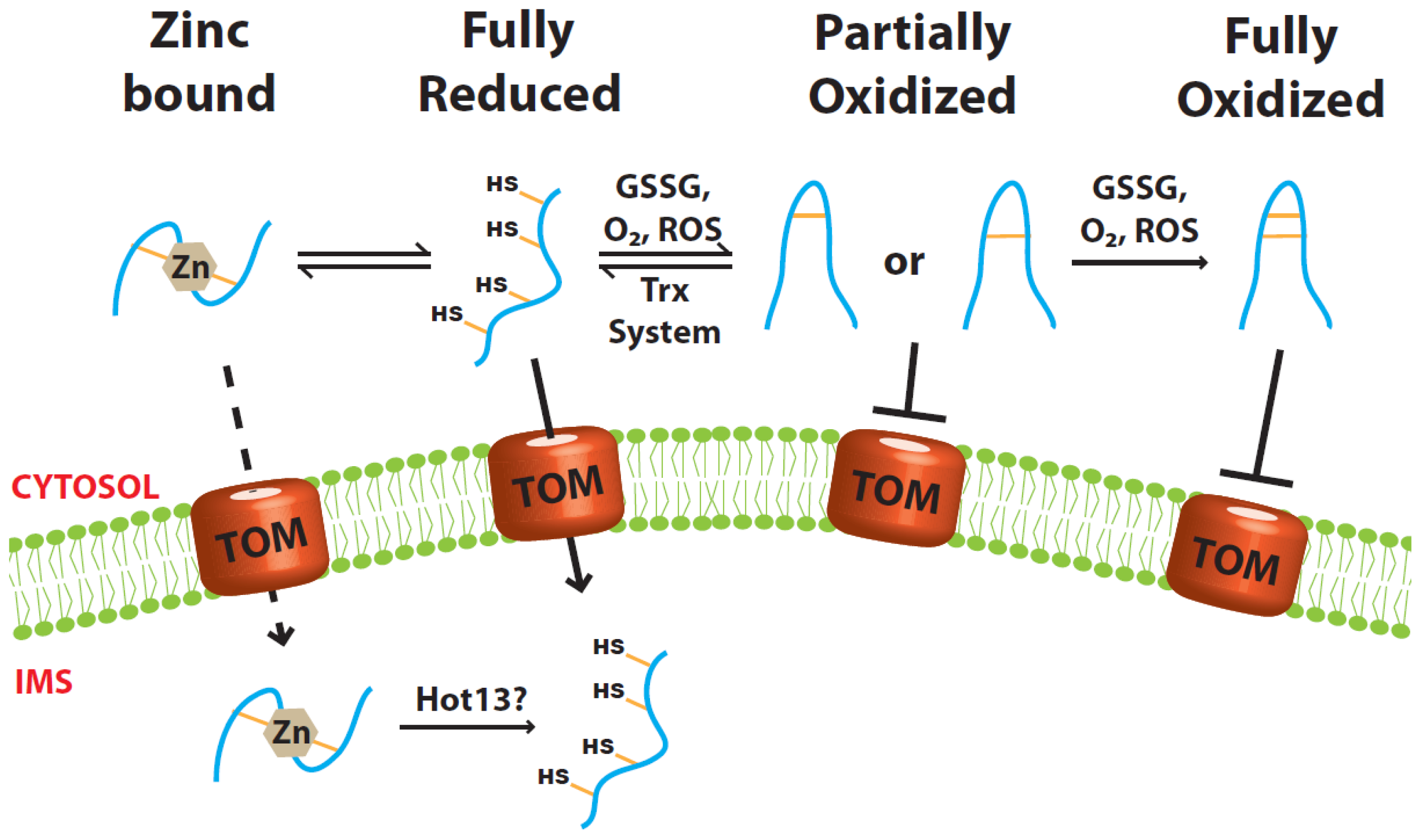
2. Keeping the Precursor Protein Reduced and Unfolded in the Cytosol
2.1. Role of Zinc Ions
2.2. Role of the Cytosolic Redoxin Systems
3. Import, Folding and Oxidation
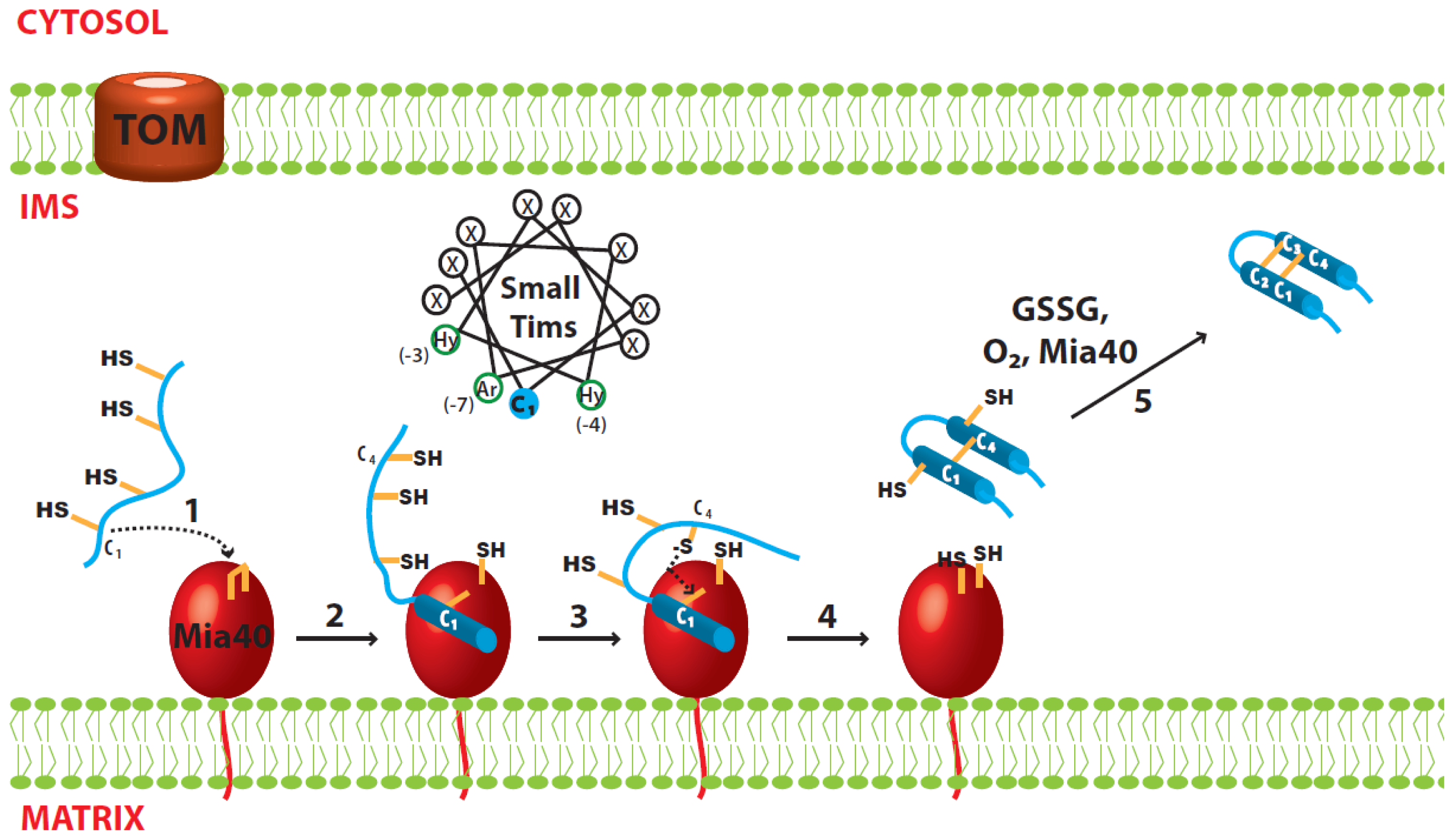
3.1. Disulfide Relay System of the IMS
3.2. Import of the Small Tim Proteins
3.3. Oxidative Folding of the Small Tim Proteins
3.4. Oxidized Tim9 and Tim10
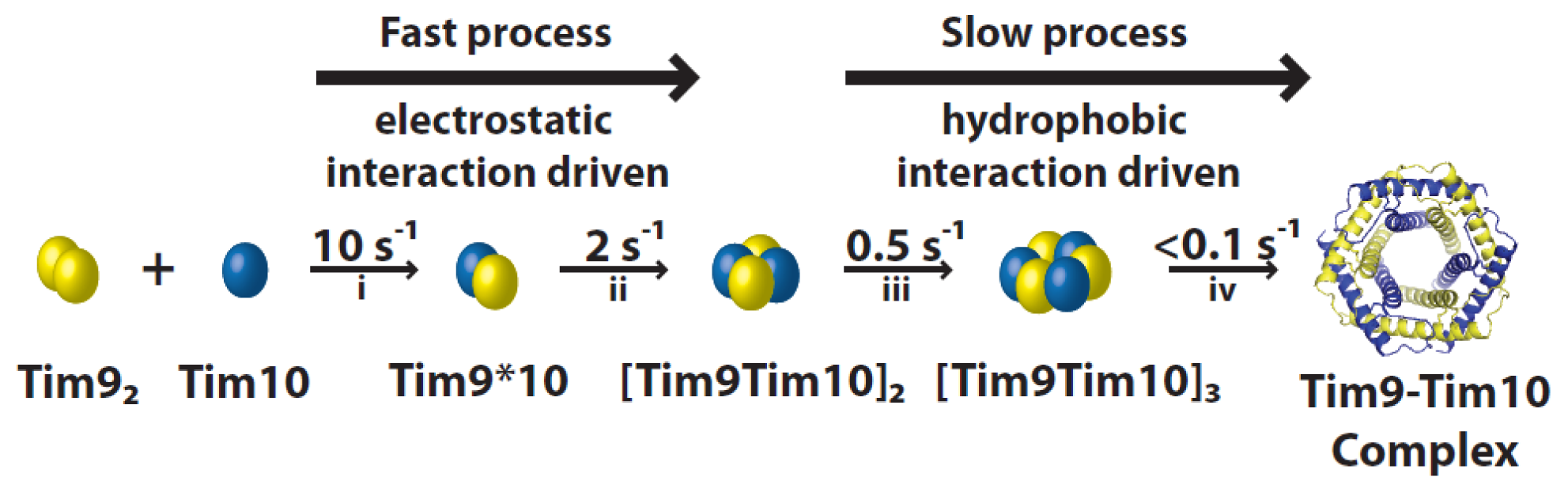
4. Complex Assembly
4.1. Assembly Process of the Tim9–Tim10 Complex
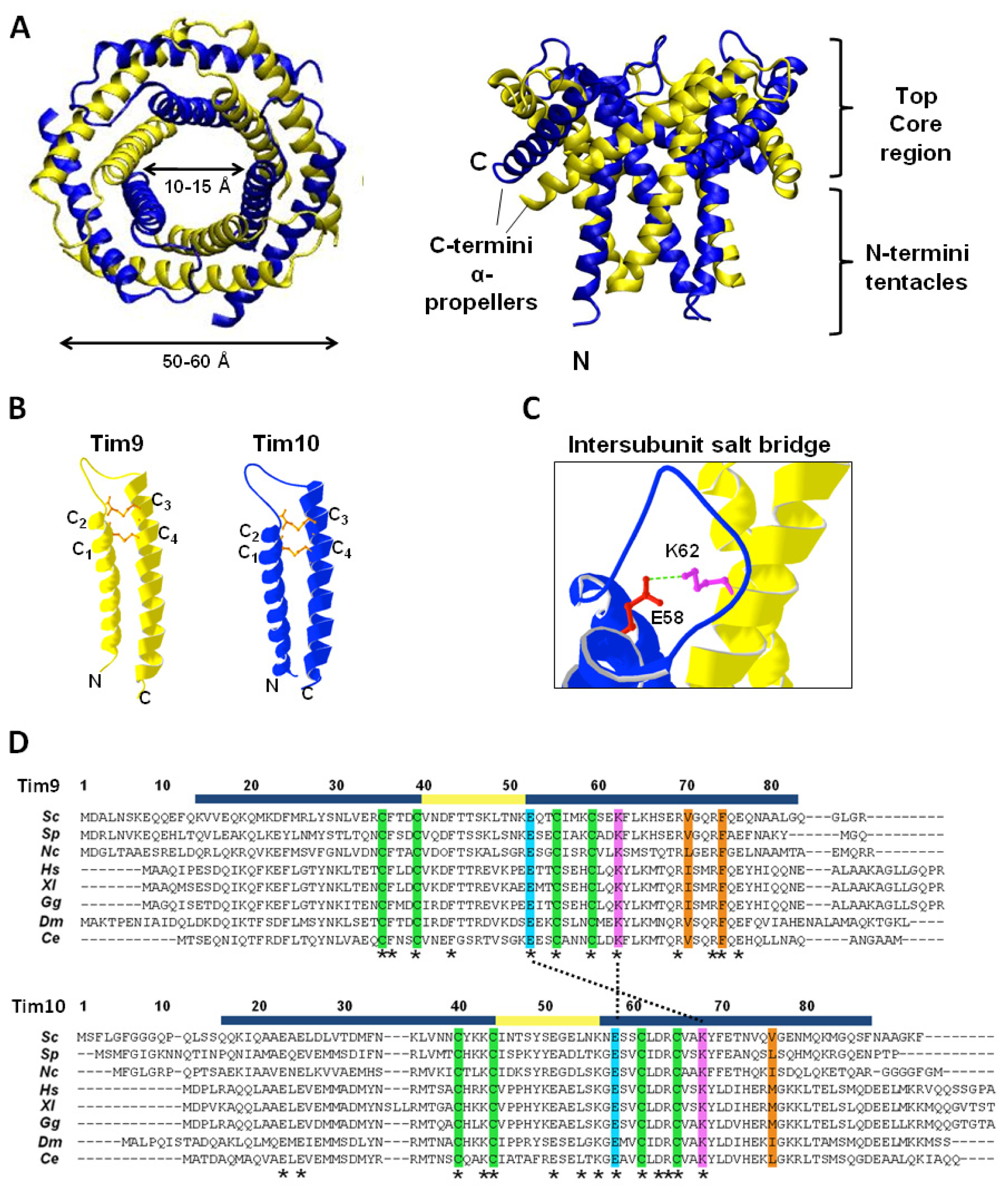
4.2. Structure of the Tim9–Tim10 Complex
4.3. Stability of the Tim9–Tim10 Complex
5. Functional Mechanism of the Small Tim Proteins
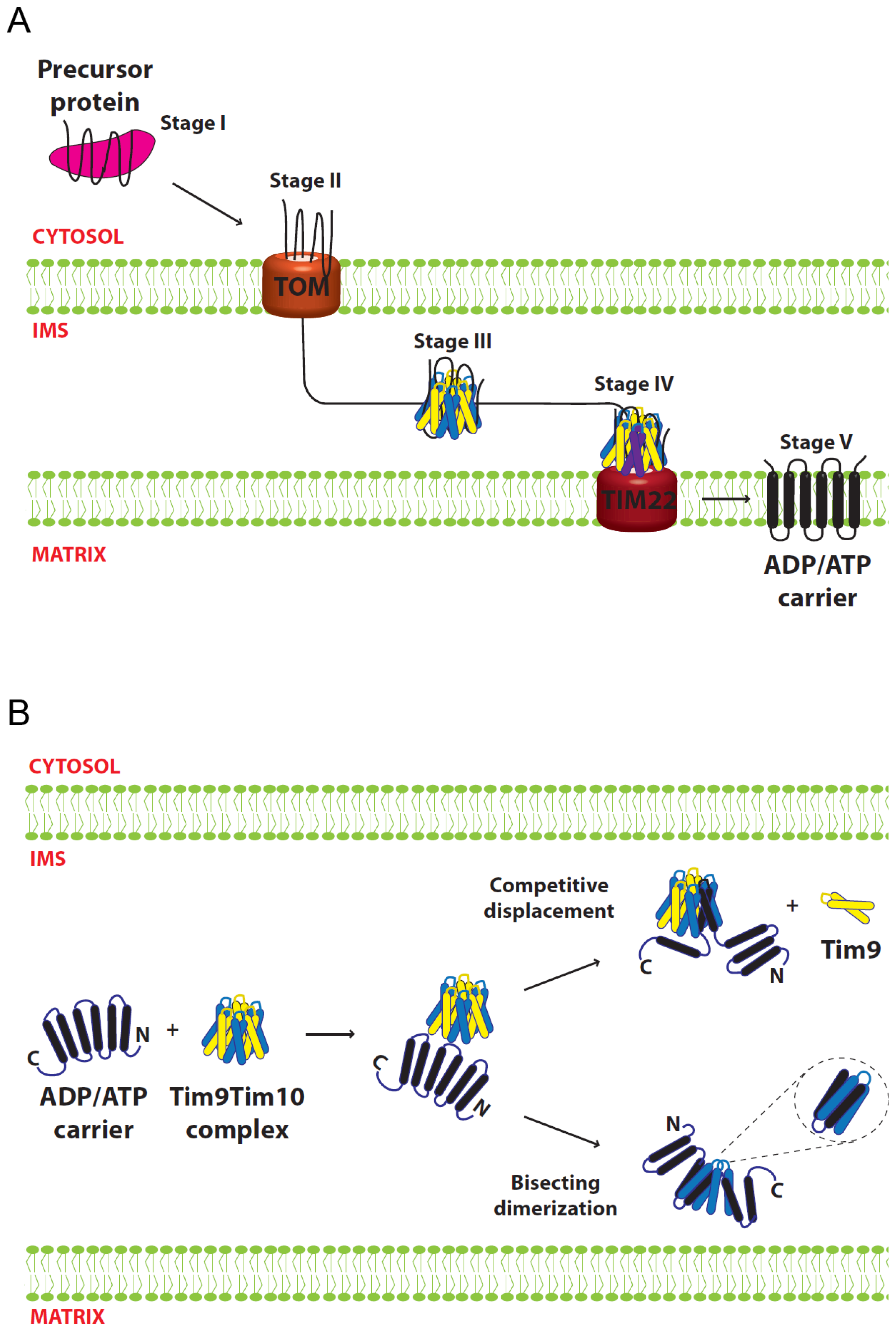
5.1. Import of AAC
5.2. Models for Tim9–Tim10 Chaperone Function
6. Conclusions
Acknowledgments
Conflicts of Interest
References
- Koehler, C.M. The small Tim proteins and the twin Cx3C motif. Trends Biochem. Sci 2004, 29, 1–4. [Google Scholar]
- Sirrenberg, C.; Endres, M.; Folsch, H.; Stuart, R.A.; Neupert, W.; Brunner, M. Carrier protein import into mitochondria mediated by the intermembrane proteins Tim10/Mrs11 and Tim12/Mrs5. Nature 1998, 391, 912–915. [Google Scholar]
- Koehler, C.M.; Merchant, S.; Oppliger, W.; Schmid, K.; Jarosch, E.; Dolfini, L.; Junne, T.; Schatz, G.; Tokatlidis, K. Tim9p, an essential partner subunit of Tim10p for the import of mitochondrial carrier proteins. EMBO J 1998, 17, 6477–6486. [Google Scholar]
- Adam, A.; Endres, M.; Sirrenberg, C.; Lottspeich, F.; Neupert, W.; Brunner, M. Tim9, a new component of the TIM22.54 translocase in mitochondria. EMBO J 1999, 18, 313–319. [Google Scholar]
- Gentle, I.E.; Perry, A.J.; Alcock, F.H.; Likic, V.A.; Dolezal, P.; Ng, E.T.; Purcell, A.W.; McConnville, M.; Naderer, T.; Chanez, A.L.; et al. Conserved motifs reveal details of ancestry and structure in the small TIM chaperones of the mitochondrial intermembrane space. Mol. Biol. Evol 2007, 24, 1149–1160. [Google Scholar]
- Wiedemann, N.; Pfanner, N.; Chacinska, A. Chaperoning through the mitochondrial intermembrane space. Mol. Cell 2006, 21, 145–148. [Google Scholar]
- Hofmann, S.; Rothbauer, U.; Muhlenbein, N.; Neupert, W.; Gerbitz, K.D.; Brunner, M.; Bauer, M.F. The C66W mutation in the deafness dystonia peptide 1 (DDP1) affects the formation of functional DDP1.TIM13 complexes in the mitochondrial intermembrane space. J. Biol. Chem 2002, 277, 23287–23293. [Google Scholar]
- Rothbauer, U.; Hofmann, S.; Muhlenbein, N.; Paschen, S.A.; Gerbitz, K.D.; Neupert, W.; Brunner, M.; Bauer, M.F. Role of the deafness dystonia peptide 1 (DDP1) in import of human Tim23 into the inner membrane of mitochondria. J. Biol. Chem 2001, 276, 37327–37334. [Google Scholar]
- Beverly, K.N.; Sawaya, M.R.; Schmid, E.; Koehler, C.M. The Tim8–Tim13 complex has multiple substrate binding sites and binds cooperatively to Tim23. J. Mol. Biol 2008, 382, 1144–1156. [Google Scholar]
- Webb, C.T.; Gorman, M.A.; Lazarou, M.; Ryan, M.T.; Gulbis, J.M. Crystal structure of the mitochondrial chaperone TIM9.10 reveals a six-bladed alpha-propeller. Mol. Cell 2006, 21, 123–133. [Google Scholar]
- Herrmann, J.M.; Riemer, J. Mitochondrial disulfide relay: Redox-regulated protein import into the intermembrane space. J. Biol. Chem 2012, 287, 4426–4433. [Google Scholar]
- Sideris, D.P.; Tokatlidis, K. Oxidative protein folding in the mitochondrial intermembrane space. Antioxid. Redox Signal 2010, 13, 1189–1204. [Google Scholar]
- Morgan, B.; Ang, S.K.; Yan, G.; Lu, H. Zinc can play chaperone-like and inhibitor roles during import of mitochondrial small Tim proteins. J. Biol. Chem 2009, 284, 6818–6825. [Google Scholar]
- Durigon, R.; Wang, Q.; Ceh Pavia, E.; Grant, C.M.; Lu, H. Cytosolic thioredoxin system facilitates the import of mitochondrial small Tim proteins. EMBO Rep 2012, 13, 916–922. [Google Scholar]
- Lutz, T.; Neupert, W.; Herrmann, J.M. Import of small Tim proteins into the mitochondrial intermembrane space. EMBO J 2003, 22, 4400–4408. [Google Scholar]
- Milenkovic, D.; Gabriel, K.; Guiard, B.; Schulze-Specking, A.; Pfanner, N.; Chacinska, A. Biogenesis of the essential Tim9–Tim10 chaperone complex of mitochondria: Site-specific recognition of cysteine residues by the intermembrane space receptor Mia40. J. Biol. Chem 2007, 282, 22472–22480. [Google Scholar]
- Sideris, D.P.; Tokatlidis, K. Oxidative folding of small Tims is mediated by site-specific docking onto Mia40 in the mitochondrial intermembrane space. Mol. Microbiol 2007, 65, 1360–1373. [Google Scholar]
- Banci, L.; Bertini, I.; Cefaro, C.; Cenacchi, L.; Ciofi-Baffoni, S.; Felli, I.C.; Gallo, A.; Gonnelli, L.; Luchinat, E.; Sideris, D.; Tokatlidis, K. Molecular chaperone function of Mia40 triggers consecutive induced folding steps of the substrate in mitochondrial protein import. Proc. Natl. Acad. Sci. USA 2010, 107, 20190–20195. [Google Scholar]
- Lu, H.; Allen, S.; Wardleworth, L.; Savory, P.; Tokatlidis, K. Functional TIM10 chaperone assembly is redox-regulated in vivo. J. Biol. Chem 2004, 279, 18952–18958. [Google Scholar]
- Ivanova, E.; Jowitt, T.A.; Lu, H. Assembly of the mitochondrial Tim9–Tim10 complex: A multi-step reaction with novel intermediates. J. Mol. Biol 2008, 375, 229–239. [Google Scholar]
- Lu, H.; Woodburn, J. Zinc binding stabilizes mitochondrial Tim10 in a reduced and import-competent state kinetically. J. Mol. Biol 2005, 353, 897–910. [Google Scholar]
- Morgan, B.; Lu, H. Oxidative folding competes with mitochondrial import of the small Tim proteins. Biochem. J 2008, 411, 115–122. [Google Scholar]
- Morgan, B.; Ezerina, D.; Amoako, T.N.; Riemer, J.; Seedorf, M.; Dick, T.P. Multiple glutathione disulfide removal pathways mediate cytosolic redox homeostasis. Nat. Chem. Biol 2013, 9, 119–125. [Google Scholar]
- Kojer, K.; Bien, M.; Gangel, H.; Morgan, B.; Dick, T.P.; Riemer, J. Glutathione redox potential in the mitochondrial intermembrane space is linked to the cytosol and impacts the Mia40 redox state. EMBO J 2012, 31, 3169–3182. [Google Scholar]
- Fischer, M.; Horn, S.; Belkacemi, A.; Kojer, K.; Petrungaro, C.; Habich, M.; Ali, M.; Kuttner, V.; Bien, M.; Kauff, F.; et al. Protein import and oxidative folding in the mitochondrial intermembrane space of intact mammalian cells. Mol. Biol. Cell 2013, 24, 2160–2170. [Google Scholar]
- Bragoszewski, P.; Gornicka, A.; Sztolsztener, M.E.; Chacinska, A. The ubiquitin-proteasome system regulates mitochondrial intermembrane space proteins. Mol. Cell Biol 2013, 33, 2136–2148. [Google Scholar]
- Deshaies, R.J.; Koch, B.D.; Werner-Washburne, M.; Craig, E.A.; Schekman, R. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature 1988, 332, 800–805. [Google Scholar]
- Murakami, H.; Pain, D.; Blobel, G. 70-kD heat shock-related protein is one of at least two distinct cytosolic factors stimulating protein import into mitochondria. J. Cell Biol 1988, 107, 2051–2057. [Google Scholar]
- Becker, J.; Walter, W.; Yan, W.; Craig, E.A. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol. Cell Biol 1996, 16, 4378–4386. [Google Scholar]
- Young, J.C.; Hoogenraad, N.J.; Hartl, F.U. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 2003, 112, 41–50. [Google Scholar]
- Lu, H.; Golovanov, A.P.; Alcock, F.; Grossmann, J.G.; Allen, S.; Lian, L.Y.; Tokatlidis, K. The structural basis of the TIM10 chaperone assembly. J. Biol. Chem 2004, 279, 18959–18966. [Google Scholar]
- Ivanova, E.; Ball, M.; Lu, H. Zinc binding of Tim10: Evidence for existence of an unstructured binding intermediate for a zinc finger protein. Proteins 2008, 71, 467–475. [Google Scholar]
- Frausto, J.J.R.; Williams, R.J.P. The Biological Chemistry of the Elements; Oxford University Press: Oxford, UK; p. 2001.
- Sensi, S.L.; Canzoniero, L.M.; Yu, S.P.; Ying, H.S.; Koh, J.Y.; Kerchner, G.A.; Choi, D.W. Measurement of intracellular free zinc in living cortical neurons: Routes of entry. J. Neurosci 1997, 17, 9554–9564. [Google Scholar]
- Sensi, S.L.; Ton-That, D.; Sullivan, P.G.; Jonas, E.A.; Gee, K.R.; Kaczmarek, L.K.; Weiss, J.H. Modulation of mitochondrial function by endogenous Zn2+ pools. Proc. Natl. Acad. Sci. USA 2003, 100, 6157–6162. [Google Scholar]
- Eide, D.J. Zinc transporters and the cellular trafficking of zinc. Biochim. Biophys. Acta 2006, 1763, 711–722. [Google Scholar]
- Haase, H.; Mocchegiani, E.; Rink, L. Correlation between zinc status and immune function in the elderly. Biogerontology 2006, 7, 421–428. [Google Scholar]
- Mesecke, N.; Bihlmaier, K.; Grumbt, B.; Longen, S.; Terziyska, N.; Hell, K.; Herrmann, J.M. The zinc-binding protein Hot13 promotes oxidation of the mitochondrial import receptor Mia40. EMBO Rep 2008, 9, 1107–1113. [Google Scholar]
- Curran, S.P.; Leuenberger, D.; Leverich, E.P.; Hwang, D.K.; Beverly, K.N.; Koehler, C.M. The role of Hot13p and redox chemistry in the mitochondrial TIM22 import pathway. J. Biol. Chem 2004, 279, 43744–43751. [Google Scholar]
- Berndt, C.; Lillig, C.H.; Holmgren, A. Thioredoxins and glutaredoxins as facilitators of protein folding. Biochim. Biophys. Acta 2008, 1783, 641–650. [Google Scholar]
- Banci, L.; Barbieri, L.; Luchinat, E.; Secci, E. Visualization of redox-controlled protein fold in living cells. Chem. Biol 2013, 20, 747–752. [Google Scholar]
- Spiller, M.P.; Ang, S.K.; Ceh-Pavia, E.; Fisher, K.; Wang, Q.; Rigby, S.E.; Lu, H. Identification and characterisation of mitochondrial Mia40 as an iron-sulphur protein. Biochem. J. 2013. [Google Scholar] [CrossRef]
- Chacinska, A.; Guiard, B.; Muller, J.M.; Schulze-Specking, A.; Gabriel, K.; Kutik, S.; Pfanner, N. Mitochondrial biogenesis, switching the sorting pathway of the intermembrane space receptor Mia40. J. Biol. Chem 2008, 283, 29723–29729. [Google Scholar]
- Chacinska, A.; Pfannschmidt, S.; Wiedemann, N.; Kozjak, V.; Sanjuan Szklarz, L.K.; Schulze-Specking, A.; Truscott, K.N.; Guiard, B.; Meisinger, C.; Pfanner, N. Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J 2004, 23, 3735–3746. [Google Scholar]
- Naoe, M.; Ohwa, Y.; Ishikawa, D.; Ohshima, C.; Nishikawa, S.; Yamamoto, H.; Endo, T. Identification of Tim40 that mediates protein sorting to the mitochondrial intermembrane space. J. Biol. Chem 2004, 279, 47815–47821. [Google Scholar]
- Hofmann, S.; Rothbauer, U.; Muhlenbein, N.; Baiker, K.; Hell, K.; Bauer, M.F. Functional and mutational characterization of human MIA40 acting during import into the mitochondrial intermembrane space. J. Mol. Biol 2005, 353, 517–528. [Google Scholar]
- Porras, P.; Padilla, C.A.; Krayl, M.; Voos, W.; Barcena, J.A. One single in-frame AUG codon is responsible for a diversity of subcellular localizations of glutaredoxin 2 in Saccharomyces cerevisiae. J. Biol. Chem 2006, 281, 16551–16562. [Google Scholar]
- Koehler, C.M.; Tienson, H.L. Redox regulation of protein folding in the mitochondrial intermembrane space. Biochim. Biophys. Acta 2009, 1793, 139–145. [Google Scholar]
- Endo, T.; Yamano, K.; Kawano, S. Structural basis for the disulfide relay system in the mitochondrial intermembrane space. Antioxid. Redox Signal 2010, 13, 1359–1373. [Google Scholar]
- Mesecke, N.; Terziyska, N.; Kozany, C.; Baumann, F.; Neupert, W.; Hell, K.; Herrmann, J.M. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell 2005, 121, 1059–1069. [Google Scholar]
- Milenkovic, D.; Ramming, T.; Muller, J.M.; Wenz, L.S.; Gebert, N.; Schulze-Specking, A.; Stojanovski, D.; Rospert, S.; Chacinska, A. Identification of the signal directing Tim9 and Tim10 into the intermembrane space of mitochondria. Mol. Biol. Cell 2009, 20, 2530–2539. [Google Scholar]
- Sideris, D.P.; Petrakis, N.; Katrakili, N.; Mikropoulou, D.; Gallo, A.; Ciofi-Baffoni, S.; Banci, L.; Bertini, I.; Tokatlidis, K. A novel intermembrane space-targeting signal docks cysteines onto Mia40 during mitochondrial oxidative folding. J. Cell Biol 2009, 187, 1007–1022. [Google Scholar]
- Grumbt, B.; Stroobant, V.; Terziyska, N.; Israel, L.; Hell, K. Functional characterization of Mia40p, the central component of the disulfide relay system of the mitochondrial intermembrane space. J. Biol. Chem 2007, 282, 37461–37470. [Google Scholar]
- Bien, M.; Longen, S.; Wagener, N.; Chwalla, I.; Herrmann, J.M.; Riemer, J. Mitochondrial disulfide bond formation is driven by intersubunit electron transfer in Erv1 and proofread by glutathione. Mol. Cell 2010, 37, 516–528. [Google Scholar]
- Banci, L.; Bertini, I.; Cefaro, C.; Ciofi-Baffoni, S.; Gallo, A.; Martinelli, M.; Sideris, D.P.; Katrakili, N.; Tokatlidis, K. MIA40 is an oxidoreductase that catalyzes oxidative protein folding in mitochondria. Nat. Struct. Mol. Biol 2009, 16, 198–206. [Google Scholar]
- Stojanovski, D.; Milenkovic, D.; Muller, J.M.; Gabriel, K.; Schulze-Specking, A.; Baker, M.J.; Ryan, M.T.; Guiard, B.; Pfanner, N.; Chacinska, A. Mitochondrial protein import: Precursor oxidation in a ternary complex with disulfide carrier and sulfhydryl oxidase. J. Cell Biol 2008, 183, 195–202. [Google Scholar]
- Allen, S.; Lu, H.; Thornton, D.; Tokatlidis, K. Juxtaposition of the two distal CX3C motifs via intrachain disulfide bonding is essential for the folding of Tim10. J. Biol. Chem 2003, 278, 38505–38513. [Google Scholar]
- Vial, S.; Lu, H.; Allen, S.; Savory, P.; Thornton, D.; Sheehan, J.; Tokatlidis, K. Assembly of Tim9 and Tim10 into a functional chaperone. J. Biol. Chem 2002, 277, 36100–36108. [Google Scholar]
- Baker, M.J.; Webb, C.T.; Stroud, D.A.; Palmer, C.S.; Frazier, A.E.; Guiard, B.; Chacinska, A.; Gulbis, J.M.; Ryan, M.T. Structural and functional requirements for activity of the Tim9–Tim10 complex in mitochondrial protein import. Mol. Biol. Cell 2009, 20, 769–779. [Google Scholar]
- Ivanova, E.; Lu, H. Allosteric and electrostatic protein-protein interactions regulate the assembly of the heterohexameric Tim9–Tim10 complex. J. Mol. Biol 2008, 379, 609–616. [Google Scholar]
- Walton, T.A.; Sousa, M.C. Crystal structure of Skp, a prefoldin-like chaperone that protects soluble and membrane proteins from aggregation. Mol. Cell 2004, 15, 367–374. [Google Scholar]
- Siegert, R.; Leroux, M.R.; Scheufler, C.; Hartl, F.U.; Moarefi, I. Structure of the molecular chaperone prefoldin: Unique interaction of multiple coiled coil tentacles with unfolded proteins. Cell 2000, 103, 621–632. [Google Scholar]
- Korndorfer, I.P.; Dommel, M.K.; Skerra, A. Structure of the periplasmic chaperone Skp suggests functional similarity with cytosolic chaperones despite differing architecture. Nat. Struct. Mol. Biol 2004, 11, 1015–1020. [Google Scholar]
- Ivanova, E.; Pang, J.; Jowitt, T.A.; Yan, G.; Warwicker, J.; Sutcliffe, M.J.; Lu, H. Temperature-dependent study reveals that dynamics of hydrophobic residues plays an important functional role in the mitochondrial Tim9–Tim10 complex. Proteins 2011. [Google Scholar] [CrossRef]
- Vergnolle, M.A.; Baud, C.; Golovanov, A.P.; Alcock, F.; Luciano, P.; Lian, L.Y.; Tokatlidis, K. Distinct domains of small Tims involved in subunit interaction and substrate recognition. J. Mol. Biol 2005, 351, 839–849. [Google Scholar]
- Roesch, K.; Curran, S.P.; Tranebjaerg, L.; Koehler, C.M. Human deafness dystonia syndrome is caused by a defect in assembly of the DDP1/TIMM8a-TIMM13 complex. Hum. Mol. Genet 2002, 11, 477–486. [Google Scholar]
- Baker, M.J.; Mooga, V.P.; Guiard, B.; Langer, T.; Ryan, M.T.; Stojanovski, D. Impaired folding of the mitochondrial small TIM chaperones induces clearance by the i-AAA protease. J. Mol. Biol 2012, 424, 227–239. [Google Scholar]
- Vergnolle, M.A.; Alcock, F.H.; Petrakis, N.; Tokatlidis, K. Mutation of conserved charged residues in mitochondrial TIM10 subunits precludes TIM10 complex assembly, but does not abolish growth of yeast cells. J. Mol. Biol 2007, 371, 1315–1324. [Google Scholar]
- Leuenberger, D.; Curran, S.P.; Wong, D.; Koehler, C.M. The role of Tim9p in the assembly of the TIM22 import complexes. Traffic 2003, 4, 144–152. [Google Scholar]
- Jarosch, E.; Tuller, G.; Daum, G.; Waldherr, M.; Voskova, A.; Schweyen, R.J. Mrs5p, an essential protein of the mitochondrial intermembrane space, affects protein import into yeast mitochondria. J. Biol. Chem 1996, 271, 17219–17225. [Google Scholar]
- Gebert, N.; Chacinska, A.; Wagner, K.; Guiard, B.; Koehler, C.M.; Rehling, P.; Pfanner, N.; Wiedemann, N. Assembly of the three small Tim proteins precedes docking to the mitochondrial carrier translocase. EMBO Rep 2008, 9, 548–554. [Google Scholar]
- Truscott, K.N.; Wiedemann, N.; Rehling, P.; Muller, H.; Meisinger, C.; Pfanner, N.; Guiard, B. Mitochondrial import of the ADP/ATP carrier: The essential TIM complex of the intermembrane space is required for precursor release from the TOM complex. Mol. Cell Biol 2002, 22, 7780–7789. [Google Scholar]
- Leuenberger, D.; Bally, N.A.; Schatz, G.; Koehler, C.M. Different import pathways through the mitochondrial intermembrane space for inner membrane proteins. EMBO J 1999, 18, 4816–4822. [Google Scholar]
- Vergnolle, M.A.; Sawney, H.; Junne, T.; Dolfini, L.; Tokatlidis, K. A cryptic matrix targeting signal of the yeast ADP/ATP carrier normally inserted by the TIM22 complex is recognized by the TIM23 machinery. Biochem. J 2005, 385, 173–180. [Google Scholar]
- Endres, M.; Neupert, W.; Brunner, M. Transport of the ADP/ATP carrier of mitochondria from the TOM complex to the TIM22.54 complex. EMBO J 1999, 18, 3214–3221. [Google Scholar]
- Pfanner, N.; Hoeben, P.; Tropschug, M.; Neupert, W. The carboxyl-terminal two-thirds of the ADP/ATP carrier polypeptide contains sufficient information to direct translocation into mitochondria. J. Biol. Chem 1987, 262, 14851–14854. [Google Scholar]
- Ryan, M.T.; Muller, H.; Pfanner, N. Functional staging of ADP/ATP carrier translocation across the outer mitochondrial membrane. J. Biol. Chem 1999, 274, 20619–20627. [Google Scholar]
- Rehling, P.; Model, K.; Brandner, K.; Kovermann, P.; Sickmann, A.; Meyer, H.E.; Kuhlbrandt, W.; Wagner, R.; Truscott, K.N.; Pfanner, N. Protein insertion into the mitochondrial inner membrane by a twin-pore translocase. Science 2003, 299, 1747–1751. [Google Scholar]
- Curran, S.P.; Leuenberger, D.; Oppliger, W.; Koehler, C.M. The Tim9p-Tim10p complex binds to the transmembrane domains of the ADP/ATP carrier. EMBO J 2002, 21, 942–953. [Google Scholar]
- Vasiljev, A.; Ahting, U.; Nargang, F.E.; Go, N.E.; Habib, S.J.; Kozany, C.; Panneels, V.; Sinning, I.; Prokisch, H.; Neupert, W.; et al. Reconstituted TOM core complex and Tim9/Tim10 complex of mitochondria are sufficient for translocation of the ADP/ATP carrier across membranes. Mol. Biol. Cell 2004, 15, 1445–1458. [Google Scholar]
- Wiedemann, N.; Pfanner, N.; Ryan, M.T. The three modules of ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. EMBO J 2001, 20, 951–960. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ceh-Pavia, E.; Spiller, M.P.; Lu, H. Folding and Biogenesis of Mitochondrial Small Tim Proteins. Int. J. Mol. Sci. 2013, 14, 16685-16705. https://doi.org/10.3390/ijms140816685
Ceh-Pavia E, Spiller MP, Lu H. Folding and Biogenesis of Mitochondrial Small Tim Proteins. International Journal of Molecular Sciences. 2013; 14(8):16685-16705. https://doi.org/10.3390/ijms140816685
Chicago/Turabian StyleCeh-Pavia, Efrain, Michael P. Spiller, and Hui Lu. 2013. "Folding and Biogenesis of Mitochondrial Small Tim Proteins" International Journal of Molecular Sciences 14, no. 8: 16685-16705. https://doi.org/10.3390/ijms140816685



