3D Structure Elucidation of Thermostable L2 Lipase from Thermophilic Bacillus sp. L2
Abstract
:1. Introduction
2. Results and Discussion
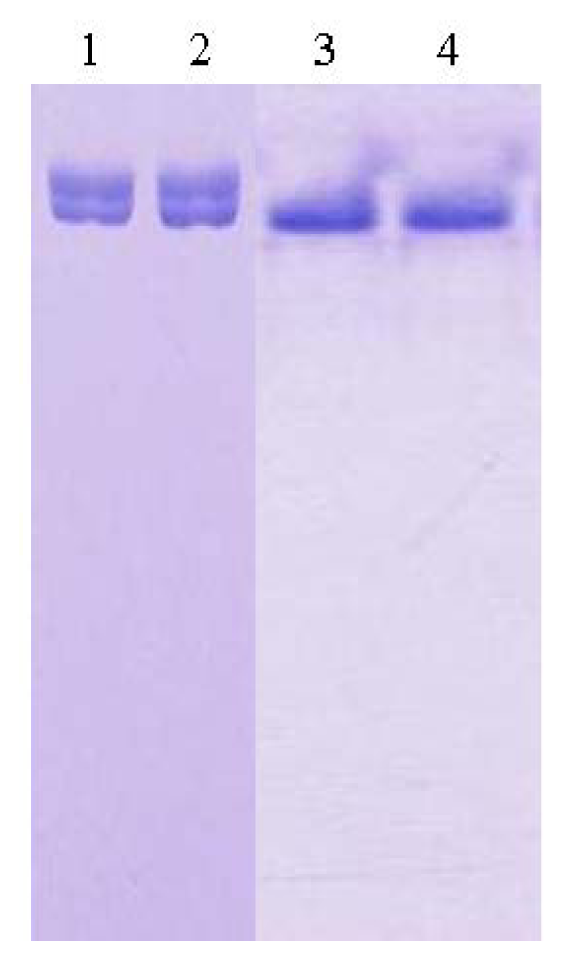
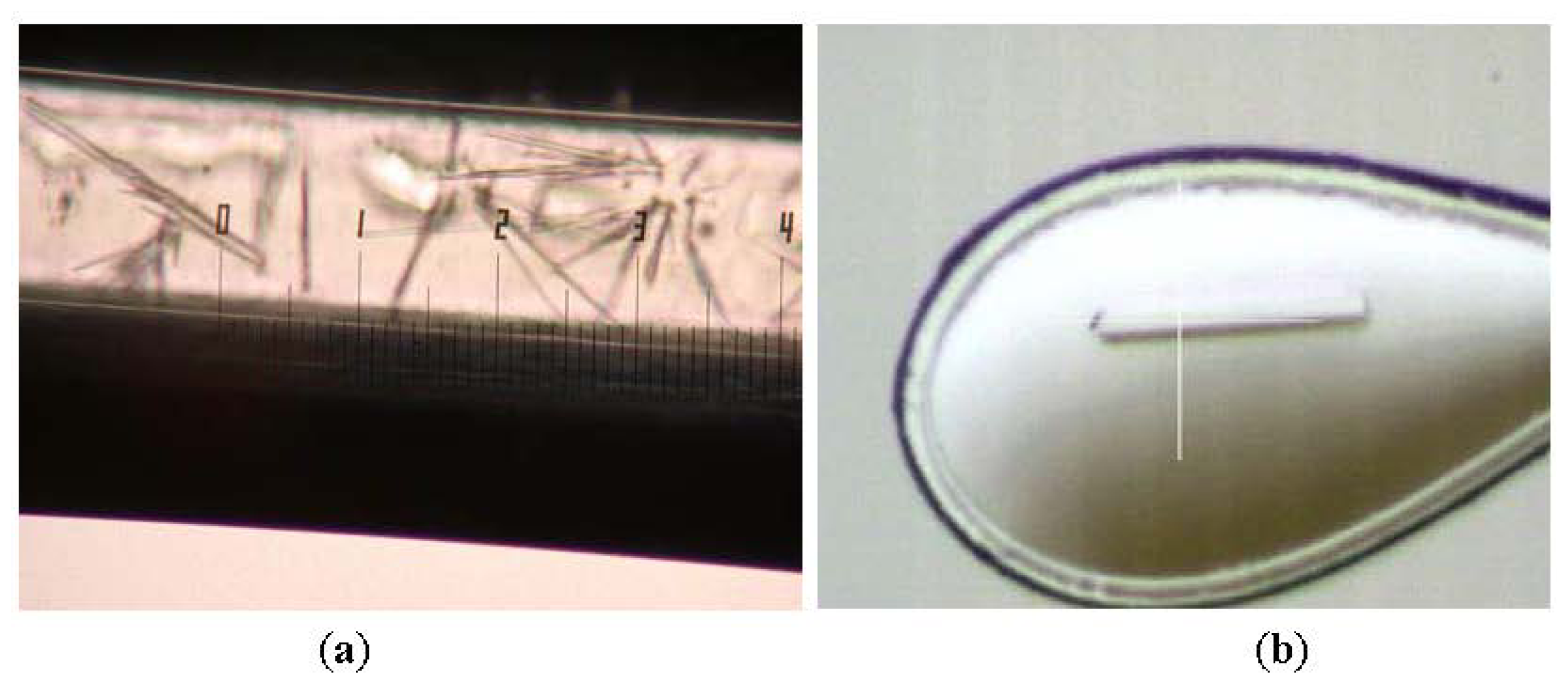
2.1. Purification and Crystallization
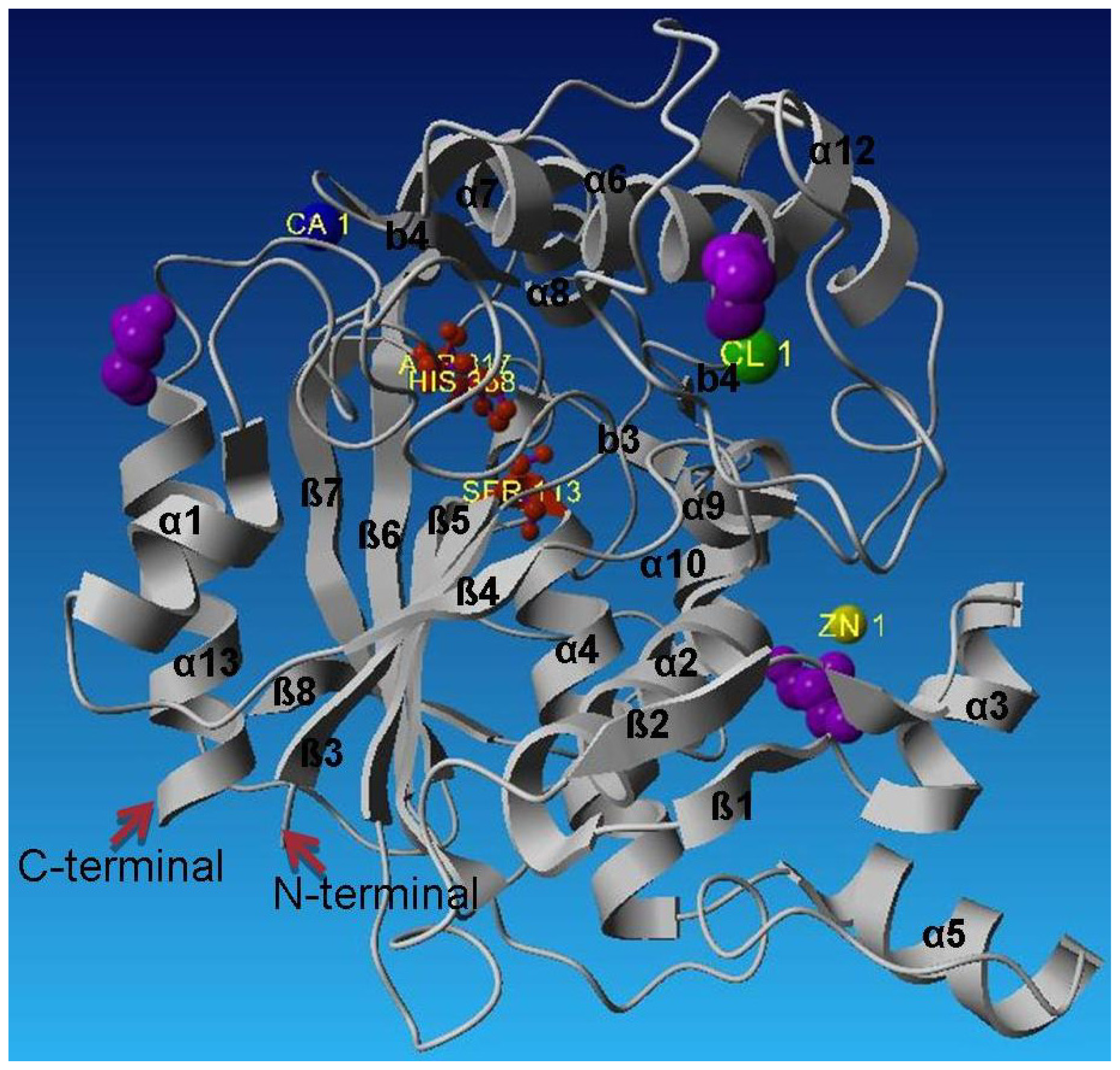
2.2. Description of Crystal Structure of L2 Lipase
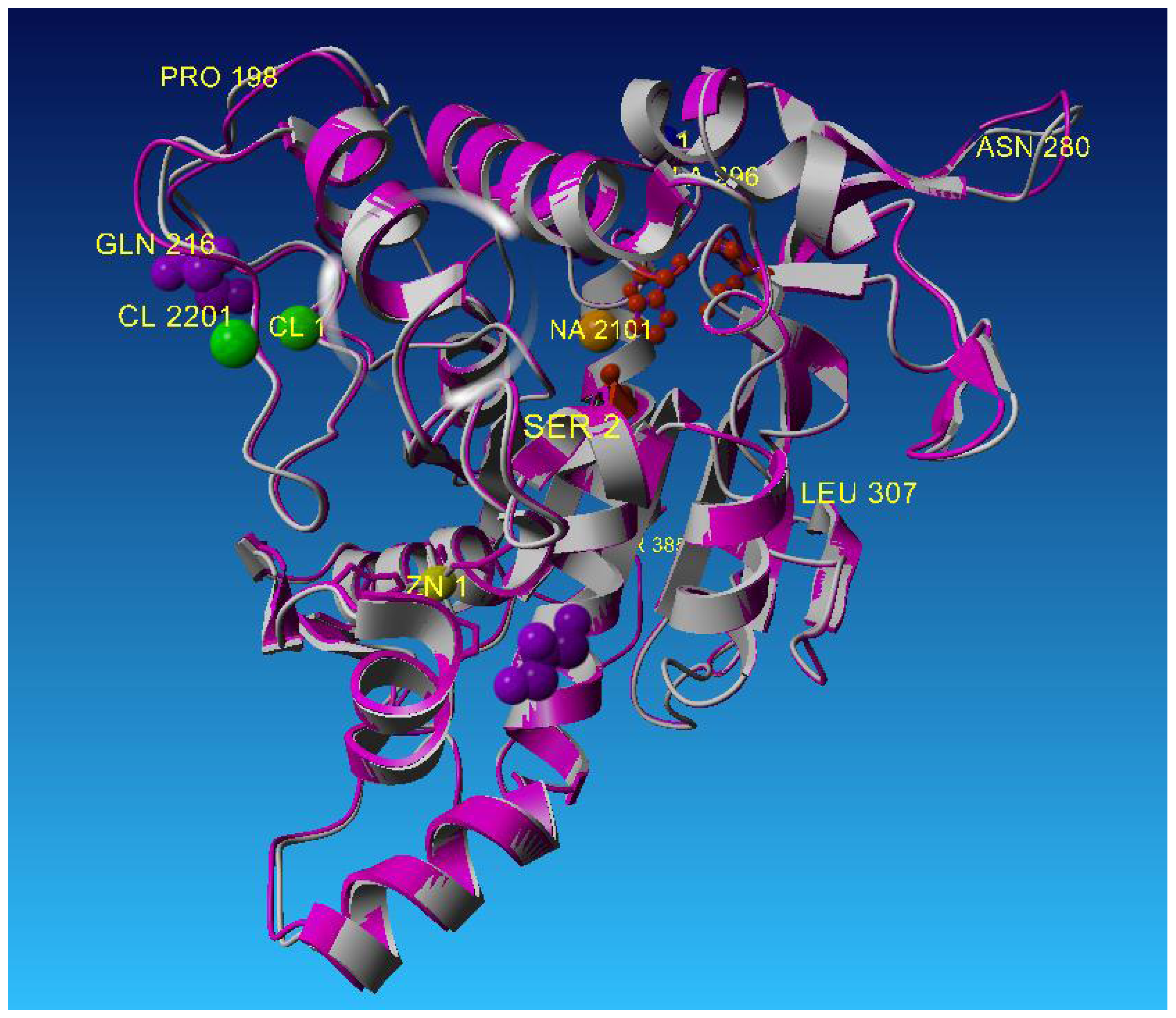
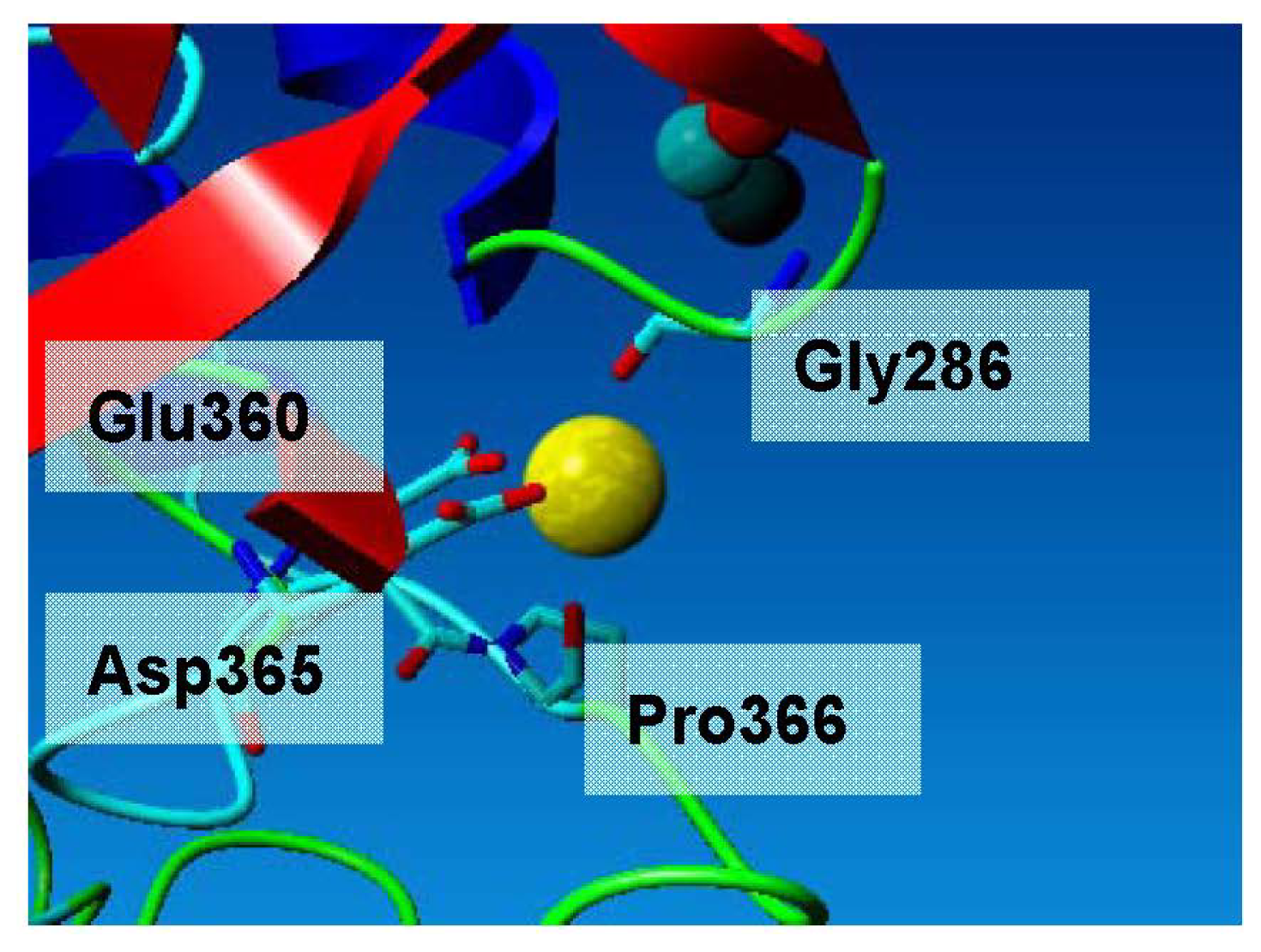
2.3. Metal-Binding Sites
3. Experimental Section
3.1. Purification of L2 Lipase
3.2. Crystallization and Data Collection
3.3. Structure Solution and Crystallographic Refinement
3.3. Protein Data Bank ID Code
4. Conclusions
Acknowledgments
References
- Triechel, H.; de Oliveira, D.; Mazutti, A.M.; Luccio, M.D.; Oliveira, J.V. A review on microbial lipases production. Food Bioproc. Technol 2010, 3, 182–196. [Google Scholar]
- Bornscheuer, U.T.; Bessler, C.; Srinivas, R.; Krishna, S.H. Optimizing lipases and related enzymes for efficient application. Trends Biotechnol 2002, 20, 433–437. [Google Scholar]
- Menoncin, S.; Domingues, N.M.; Freire, D.M.G.; Toniazzo, G.; Cansian, R.L.; Oliveira, J.V. Study of the extraction, concentration, and partial characterization of lipases obtained from Penicillium verrucosum using solid-state fermentation of soybean bran. Food Bioproc. Technol 2008, 3, 537–544. [Google Scholar]
- Sharma, R.; Soni, S.; Vohra, R.; Gupta, L.; Gupta, J. Purification and characterization of a thermostable alkaline lipase from a new thermophilic Bacillus sp. RSJ-1. Proc. Biochem 2002, 37, 1075–1084. [Google Scholar]
- Jaeger, K.E.; Ransac, S.; Dijkstra, B.W.; Clson, C.; Heuvel, M.V.; Misset, O. Bacterial lipases. FEMS Microbiol. Rev 1994, 15, 29–63. [Google Scholar]
- Ollis, D.L.; Cheah, E.; Cygler, M.; Dijkstra, B.; Frolow, F.; Ken, S.M. The α/β hydrolase fold. Protein Eng 1992, 5, 197–211. [Google Scholar]
- Arpigny, J.L.; Jaeger, K.-E. Bacterial lipolytic enzymes: Classification and properties. Biochem. J 1999, 343, 177–183. [Google Scholar]
- Shariff, F.M.; Chor, A.L.T.; Mukred, A.D.; Salleh, A.B.; Basri, M.; Rahman, R.N.Z.R.A. Production of L2 lipase by Bacillus sp. strain L2: nutritional and physical factors. J. Basic Microbiol 2007, 47, 406–412. [Google Scholar]
- Shariff, F.M.; Rahman, R.N.Z.R.A.; Basri, M.; Salleh, A.B. A newly isolated thermostable lipase from Bacillus sp. Int. J. Mol. Sci 2011, 12, 2917–2934. [Google Scholar]
- Shariff, F.M.; Rahman, R.N.Z.R.A.; Ali, M.S.M.; Chor, A.L.T.; Basri, M.; Salleh, A.B. Crystallization and preliminary X-ray crystallographic analysis of a highly thermostable L2 lipase from a newly isolated Bacillus sp. L2. Acta Crystallogr. F 2010, 66, 715–717. [Google Scholar]
- Dodson, G.; Wlodawer, A. Catalytic triads and their relatives. Trends Biochem. Sci 1998, 23, 347–352. [Google Scholar]
- Joel, D.A.; Tyndall, J.D.A.; Sinchaikul, S.; Fothergill-Gilmorel, L.A.; Tayor, P.; Walkinshaw, M.D. Crystal structure of a thermostable lipase from Bacillus stearothermophilus P1. J. Mol. Biol 2002, 323, 859–869. [Google Scholar]
- Matsumura, H.; Yamamoto, T.; Chor, A.L.T.; Mori, T.; Salleh, A.B.; Basri, M.; Inoue, T.; Kai, Y.; Rahman, R.N.Z.R.A. Novel cation-pi interaction revealed by crystal structure of thermoalkalophilic lipase. Proteins 2007, 70, 592–598. [Google Scholar]
- Lewit-Bentley, A.; Réty, S. EF-hand calcium-binding proteins. Curr. Opin. Struct 2000, 10, 637–643. [Google Scholar]
- Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr 1994, D50, 760–763.
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of coot. Acta Crystallogr. Biol. Crystallogr 2010, 66, 486–501. [Google Scholar]
- Murshudov, A.A.; Vagin, A.A.; Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 1997, 53, 240–255. [Google Scholar]
- Jeong, S.-T.; Kim, H.-K.; Kim, S.-J.; Pan, J.-G.; Oh, T.K.; Ryu, S.E. Crystallization and preliminary X-ray crystallographic analysis of a thermoalkalophilic lipase from Bacillus stearothermophilus L1. Acta Crystallogr. D 2001, 57, 1300–1302. [Google Scholar]

| Crystal Data | |
|---|---|
| Space group | P212121 |
| Resolution range Unit cell parameter (Å) | 40.00–1.49 |
| a | 71.98 |
| b | 81.78 |
| c | 83.37 |
| Mosaicity range (°) | 0.14–0.29 |
| Data Processing | |
| Temperature (K) | 100 |
| Wavelength (Å) | 0.80 |
| Resolution (Å) | 40.00–1.49 |
| Total number of reflections | 827852 |
| Oscillation angle per frame (°) | 0.3 |
| Unique data | 81336 |
| Redundancy (%) | 4.05 (4.23) |
| Data completeness (%) | 99.50 (97.90) |
| Mean I/Sigma | 20.50 (2.87) |
| Molecules per ASU | 1 |
| Matthews coefficient, Å3 Da−1 | 2.85 |
| Solvent content (%) | 56.89 |
| Total linear Rmerge | 0.098 |
| Refinement Statistics | L2 Shape 2 |
|---|---|
| Molecular per ASU | 1 |
| Resolution Range (Å) | 58.4–1.5 |
| Protein non-hydrogen atoms | 3037 |
| Ligand non-hydrogen atoms | - |
| Glycerol | 3 |
| Water molecules | 497 |
| Metal ions | 2 |
| Rfactor (%) | 0.167 |
| Rfree (%) | 0.172 |
| R.m.s.d. bond length (Å) | 0.009 |
| R.m.s.d. bond angle (°) | 1.20 |
| Average B-factor | 15.01 |

© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Raja Abd. Rahman, R.N.Z.; Mohd Shariff, F.; Basri, M.; Salleh, A.B. 3D Structure Elucidation of Thermostable L2 Lipase from Thermophilic Bacillus sp. L2. Int. J. Mol. Sci. 2012, 13, 9207-9217. https://doi.org/10.3390/ijms13079207
Raja Abd. Rahman RNZ, Mohd Shariff F, Basri M, Salleh AB. 3D Structure Elucidation of Thermostable L2 Lipase from Thermophilic Bacillus sp. L2. International Journal of Molecular Sciences. 2012; 13(7):9207-9217. https://doi.org/10.3390/ijms13079207
Chicago/Turabian StyleRaja Abd. Rahman, Raja Noor Zaliha, Fairolniza Mohd Shariff, Mahiran Basri, and Abu Bakar Salleh. 2012. "3D Structure Elucidation of Thermostable L2 Lipase from Thermophilic Bacillus sp. L2" International Journal of Molecular Sciences 13, no. 7: 9207-9217. https://doi.org/10.3390/ijms13079207
APA StyleRaja Abd. Rahman, R. N. Z., Mohd Shariff, F., Basri, M., & Salleh, A. B. (2012). 3D Structure Elucidation of Thermostable L2 Lipase from Thermophilic Bacillus sp. L2. International Journal of Molecular Sciences, 13(7), 9207-9217. https://doi.org/10.3390/ijms13079207




