Combinatorial Signal Integration by APETALA2/Ethylene Response Factor (ERF)-Transcription Factors and the Involvement of AP2-2 in Starvation Response
Abstract
:1. Introduction
2. Results
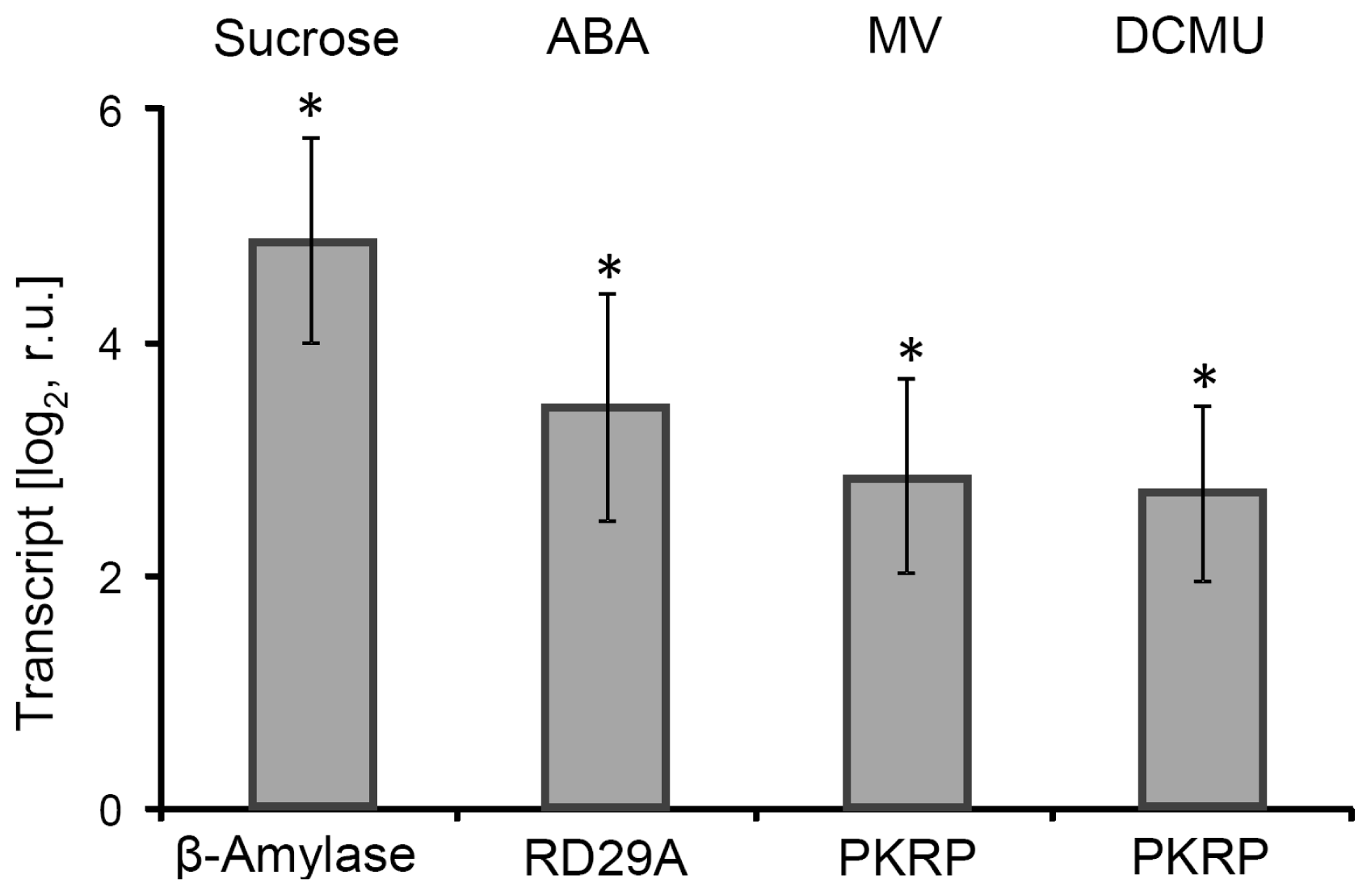
2.1. Effector Treatments with Relevance for Retrograde Signaling
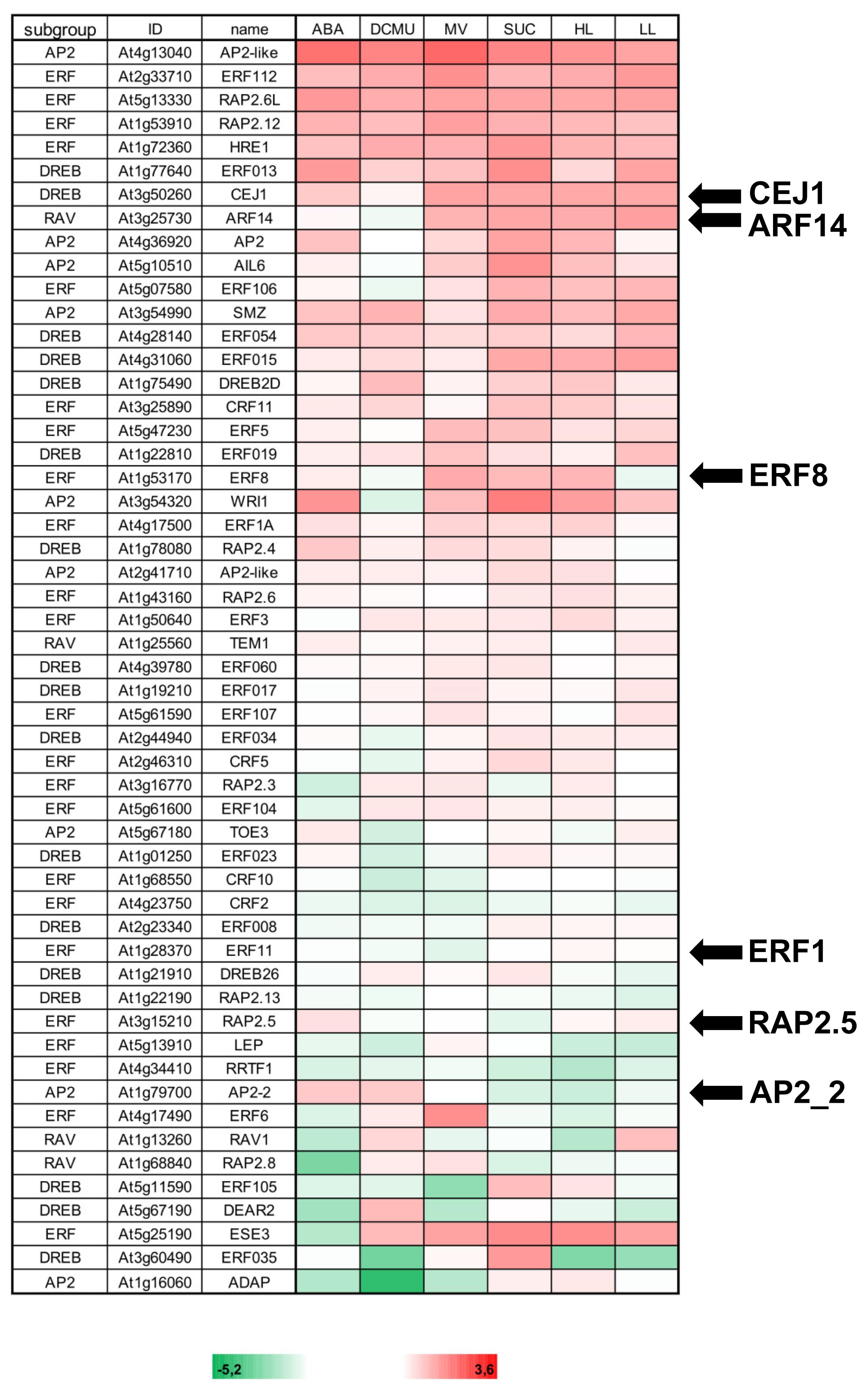
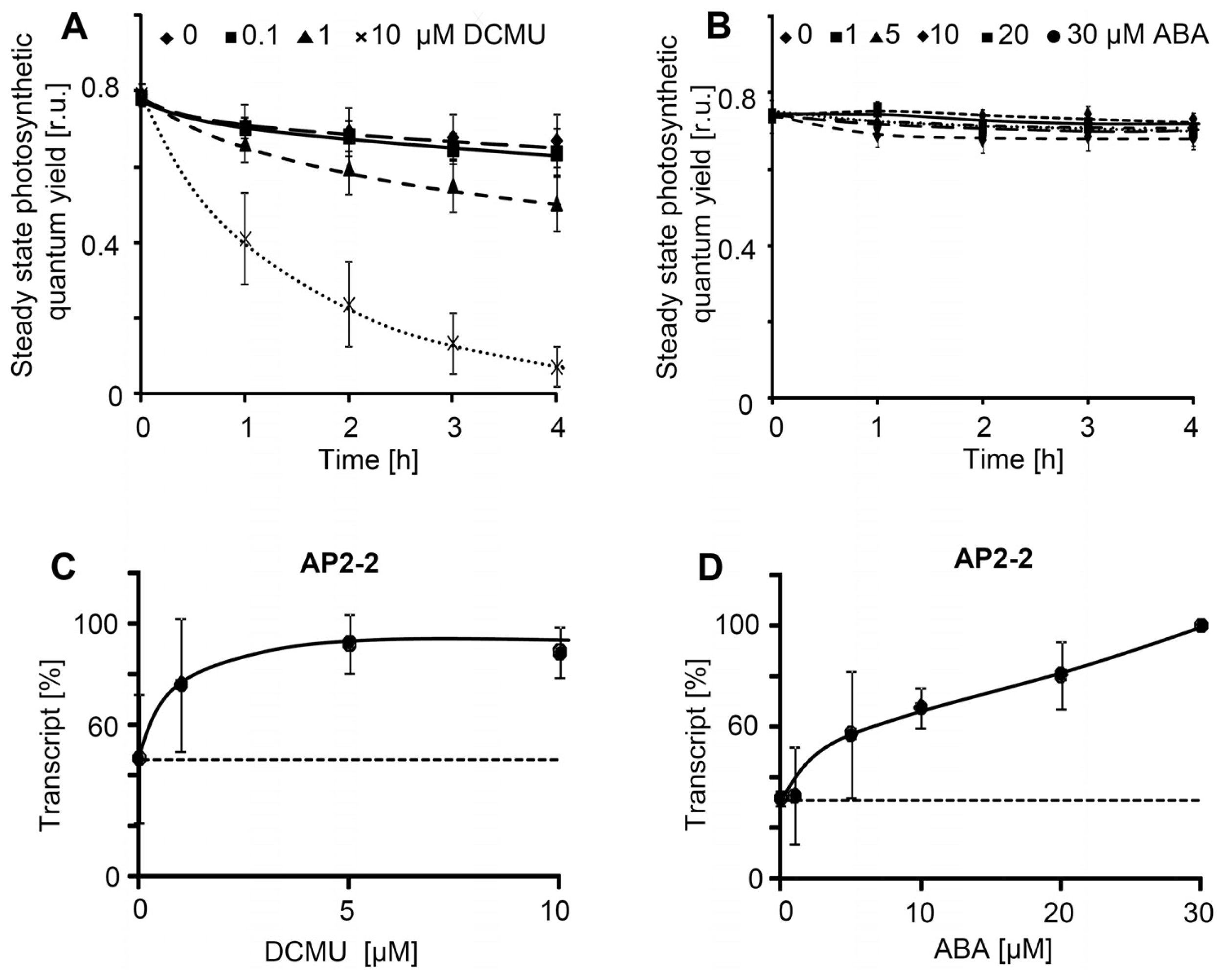
2.2. Effector Concentration-Dependent Expression of AP2/ERF-TFs
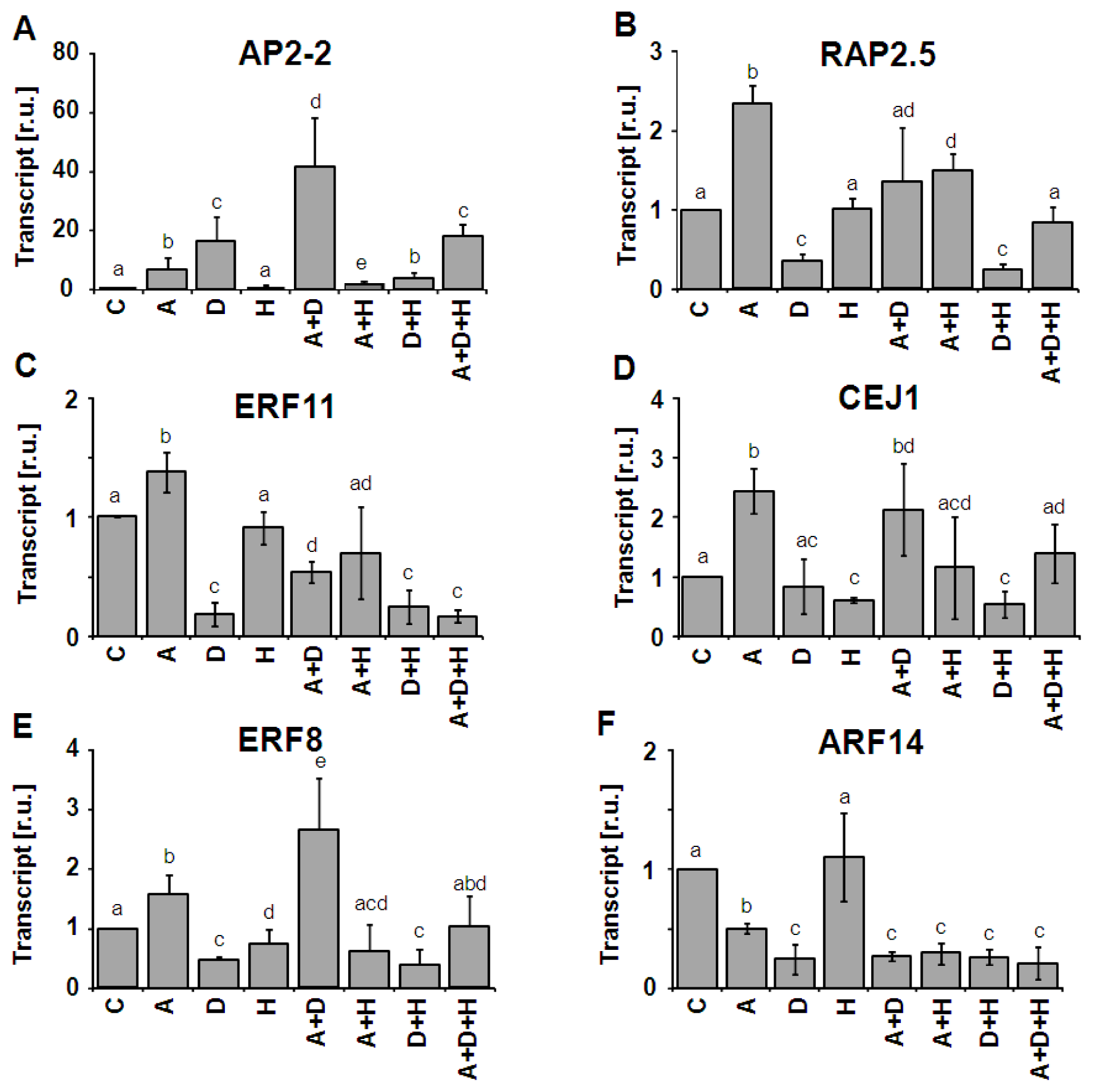
2.3. Combinatorial Effects on AP2/ERF-TF Expression
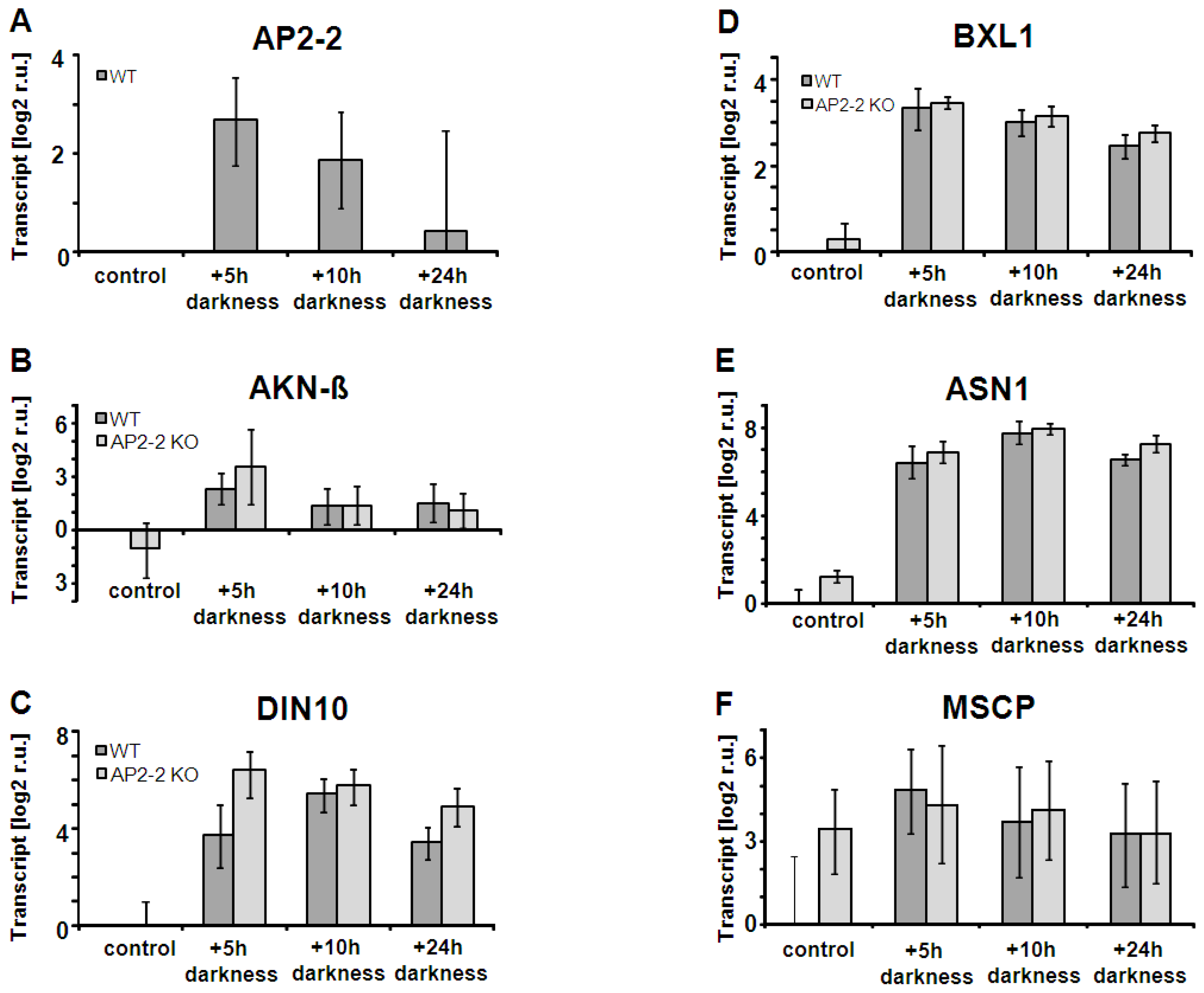
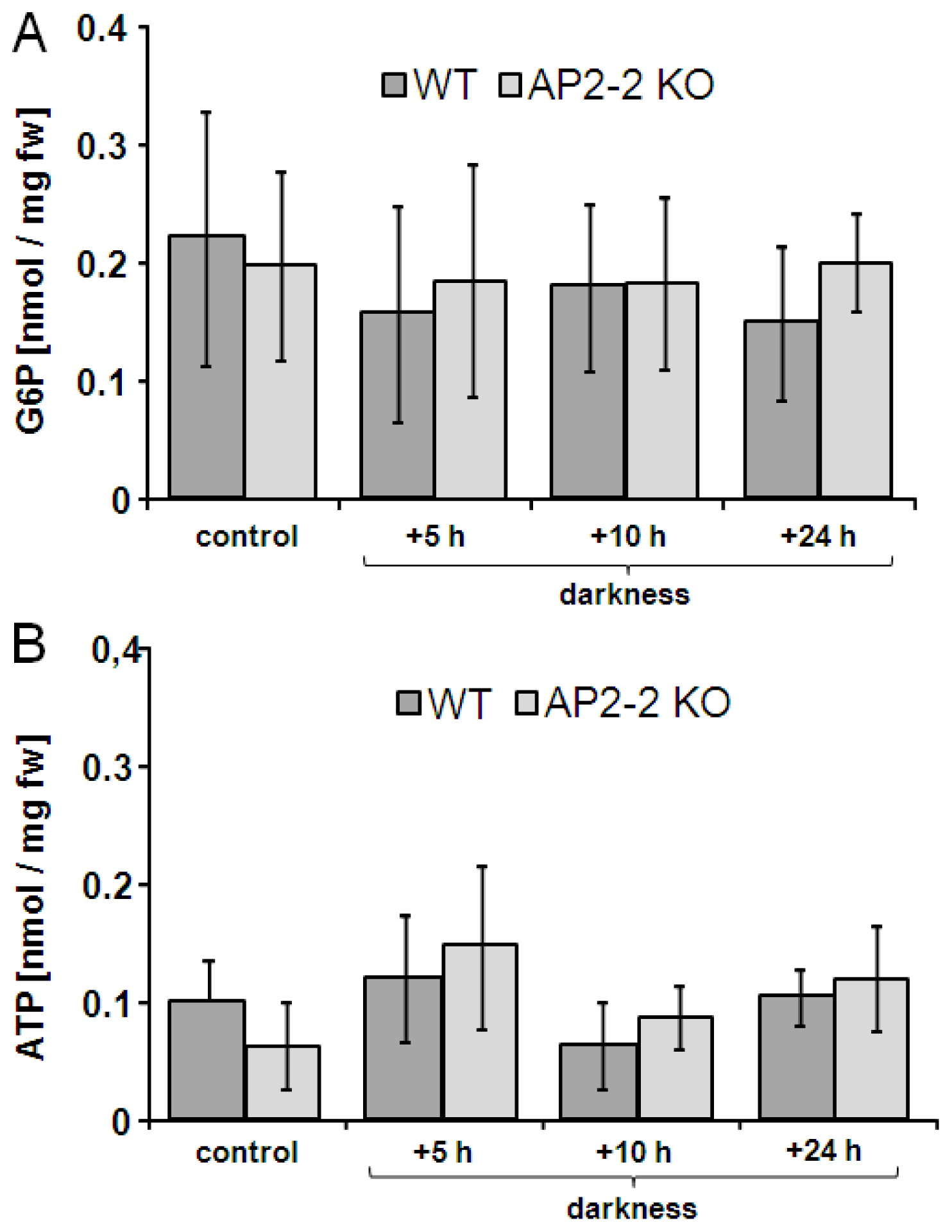
2.4. AP2-2 Transcript Regulation Relates to Starvation and Stress Versus Satiety
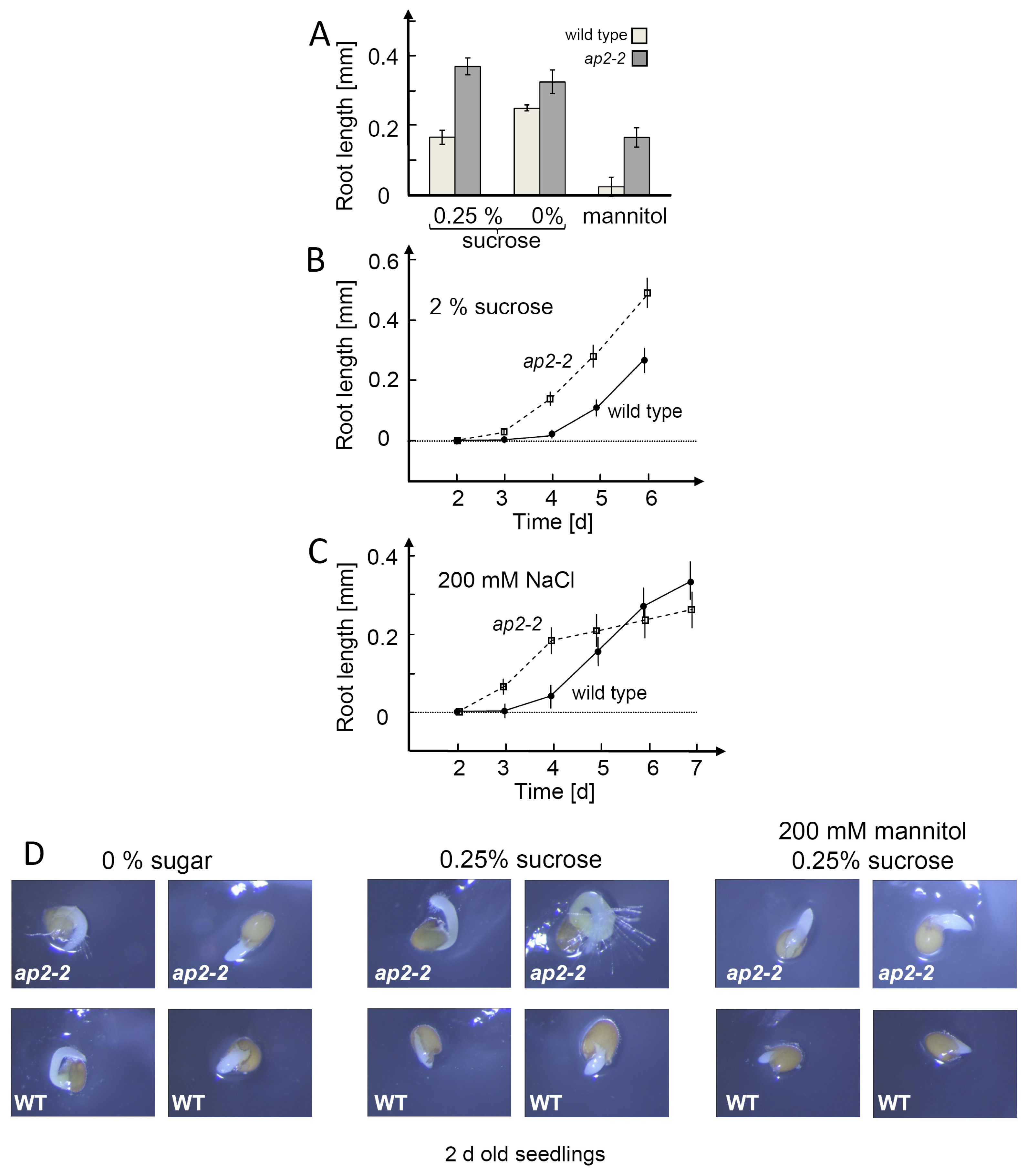
2.5. AP2-2 Knock Out Plant is Disturbed in Seeding Establishment
3. Discussion
3.1. AP2/ERF-TFs Appear to Be Involved in Retrograde Signaling at Various Levels of Regulation
3.2. Unpredictable Behavior of Transcript Regulation from Single Effectors to Combinatorial Conditions
3.3. AP2-2 as an Element in a Shunt Circuitry in Starvation Versus Satiety Acclimation
4. Experimental Section
4.1. Plant growth and Treatments
4.2. RNA-Isolation and cDNA Synthesis
4.3. Transcript Profiling
4.4. Mutant Screening
4.5. PSII Measurements
4.6. Metabolite Analysis
Supplementary Information
ijms-13-05933-s001.pdfAcknowledgments
Abbreviations
| ABA | abscisic acid |
| A. thaliana | Arabidopsis thaliana |
| AP2 | Apetala 2 transcription factor |
| Chl a | chlorophyll a |
| DCMU | 3-(3,4-dichlorophenyl)-1,1-dimethylurea |
| DREB | dehydration-response element binding protein |
| ERF | ethylene response factor |
| G6P | glucose-6-phosphate |
| H | high light (800 μmol quanta/m2s) |
| KO | knock out |
| L | low light (8 μmol quanta/m2s) |
| log-FC | logarithmic fold change |
| MV | methylviologen |
| N | normal light (80 μmol quanta/m2s) |
| RAP2 | related to apetala 2 |
| TF | transcription factor |
References
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.Z.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar]
- Mitsuda, N.; Ohme-Takagi, M. Functional analysis of transcription factors in Arabidopsis. Plant Cell Physiol 2009, 50, 1232–1248. [Google Scholar]
- Jofuku, K.D.; Denboer, B.G.W.; van Montagu, M.; Okamuro, J.K. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 1994, 6, 1211–1225. [Google Scholar]
- Ohme-Takagi, M.; Shinshi, H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 1995, 7, 173–182. [Google Scholar]
- Perez-Rodriguez, P.; Riano-Pachon, D.M.; Guedes Correa, L.G.; Rensing, S.A.; Kersten, B.; Mueller-Roeber, B. PlnTFDB: Updated content and new features of the plant transcription factor database. Nucleic Acids Res 2009, 38, 822–827. [Google Scholar]
- Dietz, K.J.; Vogel, M.O.; Viehhauser, A. AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signaling. Protoplasma 2010, 245, 3–14. [Google Scholar]
- Magnani, E.; Sjölander, K.; Hake, S. From endonucleases to transcription factors: Evolution of the AP2 DNA binding domain in plants. Plant Cell 2004, 16, 2265–2277. [Google Scholar]
- Koussevitzky, S.; Nott, A.; Mockler, T.C.; Hong, F.; Sachetto-Martins, G.; Surpin, M.; Lim, J.; Mittler, R.; Chory, J. Multiple signals from damaged chloroplasts converge on a common pathway to regulate nuclear gene expression. Science 2007, 316, 715–719. [Google Scholar]
- Khandelwal, A.; Elvitigala, T.; Ghosh, B.; Quatrano, R.S. Arabidopsis transcriptome reveals control circuits regulating redox homeostasis and the role of an AP2 transcription factor. Plant Physiol 2008, 148, 2050–2058. [Google Scholar]
- Shinwari, Z.K.; Nakashima, K.; Miura, S.; Kasuga, M.; Seki, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K. An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low-temperature responsive gene expression. Biochem. Biophys. Res. Commun 1998, 250, 161–170. [Google Scholar]
- Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Qin, F.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 2006, 18, 1292–1309. [Google Scholar]
- Knight, H.; Knight, M.R. Abiotic stress signaling pathways: Specificity and cross-talk. Trends Plant Sci 2001, 6, 262–267. [Google Scholar]
- Seki, M.; Ishida, J.; Narusaka, M.; Fujita, M.; Nanjo, T.; Umezawa, T.; Kamiya, A.; Nakajima, M.; Enju, A.; Sakurai, T.; et al. Monitoring the expression pattern of around 7000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct. Integr. Genomics 2002, 2, 282–291. [Google Scholar]
- Shinozaki, K.; Yamaguchi-Shinozaki, K.; Seki, M. Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol 2003, 6, 410–417. [Google Scholar]
- Agarwal, P.K.; Agarwal, P.; Reddy, M.K.; Sopory, S.K. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep 2006, 25, 1263–1274. [Google Scholar]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 1994, 6, 251–264. [Google Scholar]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol 2000, 3, 217–223. [Google Scholar]
- Fowler, S.; Thomashow, M.F. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 2002, 14, 1675–1690. [Google Scholar]
- Pogson, B.J.; Nick, S.W.; Förster, B.; Small, I. Plastid signaling to the nucleus and beyond. Trends Plant Sci 2008, 13, 602–609. [Google Scholar]
- Sato, S.; Nakamura, Y.; Kaneko, T.; Asamizu, E.; Tabata, S. Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res 1999, 6, 283–290. [Google Scholar]
- Abdallah, F.; Salamini, F.; Leister, D. A prediction of the size and evolutionary origin of the proteome of chloroplasts of Arabidopsis. Trends Plant Sci 2000, 5, 141–142. [Google Scholar]
- Strand, A. Plastid-to-nucleus signaling. Curr. Opin. Plant Biol 2004, 7, 621–625. [Google Scholar]
- Beck, C.F. Signaling pathways from the chloroplast to the nucleus. Planta 2005, 222, 743–756. [Google Scholar]
- Pfannschmidt, T. Plastidial retrograde signaling—A true “plastid factor” or just metabolite signatures? Trends Plant Sci 2010, 15, 427–435. [Google Scholar]
- Baier, M.; Hemmann, G.; Holman, R.; Corke, F.; Card, R.; Smith, C.; Rook, F.; Bevan, M.W. Characterization of mutants in Arabidopsis showing increased sugar-specific gene expression, growth, and developmental responses. Plant Physiol 2004, 134, 81–91. [Google Scholar]
- Sanchez, J.P.; Duque, P.; Chua, N.H. ABA activates ADPR cyclase and cADPR induces a subset of ABA-responsive genes in Arabidopsis. Plant J 2004, 38, 381–395. [Google Scholar]
- op den Camp, R.G.; Przybyla, D.; Ochsenbein, C.; Laloi, C.; Kim, C.; Danon, A.; Wagner, D.; Hideg, E.; Göbel, C.; Feussner, I.; et al. Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 2003, 15, 2320–2332. [Google Scholar]
- Feng, J.X.; Liu, D.; Pan, Y.; Gong, W.; Ma, L.G.; Luo, J.C.; Deng, X.W.; Zhu, Y.X. An annotation update via cDNA sequence analysis and comprehensive profiling of developmental, hormonal or environmental responsiveness of the Arabidopsis AP2/EREBP transcription factor gene family. Plant Mol. Biol 2005, 59, 853–868. [Google Scholar]
- Dietz, K.J.; Jacquot, J.P.; Harris, G. Hubs and bottlenecks in plant molecular signalling networks. New Phytol 2010, 188, 919–938. [Google Scholar]
- Jones, H.G. Stomatal control of photosynthesis and transpiration. J. Exp. Bot 1998, 49, 387–398. [Google Scholar]
- Li, X.F.; Li, Y.J.; An, Y.H.; Xiong, L.J.; Shao, X.H.; Wang, Y.; Sun, Y. AKINβ1 is involved in the regulation of nitrogen metabolism and sugar signaling in Arabidopsis. J. Integr. Plant Biol 2009, 51, 513–520. [Google Scholar]
- Fujiki, Y.; Ito, M.; Nishida, I.; Watanabe, A. Multiple signaling pathways in gene expression during sugar starvation. Pharmacological analysis of din gene expression in suspension-cultured cells of Arabidopsis. Plant Physiol 2000, 124, 1139–1147. [Google Scholar]
- Riechmann, J.L.; Meyerowitz, E.M. The AP2/EREBP family of plant transcription factors. Biol. Chem 1998, 379, 633–646. [Google Scholar]
- Wellmer, F.; Riechmann, J.L. Gene network analysis in plant development by genomic technologies. Int. J. Dev. Biol 2005, 49, 745–759. [Google Scholar]
- Nakano, T.; Suzuki, K.; Ohtsuki, N.; Tsujimoto, Y.; Fujimura, T.; Shinshi, H. Identification of genes of the plant-specific transcription-factor families cooperatively regulated by ethylene and jasmonate in Arabidopsis thaliana. J. Plant Res 2006, 119, 407–413. [Google Scholar]
- Kerchev, P.I.; Pellny, T.K.; Vivancos, P.D.; Kiddle, G.; Hedden, P.; Driscoll, S.; Vanacker, H.; Verrier, P.; Hancock, R.D.; Foyer, C.H. The transcription factor ABI4 is required for the ascorbic acid-dependent regulation of growth and regulation of jasmonate-dependent defense signaling pathways in Arabidopsis. Plant Cell 2011, 23, 3319–3334. [Google Scholar]
- Hubbard, K.E.; Nishimura, N.; Hitomi, K.; Getzoff, E.D.; Schroeder, J.I. Early abscisic acid signal transduction mechanisms: Newly discovered components and newly emerging questions. Genes Dev 2010, 24, 1695–1708. [Google Scholar]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol 2005, 56, 165–185. [Google Scholar]
- Galvez-Valdivieso, G.; Fryer, M.J.; Lawson, T.; Slattery, K.; Truman, W.; Smirnoff, N.; Asami, T.; Davies, W.J.; Jones, A.M.; Baker, N.R.; Mullineaux, P.M. The high light response in Arabidopsis involves ABA signaling between vascular and bundle sheath cells. Plant Cell 2009, 21, 2143–2162. [Google Scholar]
- Flors, C.; Fryer, M.J.; Waring, J.; Reeder, B.; Bechtold, U.; Mullineaux, P.M.; Nonell, S.; Wilson, M.T.; Baker, N.R. Imaging the production of singlet oxygen in vivo using a new fluorescent sensor, Singlet Oxygen Sensor Green. J. Exp. Bot 2006, 57, 1725–1734. [Google Scholar]
- Ikeda, M.; Ohme-Takagi, M. A novel group of transcriptional repressors in Arabidopsis. Plant Cell Physiol 2009, 50, 970–975. [Google Scholar]
- Son, G.H.; Wan, J.R.; Kim, H.J.; Nguyen, X.C.; Chung, W.S.; Hong, J.C.; Stacey, G. Ethylene-responsive element-binding factor 5, ERF5, is involved in chitin-induced innate immunity response. Mol. Plant Microbe Interact 2012, 25, 48–60. [Google Scholar]
- Contento, A.L.; Kim, S.J.; Bassham, D.C. Transcriptome profiling of the response of Arabidopsis suspension culture cells to sucrose starvation. Plant Physiol 2004, 135, 2330–2347. [Google Scholar]
- Finkelstein, R.R.; Wang, M.L.; Lynch, T.J.; Rao, S.; Goodman, H.M. The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA2 domain protein. Plant Cell 1998, 10, 1043–1054. [Google Scholar]
- Arenas-Huertero, F.; Arroyo, A.; Zhou, L.; Sheen, J.; Leon, P. Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev 2000, 14, 2085–2096. [Google Scholar]
- Oelze, M.L.; Vogel, M.O.; Alsharafa, K.; Kahmann, U.; Viehhauser, A.; Maurino, V.G.; Dietz, K.J. Efficient acclimation of the chloroplast antioxidant defence of Arabidopsis thaliana leaves in response to a 10- or 100-fold light increment and the possible involvement of retrograde signals. J. Exp. Bot 2012, 63, 1297–1313. [Google Scholar]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem 1987, 162, 156–159. [Google Scholar]
- Wormuth, D.; Baier, M.; Kandlbinder, A.; Scheibe, R.; Hartung, W.; Dietz, K.J. Regulation of gene expression by photosynthetic signals triggered through modified CO2 availability. BMC Plant Biol 2006, 6, 15–39. [Google Scholar]
- Finkemeier, I.; Goodman, M.; Lamkemeyer, P.; Kandlbinder, A.; Sweetlove, L.J.; Dietz, K.J. The mitochondrial type II peroxiredoxin F is essential for redox homeostasis and root growth of Arabidopsis thaliana under stress. J. Biol. Chem 2005, 280, 12168–12180. [Google Scholar]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R.; et al. Genome-wide insertional mutagenesis, of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar]
- Edwards, K.; Johnstone, C.; Thompson, C. A simple and rapid method for preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 1991, 6. [Google Scholar] [CrossRef]
- Barsch, A.; Patschkowski, T.; Niehaus, K. Comprehensive metabolite profiling of Sinorhizobium meliloti using gas chromatography-mass spectrometry. Funct. Integr. Genomics 2004, 4, 219–230. [Google Scholar]
- Dietz, K.J.; Heber, U. Rate-limiting factors in leaf photosynthesis. I. Carbon fluxes in the Calvin cycle. Biochim. Biophys. Acta 1984, 767, 432–443. [Google Scholar]

© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Vogel, M.O.; Gomez-Perez, D.; Probst, N.; Dietz, K.-J. Combinatorial Signal Integration by APETALA2/Ethylene Response Factor (ERF)-Transcription Factors and the Involvement of AP2-2 in Starvation Response. Int. J. Mol. Sci. 2012, 13, 5933-5951. https://doi.org/10.3390/ijms13055933
Vogel MO, Gomez-Perez D, Probst N, Dietz K-J. Combinatorial Signal Integration by APETALA2/Ethylene Response Factor (ERF)-Transcription Factors and the Involvement of AP2-2 in Starvation Response. International Journal of Molecular Sciences. 2012; 13(5):5933-5951. https://doi.org/10.3390/ijms13055933
Chicago/Turabian StyleVogel, Marc Oliver, Deborah Gomez-Perez, Nina Probst, and Karl-Josef Dietz. 2012. "Combinatorial Signal Integration by APETALA2/Ethylene Response Factor (ERF)-Transcription Factors and the Involvement of AP2-2 in Starvation Response" International Journal of Molecular Sciences 13, no. 5: 5933-5951. https://doi.org/10.3390/ijms13055933




