Long Non-Coding RNAs and p53 Regulation
Abstract
:1. Introduction
2. Long Non-Coding RNAs at the CDKN1A (p21) Locus
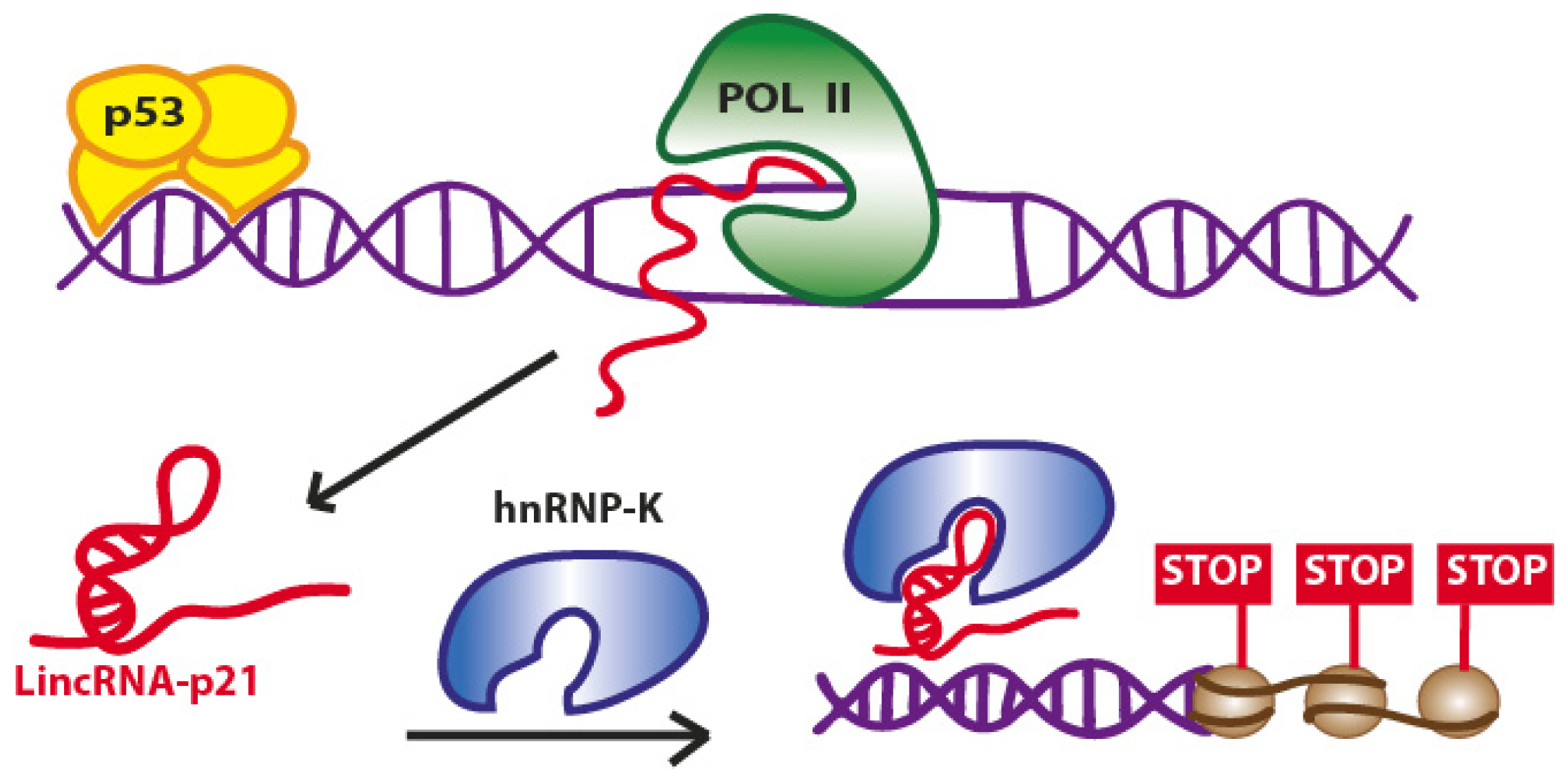
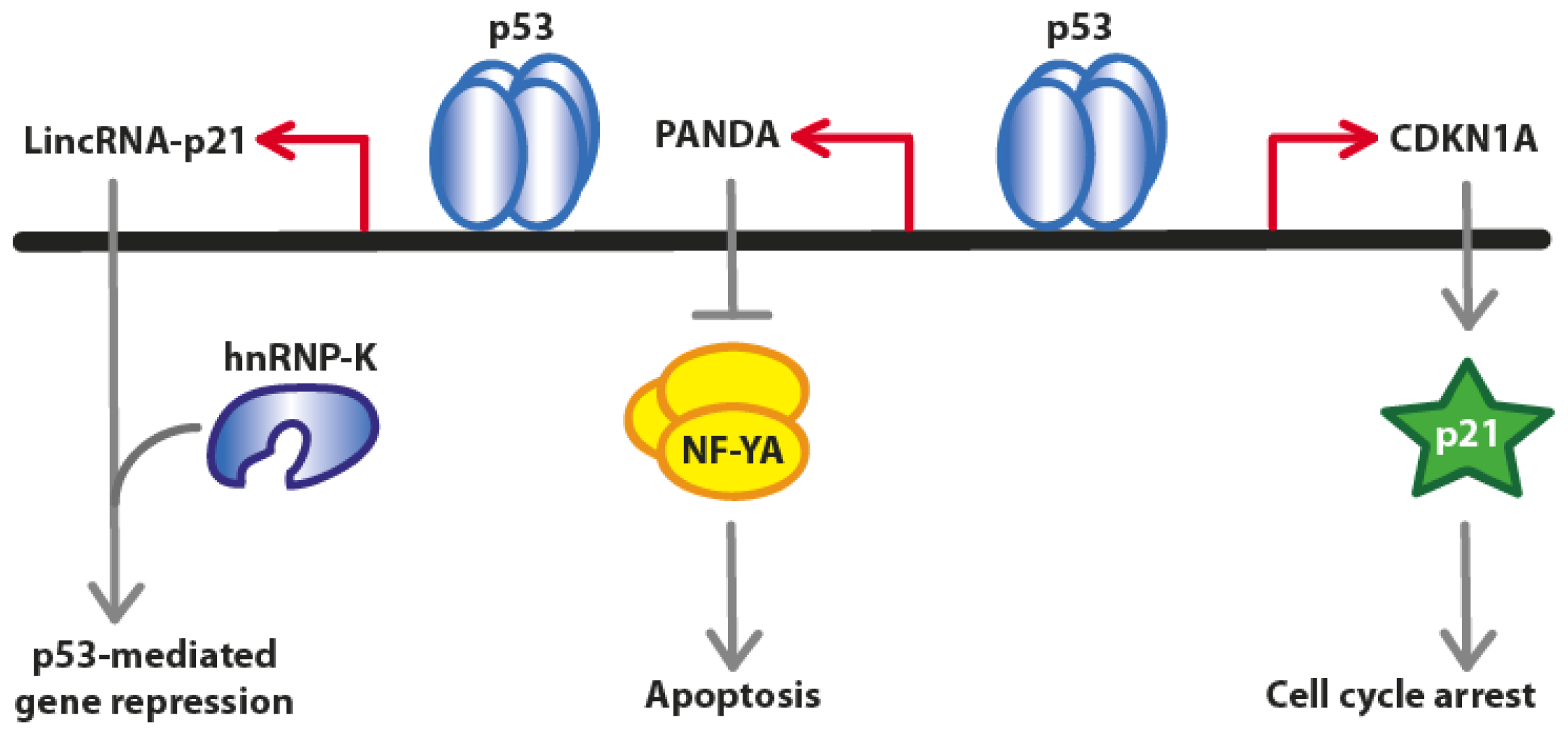
2.1. LincRNA-p21
2.2. PANDA (P21 Associated ncRNA DNA Damage Activated)
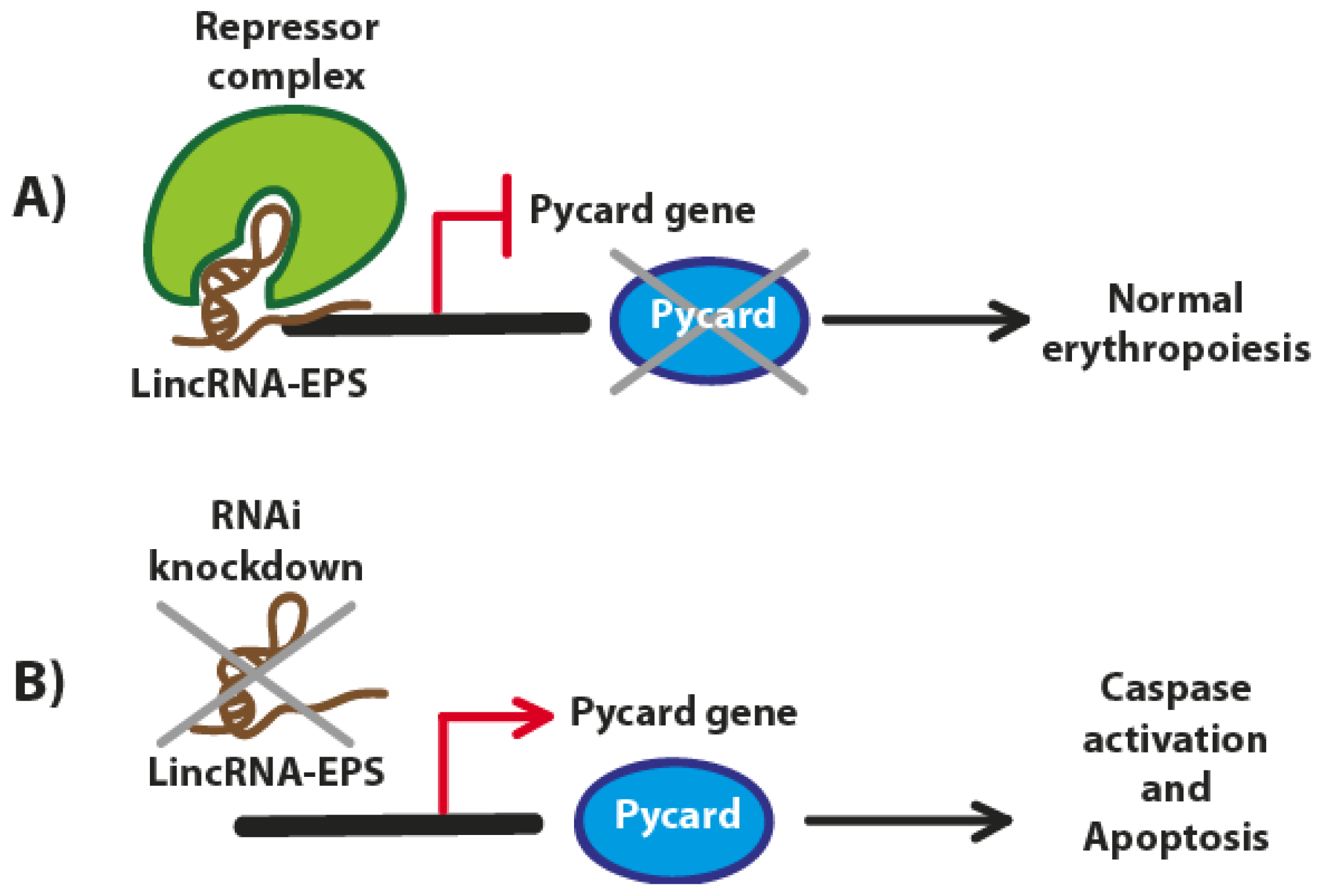
3. LincRNA-EPS
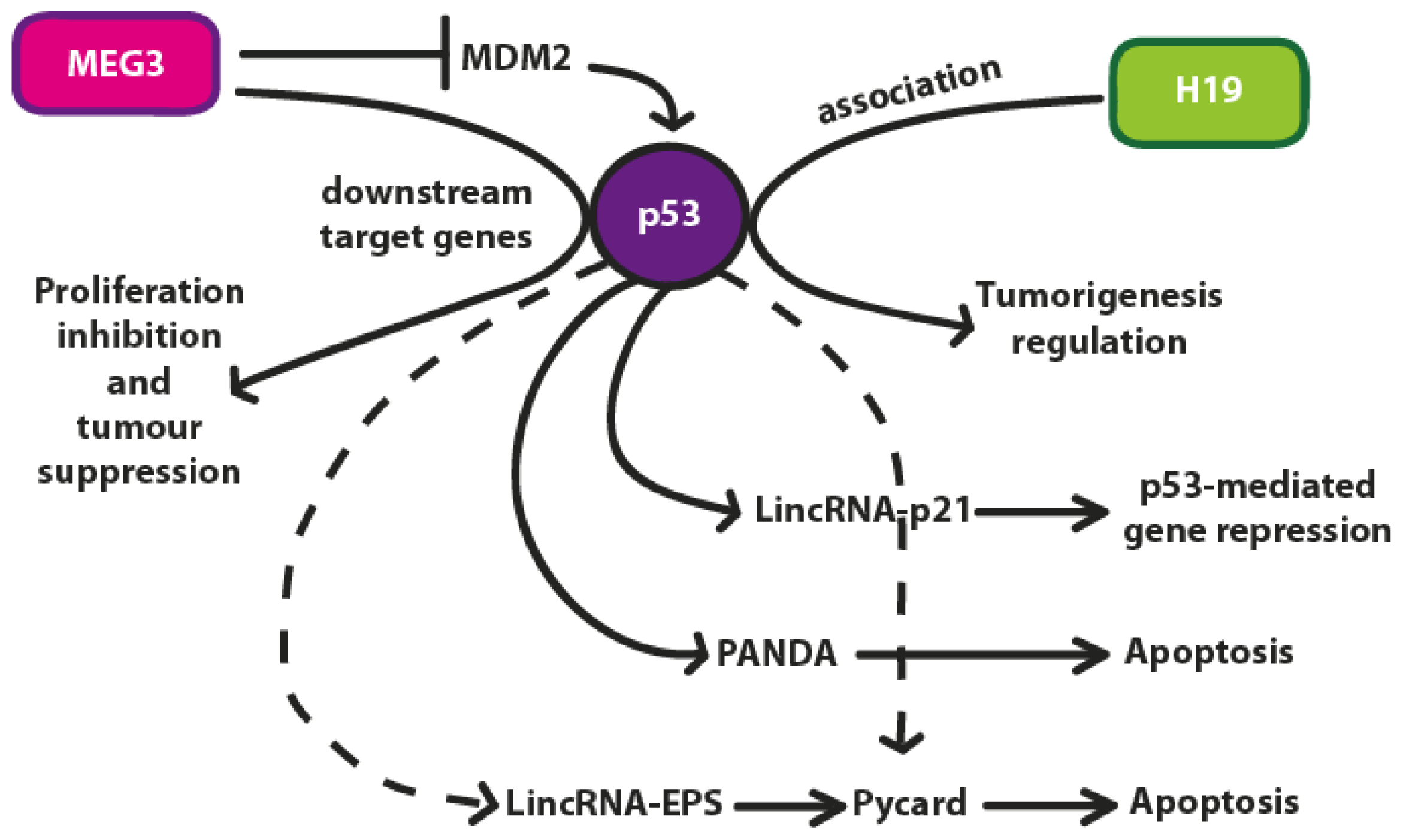
4. p53 Regulation by lncRNAs
4.1. H19
4.2. MEG3 lncRNA
5. Conclusions
Acknowledgments
References
- Mattick, J.S. RNA regulation: A new genetics? Nat. Rev. Genet 2004, 5, 316–323. [Google Scholar]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet 2009, 10, 155–159. [Google Scholar]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar]
- Wilusz, J.E.; Sunwoo, H.; Spector, D.L. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev 2009, 23, 1494–1504. [Google Scholar]
- Carninci, P. Non-coding RNA transcription: Turning on neighbours. Nat. Cell Biol 2008, 10, 1023–1024. [Google Scholar]
- The ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007, 447, 799–816.
- The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74.
- Bernstein, E.; Allis, C.D. RNA meets chromatin. Genes Dev 2005, 19, 1635–1655. [Google Scholar]
- Bracken, A.P.; Helin, K. Polycomb group proteins: Navigators of lineage pathways led astray in cancer. Nat. Rev. Cancer 2009, 9, 773–784. [Google Scholar]
- Faghihi, M.A.; Wahlestedt, C. Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell Biol 2009, 10, 637–643. [Google Scholar]
- Whitehead, J.; Pandey, G.K.; Kanduri, C. Regulation of the mammalian epigenome by long noncoding RNAs. Biochim. Biophys. Acta 2009, 1790, 936–947. [Google Scholar]
- Kanduri, C.; Whitehead, J.; Mohammad, F. The long and the short of it: RNA-directed chromatin asymmetry in mammalian X-chromosome inactivation. FEBS Lett 2009, 583, 857–864. [Google Scholar]
- Lee, J.T. Lessons from X-chromosome inactivation: Long ncRNA as guides and tethers to the epigenome. Genes Dev 2009, 23, 1831–1842. [Google Scholar]
- Mohammad, F.; Mondal, T.; Kanduri, C. Epigenetics of imprinted long noncoding RNAs. Epigenetics 2009, 4, 277–286. [Google Scholar]
- Clemson, C.M.; Hutchinson, J.N.; Sara, S.A.; Ensminger, A.W.; Fox, A.H.; Chess, A.; Lawrence, J.B. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 2009, 33, 717–726. [Google Scholar]
- Sasaki, Y.T.; Ideue, T.; Sano, M.; Mituyama, T.; Hirose, T. MENepsilon/β noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. USA 2009, 106, 2525–2530. [Google Scholar]
- Schoeftner, S.; Blasco, M.A. A “higher order” of telomere regulation: Telomere heterochromatin and telomeric RNAs. EMBO J 2009, 28, 2323–2336. [Google Scholar]
- Wong, L.H.; Brettingham-Moore, K.H.; Chan, L.; Quach, J.M.; Anderson, M.A.; Northrop, E.L.; Hannan, R.; Saffery, R.; Shaw, M.L.; Williams, E.; Choo, K.H. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res 2007, 17, 1146–1160. [Google Scholar]
- Ferri, F.; Bouzinba-Segard, H.; Velasco, G.; Hube, F.; Francastel, C. Non-coding murine centromeric transcripts associate with and potentiate Aurora B kinase. Nucleic Acids Res 2009, 37, 5071–5080. [Google Scholar]
- Willingham, A.T.; Orth, A.P.; Batalov, S.; Peters, E.C.; Wen, B.G.; Aza-Blanc, P.; Hogenesch, J.B.; Schultz, P.G. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 2005, 309, 1570–1573. [Google Scholar]
- Ponjavic, J.; Ponting, C.P.; Lunter, G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res 2007, 17, 556–565. [Google Scholar]
- Mattick, J.S. The genetic signatures of noncoding RNAs. PLoS Genet 2009, 5, e1000459. [Google Scholar]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar]
- Huarte, M.; Guttman, M.; Feldser, D.; Garber, M.; Koziol, M.J.; Kenzelmann-Broz, D.; Khalil, A.M.; Zuk, O.; Amit, I.; Rabani, M.; et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010, 142, 409–419. [Google Scholar]
- Levine, A.J.; Hu, W.; Feng, Z. The P53 pathway: What questions remain to be explored? Cell Death Differ 2006, 13, 1027–1036. [Google Scholar]
- Hung, T.; Wang, Y.; Lin, M.F.; Koegel, A.K.; Kotake, Y.; Grant, G.D.; Horlings, H.M.; Shah, N.; Umbricht, C.; Wang, P.; et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet 2011, 43, 621–629. [Google Scholar]
- Morachis, J.M.; Murawsky, C.M.; Emerson, B.M. Regulation of the p53 transcriptional response by structurally diverse core promoters. Genes Dev 2010, 24, 135–147. [Google Scholar]
- Hu, W.; Yuan, B.; Flygare, J.; Lodish, H.F. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev 2011, 25, 2573–2578. [Google Scholar]
- Ohtsuka, T.; Ryu, H.; Minamishima, Y.A.; Macip, S.; Sagara, J.; Nakayama, K.I.; Aaronson, S.A.; Lee, S.W. ASC is a Bax adaptor and regulates the p53-Bax mitochondrial apoptosis pathway. Nat. Cell Biol 2004, 6, 121–128. [Google Scholar]
- Ohtsuka, T.; Mitsuno, M.; Kitajima, Y.; Ide, T.; Lee, S.W.; Miyazaki, K. Role of ASC in hypoxia-mediated cell death in pancreatic cancer. Mol. Med. Report 2008, 1, 827–831. [Google Scholar]
- Paralkar, V.R.; Weiss, M.J. A new “Linc” between noncoding RNAs and blood development. Genes Dev 2011, 25, 2555–2558. [Google Scholar]
- Flygare, J.; Rayon Estrada, V.; Shin, C.; Gupta, S.; Lodish, H.F. HIF1alpha synergizes with glucocorticoids to promote BFU-E progenitor self-renewal. Blood 2011, 117, 3435–3444. [Google Scholar]
- Matouk, I.J.; DeGroot, N.; Mezan, S.; Ayesh, S.; Abu-lail, R.; Hochberg, A.; Galun, E. The H19 non-coding RNA is essential for human tumor growth. PLoS One 2007, 2, e845. [Google Scholar]
- Barsyte-Lovejoy, D.; Lau, S.K.; Boutros, P.C.; Khosravi, F.; Jurisica, I.; Andrulis, I.L.; Tsao, M.S.; Penn, L.Z. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res 2006, 66, 5330–5337. [Google Scholar]
- Yang, F.; Bi, J.; Xue, X.; Zheng, L.; Zhi, K.; Hua, J.; Fang, G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J 2012, 279, 3159–3165. [Google Scholar]
- Park, I.Y.; Sohn, B.H.; Choo, J.H.; Joe, C.O.; Seong, J.K.; Lee, Y.I.; Chung, J.H. Deregulation of DNA methyltransferases and loss of parental methylation at the insulin-like growth factor II (Igf2)/H19 loci in p53 knockout mice prior to tumor development. J. Cell. Biochem 2005, 94, 585–596. [Google Scholar]
- Dugimont, T.; Montpellier, C.; Adriaenssens, E.; Lottin, S.; Dumont, L.; Iotsova, V.; Lagrou, C.; Stehelin, D.; Coll, J.; Curgy, J.J. The H19 TATA-less promoter is efficiently repressed by wild-type tumor suppressor gene product p53. Oncogene 1998, 16, 2395–2401. [Google Scholar]
- Zhang, X.; Rice, K.; Wang, Y.; Chen, W.; Zhong, Y.; Nakayama, Y.; Zhou, Y.; Klibanski, A. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: Isoform structure, expression, and functions. Endocrinology 2010, 151, 939–947. [Google Scholar]
- Zhang, X.; Zhou, Y.; Mehta, K.R.; Danila, D.C.; Scolavino, S.; Johnson, S.R.; Klibanski, A. A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J. Clin. Endocrinol. Metab 2003, 88, 5119–5126. [Google Scholar]
- Zhang, X.; Gejman, R.; Mahta, A.; Zhong, Y.; Rice, K.A.; Zhou, Y.; Cheunsuchon, P.; Louis, D.N.; Klibanski, A. Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res 2010, 70, 2350–2358. [Google Scholar]
- Schmidt, J.V.; Matteson, P.G.; Jones, B.K.; Guan, X.J.; Tilghman, S.M. The Dlk1 and Gtl2 genes are linked and reciprocally imprinted. Genes Dev 2000, 14, 1997–2002. [Google Scholar]
- Zhou, Y.; Zhong, Y.; Wang, Y.; Zhang, X.; Batista, D.L.; Gejman, R.; Ansell, P.J.; Zhao, J.; Weng, C.; Klibanski, A. Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem 2007, 282, 24731–24742. [Google Scholar]
- Zhou, Y.; Zhang, X.; Klibanski, A. MEG3 noncoding RNA: A tumor suppressor. J. Mol. Endocrinol 2012, 48, R45–R53. [Google Scholar]
- Matouk, I.J.; Mezan, S.; Mizrahi, A.; Ohana, P.; Abu-Lail, R.; Fellig, Y.; Degroot, N.; Galun, E.; Hochberg, A. The oncofetal H19 RNA connection: Hypoxia, p53 and cancer. Biochim. Biophys. Acta 2010, 1803, 443–451. [Google Scholar]
- Braconi, C.; Kogure, T.; Valeri, N.; Huang, N.; Nuovo, G.; Costinean, S.; Negrini, M.; Miotto, E.; Croce, C.M.; Patel, T. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene 2011, 30, 4750–4756. [Google Scholar]
- Kogo, R.; Shimamura, T.; Mimori, K.; Kawahara, K.; Imoto, S.; Sudo, T.; Tanaka, F.; Shibata, K.; Suzuki, A.; Komune, S.; et al. Long noncoding RNA HOTAIR regulates polycombdependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 2011, 71, 6320–6326. [Google Scholar]
- Lai, M.C.; Yang, Z.; Zhou, L.; Zhu, Q.Q.; Xie, H.Y.; Zhang, F.; Wu, L.M.; Chen, L.M.; Zheng, S.S. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med. Oncol 2012, 29, 1810–1816. [Google Scholar]
- Benetatos, L.; Vartholomatos, G.; Hatzimichael, E. MEG3 imprinted gene contribution in tumorigenesis. Int. J. Cancer 2011, 129, 773–779. [Google Scholar]
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 2011, 25, 1915–1927. [Google Scholar]
- Cabili, M.N. Human Body Map lincRNAs. Available online: http://www.broadinstitute.org/genome_bio/human_lincrnas/ accessed on 5 December 2012.
- Da Sacco, L.; Baldassarre, A.; Masotti, A. Bioinformatics tools and novel challenges in long non-coding RNAs (lncRNAs) functional analysis. Int. J. Mol. Sci 2012, 13, 97–114. [Google Scholar]
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Baldassarre, A.; Masotti, A. Long Non-Coding RNAs and p53 Regulation. Int. J. Mol. Sci. 2012, 13, 16708-16717. https://doi.org/10.3390/ijms131216708
Baldassarre A, Masotti A. Long Non-Coding RNAs and p53 Regulation. International Journal of Molecular Sciences. 2012; 13(12):16708-16717. https://doi.org/10.3390/ijms131216708
Chicago/Turabian StyleBaldassarre, Antonella, and Andrea Masotti. 2012. "Long Non-Coding RNAs and p53 Regulation" International Journal of Molecular Sciences 13, no. 12: 16708-16717. https://doi.org/10.3390/ijms131216708



