Caffeine Abolishes the Ultraviolet-Induced REV3 Translesion Replication Pathway in Mouse Cells
Abstract
:1. Introduction
2. Results and Discussion
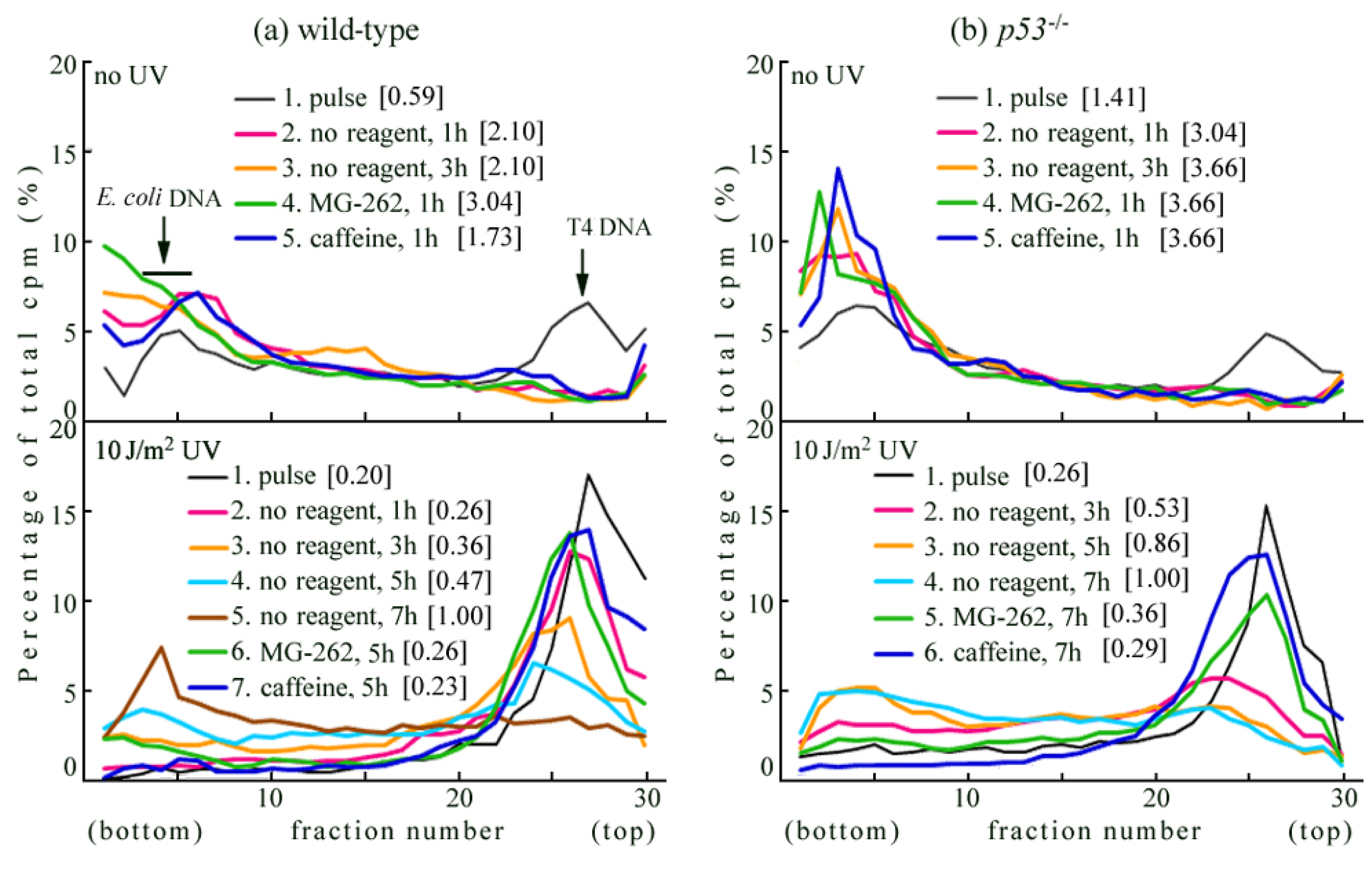
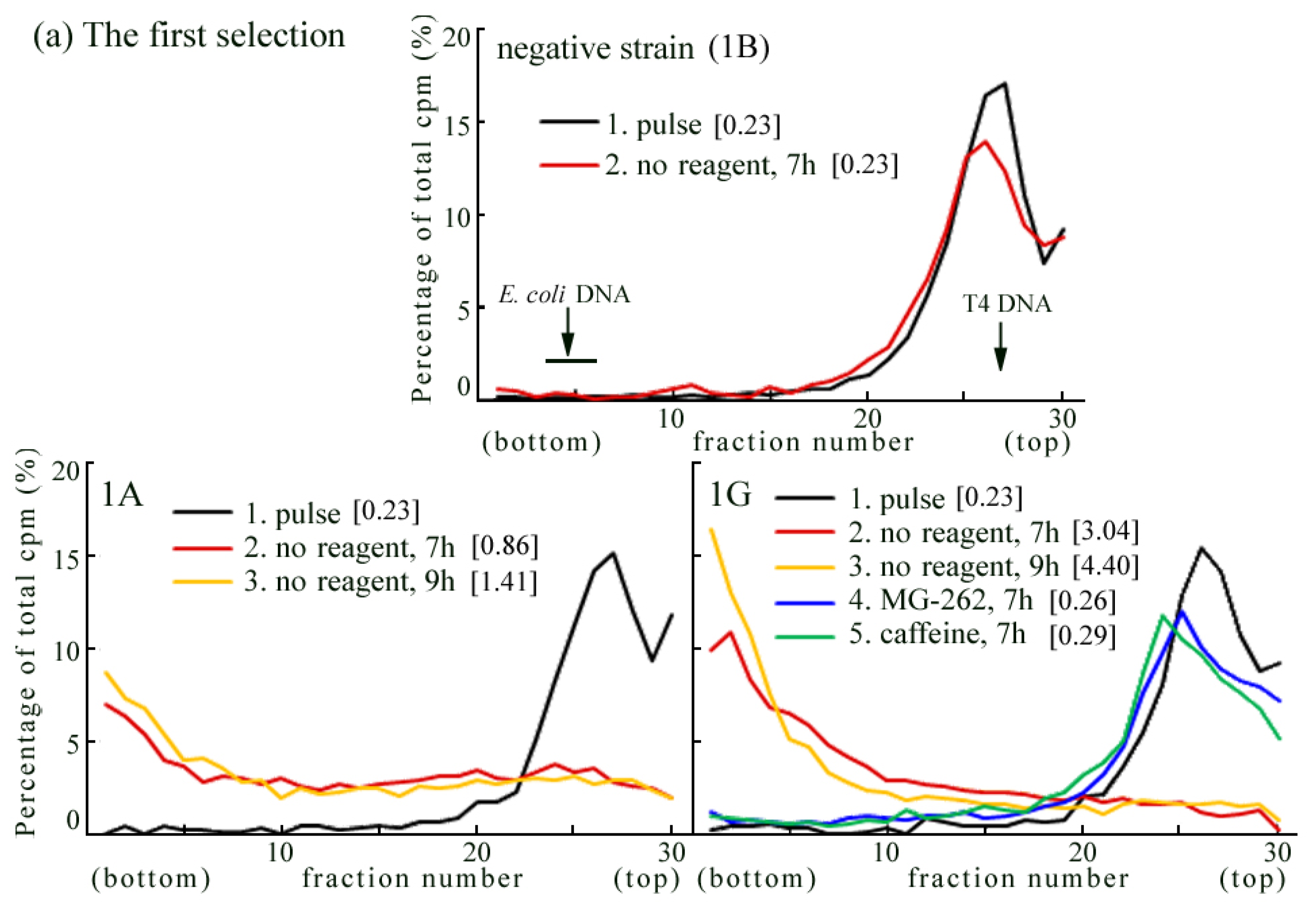
2.1. Detection of UV-Induced Translesion Replication in Mouse Cells
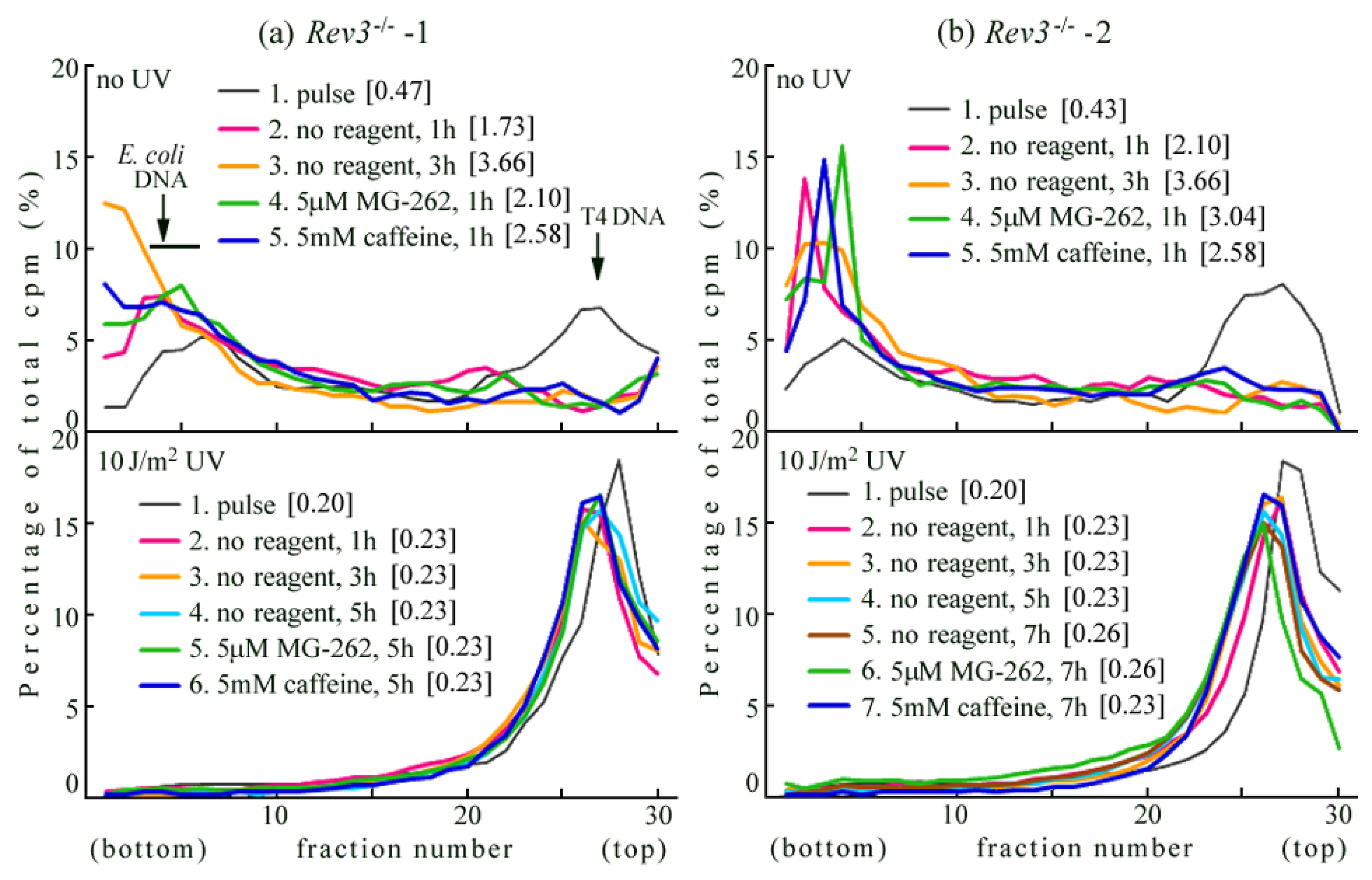
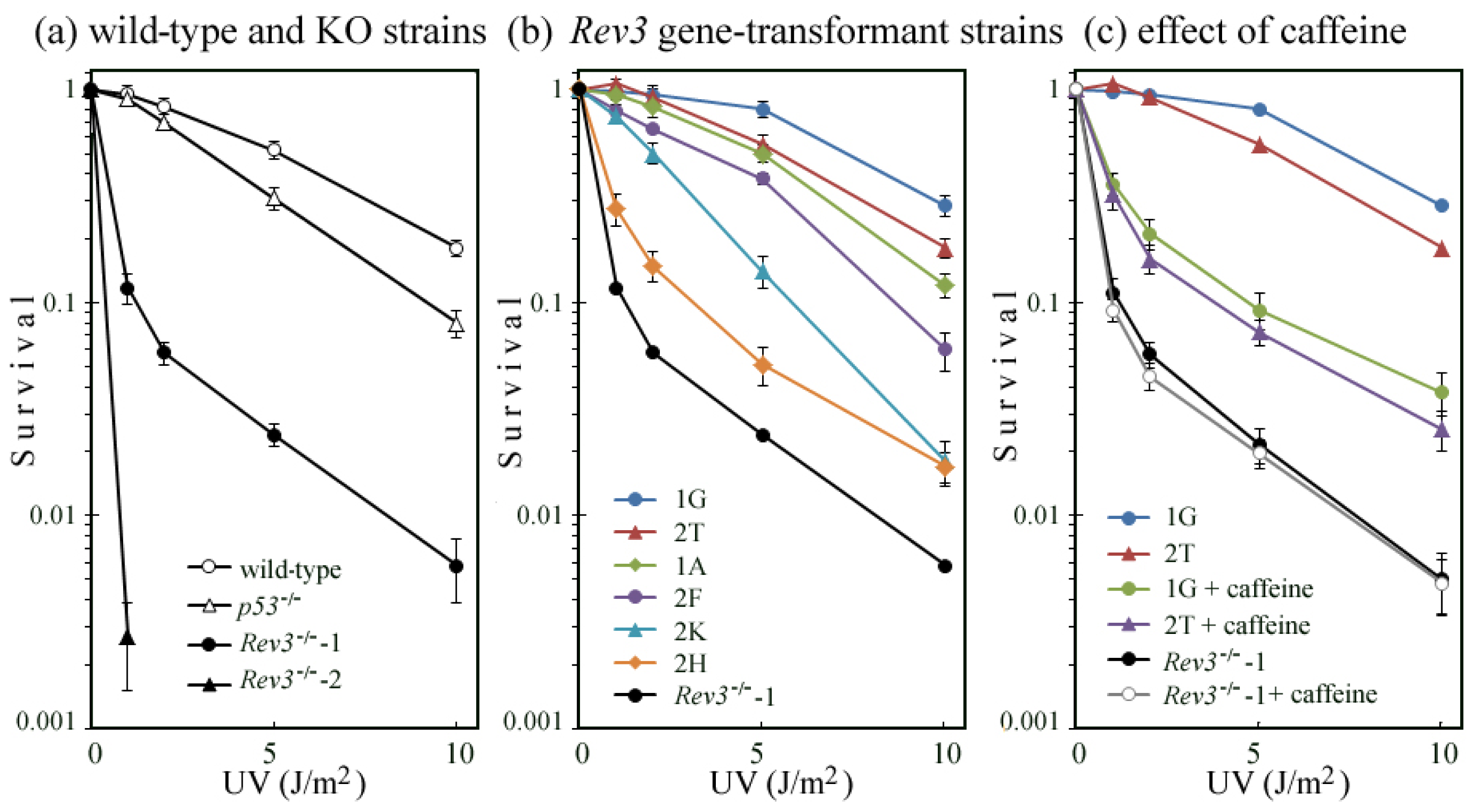
2.2. UV-TLS Was Mostly Abolished in Rev3 Knockout MEFs
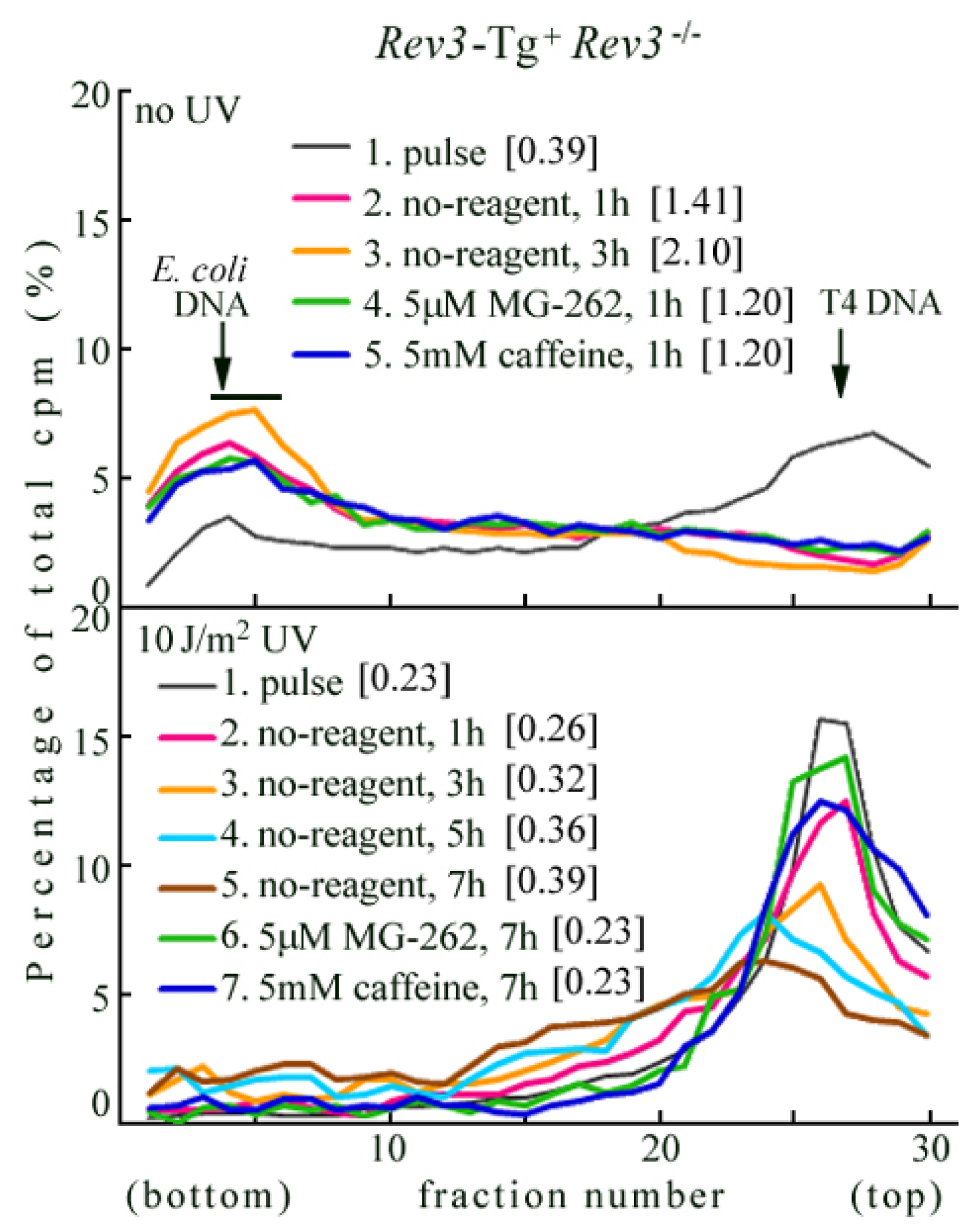
2.3. Rev3 Transgene Restored UV-TLS in Rev3 Knockout Mouse Cells
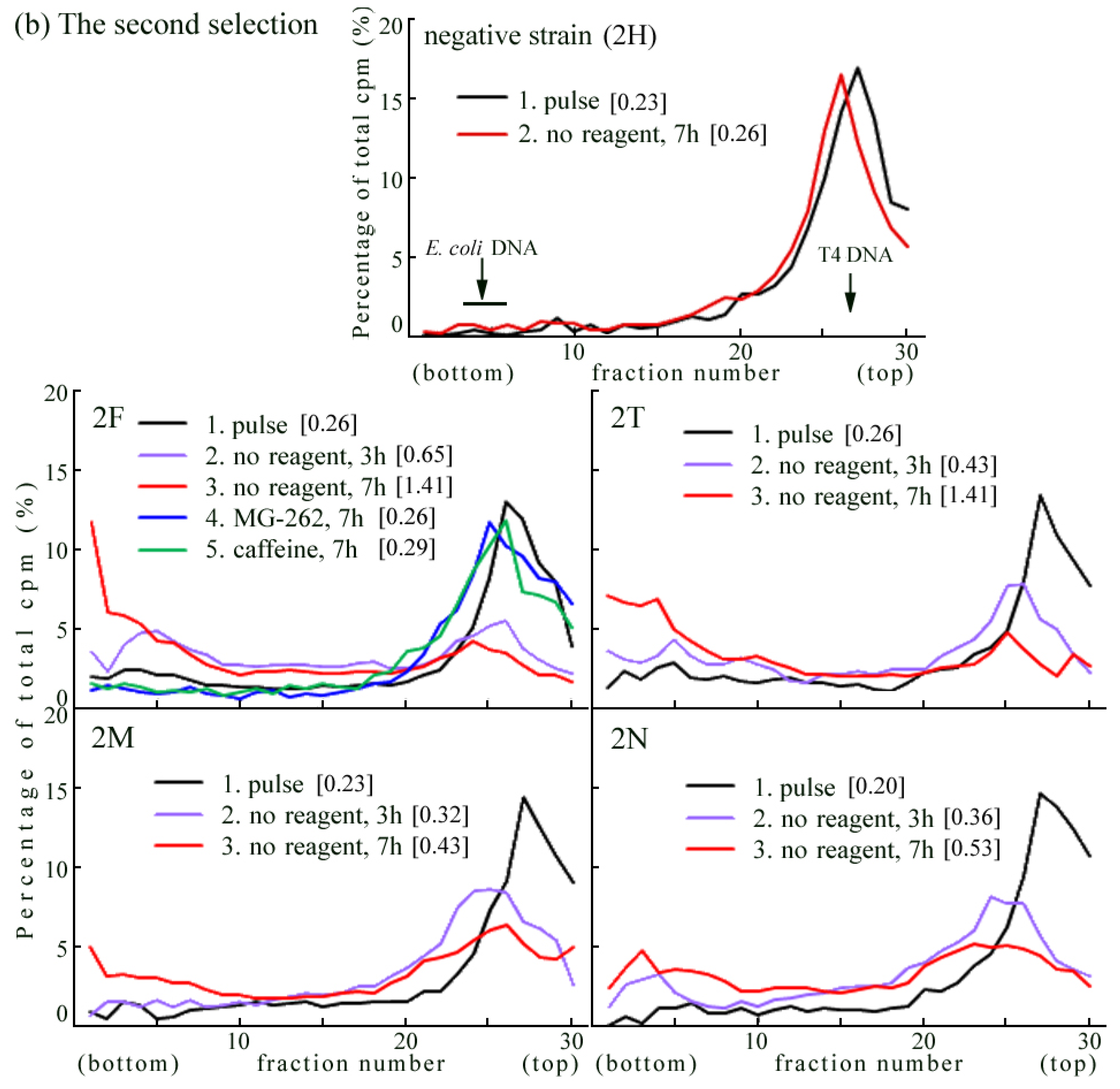
2.4. Introduction of the Rev3 Expression Vector Restored UV-TLS in Rev3 Knockout MEFs
2.5. Correlation of UV Survival with UV-TLS
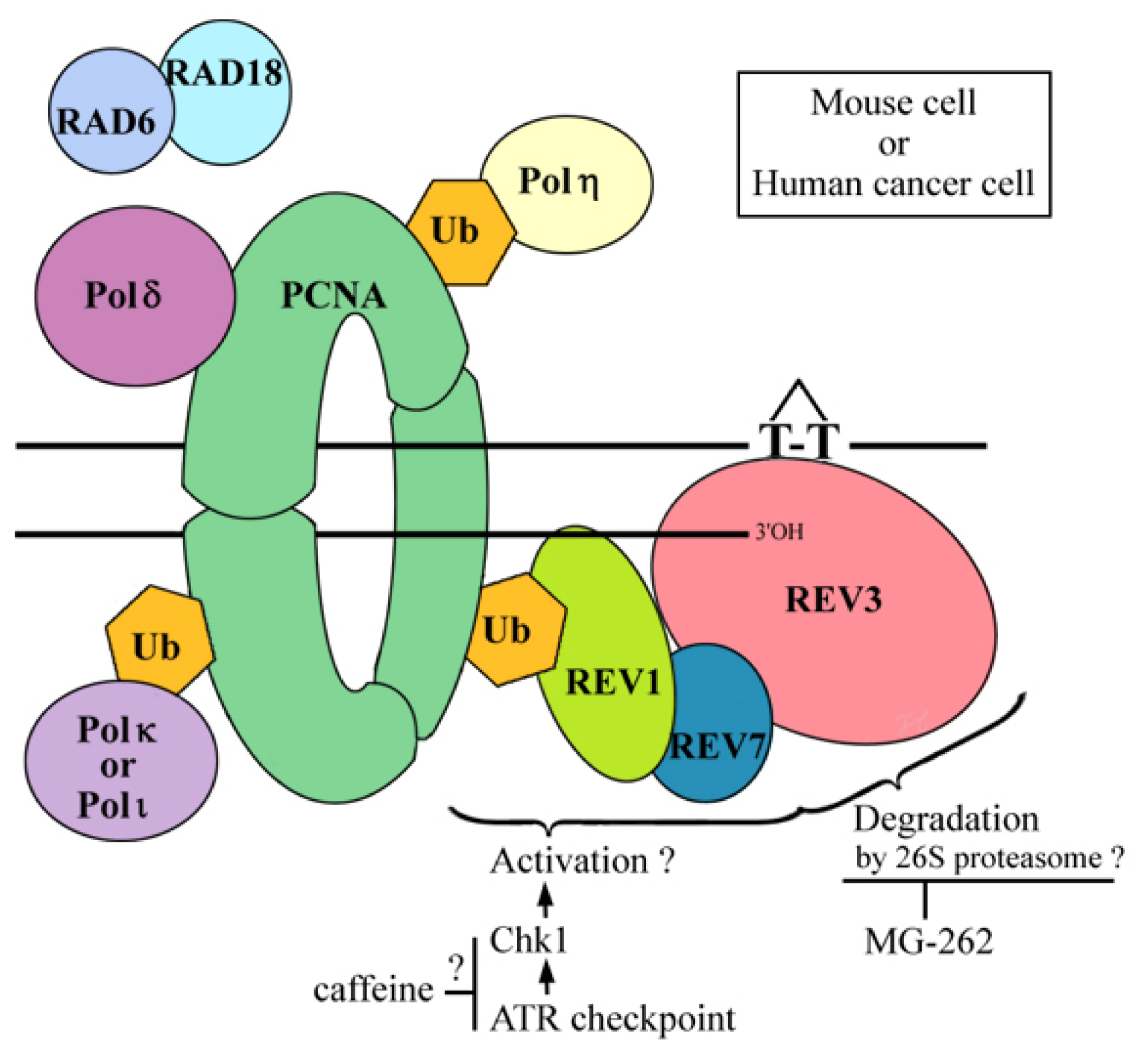
2.6. Where Is the Target for UV-TLS Inhibition by Caffeine or Proteasome Inhibitors?
3. Experimental Section
3.1. Mouse Cell Culture
3.2. UV Irradiation and Translesion Replication
3.3. Alkaline Sucrose Density Gradient Centrifugation
3.4. Construction of the Mouse Rev3-Expressing Plasmid
3.5. Transfection with the Plasmid and Selection of Stable Transformant Strains
3.6. Survival Assay of UV-Irradiated Cells
4. Conclusions
Acknowledgments
References and Notes
- Lehmann, A.R.; Niimi, A.; Ogi, T.; Brown, S.; Sabbioneda, S.; Wing, J.F.; Kannouche, P.L.; Green, C.M. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst.) 2007, 6, 891–899. [Google Scholar]
- Prakash, S.; Prakash, L. Translesion DNA synthesis in eukaryotes: A one- or two-polymerase affair. Genes Dev 2002, 16, 1872–1883. [Google Scholar]
- Prakash, S.; Johnson, R.E.; Prakash, L. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu. Rev. Biochem 2005, 74, 317–353. [Google Scholar]
- Takezawa, J.; Ishimi, Y.; Aiba, N.; Yamada, K. Rev1, Rev3, or Rev7 siRNA abolishes ultraviolet light-induced translesion replication in HeLa cells: A comprehensive study using alkaline sucrose density gradient sedimentation. J. Nucleic Acids 2010, 2010, 750296:1–750296:12. [Google Scholar]
- Hirota, K.; Sonoda, E.; Kawamoto, T.; Motegi, A.; Masutani, C.; Hanaoka, F.; Szuts, D.; Iwai, S.; Sale, J.E.; Lehmann, A.; et al. Simultaneous disruption of two DNA polymerases, Polη and Polζ, in avian DT40 cells unmasks the role of Polη in cellular response to various DNA lesions. PLoS Genet 2010, 6, e1001151:1–e1001151:13. [Google Scholar]
- Lawrence, C.W.; Christensen, R.B. Ultraviolet-induced reversion of cyc1 alleles in radiation-sensitive strains of yeast. III. Rev3 mutant strains. Genetics 1979, 92, 397–408. [Google Scholar]
- Gibbs, P.E.; Wang, X.D.; Li, Z.; McManus, T.P.; McGregor, W.G.; Lawrence, C.W.; Maher, V.M. The function of the human homolog of Saccharomyces cerevisiae REV1 is required for mutagenesis induced by UV light. Proc. Natl. Acad. Sci. USA 2000, 97, 4186–4191. [Google Scholar]
- Gibbs, P.E.; McGregor, W.G.; Maher, V.M.; Nisson, P.; Lawrence, C.W. A human homolog of the Saccharomyces cerevisiae REV3 gene, which encodes the catalytic subunit of DNA polymerase ζ. Proc. Natl. Acad. Sci. USA 1998, 95, 6876–6880. [Google Scholar]
- Murakumo, Y.; Roth, T.; Ishii, H.; Rasio, D.; Numata, S.; Croce, C.M.; Fishel, R. A human REV7 homolog that interacts with the polymerase ζ catalytic subunit hREV3 and the spindle assembly checkpoint protein hMAD2. J. Biol. Chem 2000, 275, 4391–4397. [Google Scholar]
- Lawrence, C.W. Cellular roles of DNA polymerase ζ and Rev1 protein. DNA Repair (Amst.) 2002, 1, 425–435. [Google Scholar]
- Li, Z.; Zhang, H.; McManus, T.P.; McCormick, J.J.; Lawrence, C.W.; Maher, V.M. hREV3 is essential for error-prone translesion synthesis past UV or benzo[a]pyrene diol epoxide-induced DNA lesions in human fibroblasts. Mutat. Res 2002, 510, 71–80. [Google Scholar]
- McNally, K.; Neal, J.A.; McManus, T.P.; McCormick, J.J.; Maher, V.M. hRev7, putative subunit of hPolζ, plays a critical role in survival, induction of mutations, and progression through S-phase, of UV(254 nm)-irradiated human fibroblasts. DNA Repair (Amst.) 2008, 7, 597–604. [Google Scholar]
- Diaz, M.; Watson, N.B.; Turkington, G.; Verkoczy, L.K.; Klinman, N.R.; McGregor, W.G. Decreased frequency and highly aberrant spectrum of ultraviolet-induced mutations in the hprt gene of mouse fibroblasts expressing antisense RNA to DNA polymerase ζ. Mol. Cancer Res 2003, 1, 836–847. [Google Scholar]
- Masutani, C.; Kusumoto, R.; Iwai, S.; Hanaoka, F. Mechanisms of accurate translesion synthesis by human DNA polymerase η. EMBO J 2000, 19, 3100–3109. [Google Scholar]
- Cleaver, J.E.; Kraemer, K.H. Xeroderma Pigmentosum and Cockayne Syndrome. In The Metabolic and Molecular Basis of Inherited Disease, 7th ed; Scriver, C.R., Beaudet, A.L., Sly, W.S., Valle, D., Eds.; McGraw-Hill: New York, NY, USA, 1995; pp. 4393–4419. [Google Scholar]
- Prakash, S.; Johnson, R.E.; Washington, M.T.; Haracska, L.; Kondratick, C.M.; Prakash, L. Role of yeast and human DNA polymerase η in error-free replication of damaged DNA. Cold Spring Harb. Symp. Quant. Biol 2000, 65, 51–59. [Google Scholar]
- Masutani, C.; Kusumoto, R.; Yamada, A.; Dohmae, N.; Yokoi, M.; Yuasa, M.; Araki, M.; Iwai, S.; Takio, K.; Hanaoka, F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature 1999, 399, 700–704. [Google Scholar]
- Masutani, C.; Kusumoto, R.; Yamada, A.; Yuasa, M.; Araki, M.; Nogimori, T.; Yokoi, M.; Eki, T.; Iwai, S.; Hanaoka, F. Xeroderma pigmentosum variant: From a human genetic disorder to a novel DNA polymerase. Cold Spring Harb. Symp. Quant. Biol 2000, 65, 71–80. [Google Scholar]
- Matsuda, T.; Bebenek, K.; Masutani, C.; Hanaoka, F.; Kunkel, T.A. Low fidelity DNA synthesis by human DNA polymerase-η. Nature 2000, 404, 1011–1013. [Google Scholar]
- Bebenek, K.; Matsuda, T.; Masutani, C.; Hanaoka, F.; Kunkel, T.A. Proofreading of DNA polymerase η-dependent replication errors. J. Biol. Chem 2001, 276, 2317–2320. [Google Scholar]
- Shachar, S.; Ziv, O.; Avkin, S.; Adar, S.; Wittschieben, J.; Reissner, T.; Chaney, S.; Friedberg, E.C.; Wang, Z.; Carell, T.; et al. Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J 2009, 28, 383–393. [Google Scholar]
- Hicks, J.K.; Chute, C.L.; Paulsen, M.T.; Ragland, R.L.; Howlett, N.G.; Gueranger, Q.; Glover, T.W.; Canman, C.E. Differential roles for DNA polymerases η, ζ, and Rev1 in lesion bypass of intrastrand versus interstrand DNA cross-links. Mol. Cell Biol 2010, 30, 1217–1230. [Google Scholar]
- Yoon, J.H.; Prakash, L.; Prakash, S. Highly error-free role of DNA polymerase η in the replicative bypass of UV-induced pyrimidine dimers in mouse and human cells. Proc. Natl. Acad. Sci. USA 2009, 106, 18219–18224. [Google Scholar]
- Yoon, J.H.; Prakash, L.; Prakash, S. Error-free replicative bypass of (6–4) photoproducts by DNA polymerase ζ in mouse and human cells. Genes Dev 2010, 24, 123–128. [Google Scholar]
- Lehmann, A.R.; Kirk-Bell, S.; Arlett, C.F.; Paterson, M.C.; Lohman, P.H.; de Weerd-Kastelein, E.A.; Bootsma, D. Xeroderma pigmentosum cells with normal levels of excision repair have a defect in DNA synthesis after UV-irradiation. Proc. Natl. Acad. Sci. USA 1975, 72, 219–223. [Google Scholar]
- Lehmann, A.R. Postreplication repair of DNA in ultraviolet-irradiated mammalian cells. J. Mol. Biol 1972, 66, 319–337. [Google Scholar]
- Lehmann, A.R. Postreplication repair of DNA in mammalian cells. Life Sci 1974, 15, 2005–2016. [Google Scholar]
- Yamada, K.; Kameyama, Y.; Inoue, S. An improved method of alkaline sucrose density gradient sedimentation to detect less than one lesion per 1 mb DNA. Mutat. Res 1996, 364, 125–131. [Google Scholar]
- Yamada, K.; Takezawa, J.; Ezaki, O. Translesion replication in cisplatin-treated xeroderma pigmentosum variant cells is also caffeine-sensitive: Features of the error-prone DNA polymerase(s) involved in UV-mutagenesis. DNA Repair (Amst.) 2003, 2, 909–924. [Google Scholar]
- Takezawa, J.; Ishimi, Y.; Yamada, K. Proteasome inhibitors remarkably prevent translesion replication in cancer cells but not normal cells. Cancer Sci 2008, 99, 863–871. [Google Scholar]
- Kajiwara, K.; Nagasawa, H.; Shimizu-Nishikawa, S.; Ookuri, T.; Kimura, M.; Sugaya, E. Molecular characterization of seizure-related genes isolated by differential screening. Biochem. Biophys. Res. Commun 1996, 219, 795–799. [Google Scholar]
- Kajiwara, K.; O-Wang, J.; Sakurai, T.; Yamashita, S.; Tanaka, M.; Sato, M.; Tagawa, M.; Sugaya, E.; Nakamura, K.; Nakao, K.; et al. Sez4 gene encoding an elongation subunit of DNA polymerase ζ is required for normal embryogenesis. Genes Cells 2001, 6, 99–106. [Google Scholar]
- O-Wang, J.; Kajiwara, K.; Kawamura, K.; Kimura, M.; Miyagishima, H.; Koseki, H.; Tagawa, M. An essential role for REV3 in mammalian cell survival: Absence of REV3 induces p53-independent embryonic death. Biochem. Biophys. Res. Commun 2002, 293, 1132–1137. [Google Scholar]
- Zhu, F.; Zhang, M. DNA polymerase ζ: New insight into eukaryotic mutagenesis and mammalian embryonic development. World J. Gastroenterol 2003, 9, 1165–1169. [Google Scholar]
- Gan, G.N.; Wittschieben, J.P.; Wittschieben, B.O.; Wood, R.D. DNA polymerase zeta (pol ζ) in higher eukaryotes. Cell Res 2008, 18, 174–183. [Google Scholar]
- Esposito, G.; Godindagger, I.; Klein, U.; Yaspo, M.L.; Cumano, A.; Rajewsky, K. Disruption of the Rev3l-encoded catalytic subunit of polymerase ζ in mice results in early embryonic lethality. Curr. Biol 2000, 10, 1221–1224. [Google Scholar]
- Bemark, M.; Khamlichi, A.A.; Davies, S.L.; Neuberger, M.S. Disruption of mouse polymerase ζ (Rev3) leads to embryonic lethality and impairs blastocyst development in vitro. Curr. Biol 2000, 10, 1213–1216. [Google Scholar]
- Wittschieben, J.; Shivji, M.K.; Lalani, E.; Jacobs, M.A.; Marini, F.; Gearhart, P.J.; Rosewell, I.; Stamp, G.; Wood, R.D. Disruption of the developmentally regulated Rev3l gene causes embryonic lethality. Curr. Biol 2000, 10, 1217–1220. [Google Scholar]
- Van Sloun, P.P.; Varlet, I.; Sonneveld, E.; Boei, J.J.; Romeijn, R.J.; Eeken, J.C.; De Wind, N. Involvement of mouse Rev3 in tolerance of endogenous and exogenous DNA damage. Mol. Cell Biol 2002, 22, 2159–2169. [Google Scholar]
- Jansen, J.G.; Tsaalbi-Shtylik, A.; Hendriks, G.; Verspuy, J.; Gali, H.; Haracska, L.; de Wind, N. Mammalian polymerase ζ is essential for post-replication repair of UV-induced DNA lesions. DNA Repair (Amst.) 2009, 8, 1444–1451. [Google Scholar]
- Zander, L.; Bemark, M. Immortalized mouse cell lines that lack a functional Rev3 gene are hypersensitive to UV irradiation and cisplatin treatment. DNA Repair (Amst.) 2004, 3, 743–752. [Google Scholar]
- Wittschieben, J.P.; Reshmi, S.C.; Gollin, S.M.; Wood, R.D. Loss of DNA polymerase ζ causes chromosomal instability in mammalian cells. Cancer Res 2006, 66, 134–142. [Google Scholar]
- Avkin, S.; Sevilya, Z.; Toube, L.; Geacintov, N.; Chaney, S.G.; Oren, M.; Livneh, Z. p53 and p21 regulate error-prone DNA repair to yield a lower mutation load. Mol. Cell 2006, 22, 407–413. [Google Scholar]
- Soria, G.; Podhajcer, O.; Prives, C.; Gottifredi, V. P21cip1/waf1 downregulation is required for efficient PCNA ubiquitination after UV irradiation. Oncogene 2006, 25, 2829–2838. [Google Scholar]
- Kaufmann, W.K.; Heffernan, T.P.; Beaulieu, L.M.; Doherty, S.; Frank, A.R.; Zhou, Y.; Bryant, M.F.; Zhou, T.; Luche, D.D.; Nikolaishvili-Feinberg, N.; et al. Caffeine and human DNA metabolism: The magic and the mystery. Mutat. Res 2003, 532, 85–102. [Google Scholar]
- Heffernan, T.P.; Kawasumi, M.; Blasina, A.; Anderes, K.; Conney, A.H.; Nghiem, P. ATR-Chk1 pathway inhibition promotes apoptosis after UV treatment in primary human keratinocytes: Potential basis for the UV protective effects of caffeine. J. Invest. Dermatol 2009, 129, 1805–1815. [Google Scholar]
- Ishimi, Y.; Komamura-Kohno, Y.; Karasawa-Shimizu, K.; Yamada, K. Levels of MCM4 phosphorylation and DNA synthesis in DNA replication block checkpoint control. J. Struct. Biol 2004, 146, 234–241. [Google Scholar]
- Despras, E.; Daboussi, F.; Hyrien, O.; Marheineke, K.; Kannouche, P.L. ATR/Chk1 pathway is essential for resumption of DNA synthesis and cell survival in UV-irradiated XP variant cells. Hum. Mol. Genet 2010, 19, 1690–1701. [Google Scholar]
- Niimi, A.; Brown, S.; Sabbioneda, S.; Kannouche, P.L.; Scott, A.; Yasui, A.; Green, C.M.; Lehmann, A.R. Regulation of proliferating cell nuclear antigen ubiquitination in mammalian cells. Proc. Natl. Acad. Sci. USA 2008, 105, 16125–16130. [Google Scholar]
- Podlaska, A.; McIntyre, J.; Skoneczna, A.; Sledziewska-Gojska, E. The link between 20S proteasome activity and post-replication DNA repair in Saccharomyces cerevisiae. Mol. Microbiol 2003, 49, 1321–1332. [Google Scholar]
- McIntyre, J.; Podlaska, A.; Skoneczna, A.; Halas, A.; Sledziewska-Gojska, E. Analysis of the spontaneous mutator phenotype associated with 20S proteasome deficiency in S. cerevisiae. Mutat. Res 2006, 593, 153–163. [Google Scholar]
- Sivak, A.; Rudenko, L.; Teague, L.G. Variations among species and cell types in the effects of caffeine on mutagen-induced cytotoxicity and postreplication repair of DNA. Environ. Mutagen 1982, 4, 143–162. [Google Scholar]
- Johansson, F.; Lagerqvist, A.; Filippi, S.; Palitti, F.; Erixon, K.; Helleday, T.; Jenssen, D. Caffeine delays replication fork progression and enhances UV-induced homologous recombination in Chinese hamster cell lines. DNA Repair (Amst.) 2006, 5, 1449–1458. [Google Scholar]
- We are using the following rules. Human gene symbols are italicized, with all letters in uppercase. Rodent gene symbols are italicized, with only the first letter in uppercase and the remaining letters in lowercase. Protein designations are not italicized, with all letters in uppercase.
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Takezawa, J.; Aiba, N.; Kajiwara, K.; Yamada, K. Caffeine Abolishes the Ultraviolet-Induced REV3 Translesion Replication Pathway in Mouse Cells. Int. J. Mol. Sci. 2011, 12, 8513-8529. https://doi.org/10.3390/ijms12128513
Takezawa J, Aiba N, Kajiwara K, Yamada K. Caffeine Abolishes the Ultraviolet-Induced REV3 Translesion Replication Pathway in Mouse Cells. International Journal of Molecular Sciences. 2011; 12(12):8513-8529. https://doi.org/10.3390/ijms12128513
Chicago/Turabian StyleTakezawa, Jun, Naomi Aiba, Kagemasa Kajiwara, and Kouichi Yamada. 2011. "Caffeine Abolishes the Ultraviolet-Induced REV3 Translesion Replication Pathway in Mouse Cells" International Journal of Molecular Sciences 12, no. 12: 8513-8529. https://doi.org/10.3390/ijms12128513



