Detecting and Discriminating Shigella sonnei Using an Aptamer-Based Fluorescent Biosensor Platform
Abstract
:1. Introduction
2. Results
2.1. Whole Bacteria Cell SELEX for the Isolation of S. sonnei Specific DNA Aptamers
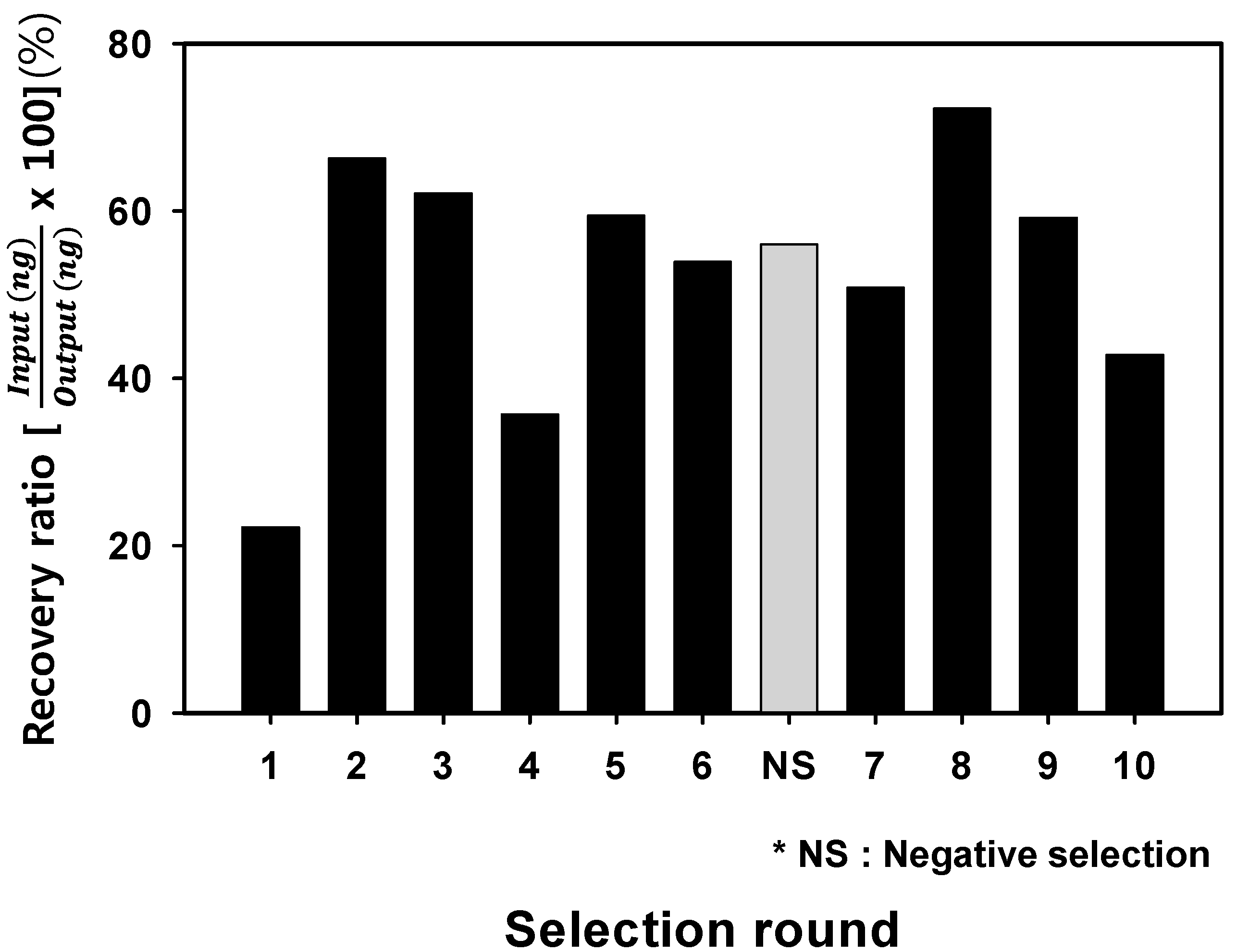
2.2. Analysis of Optimal Enrichment States for Isolating Aptamers
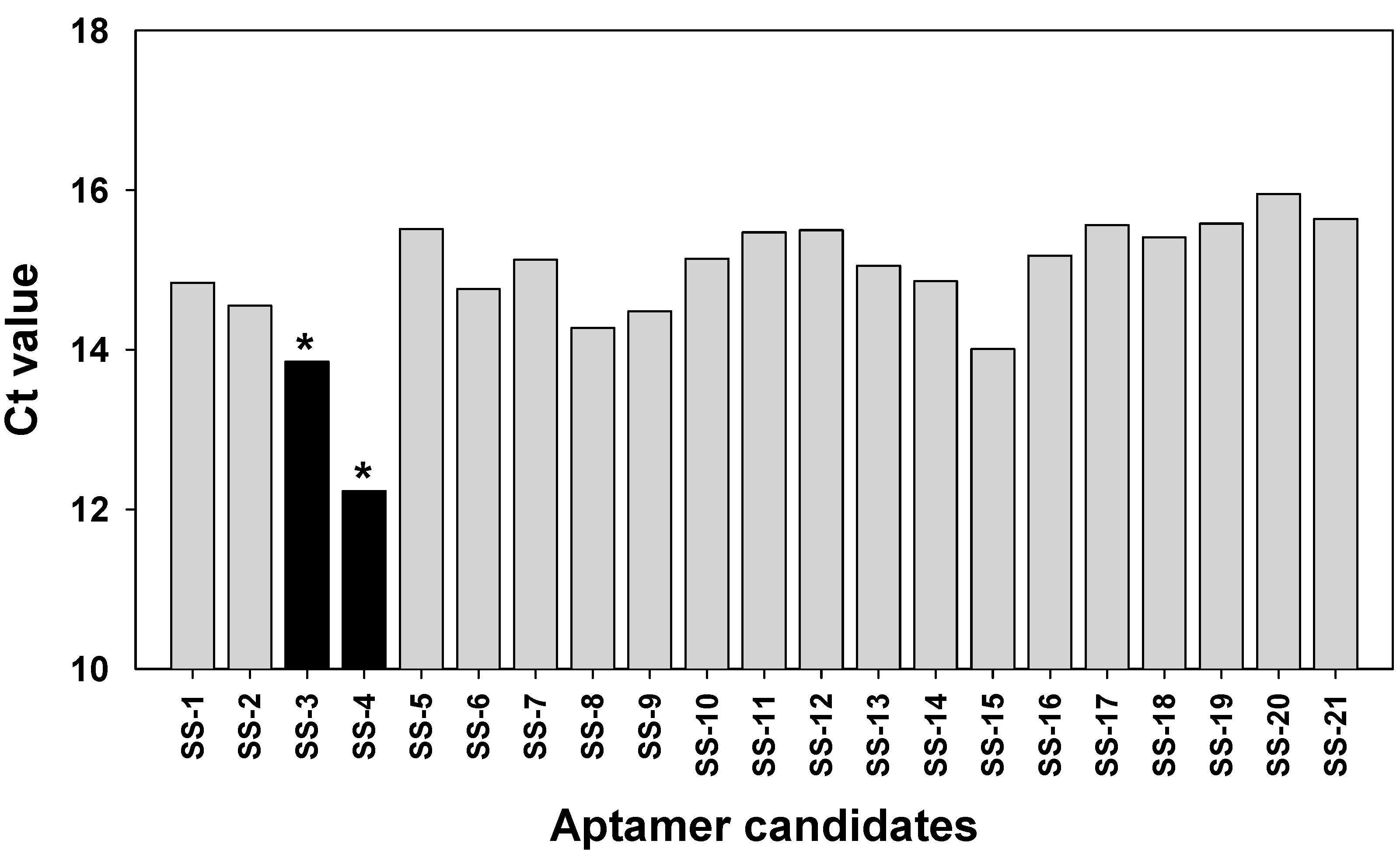
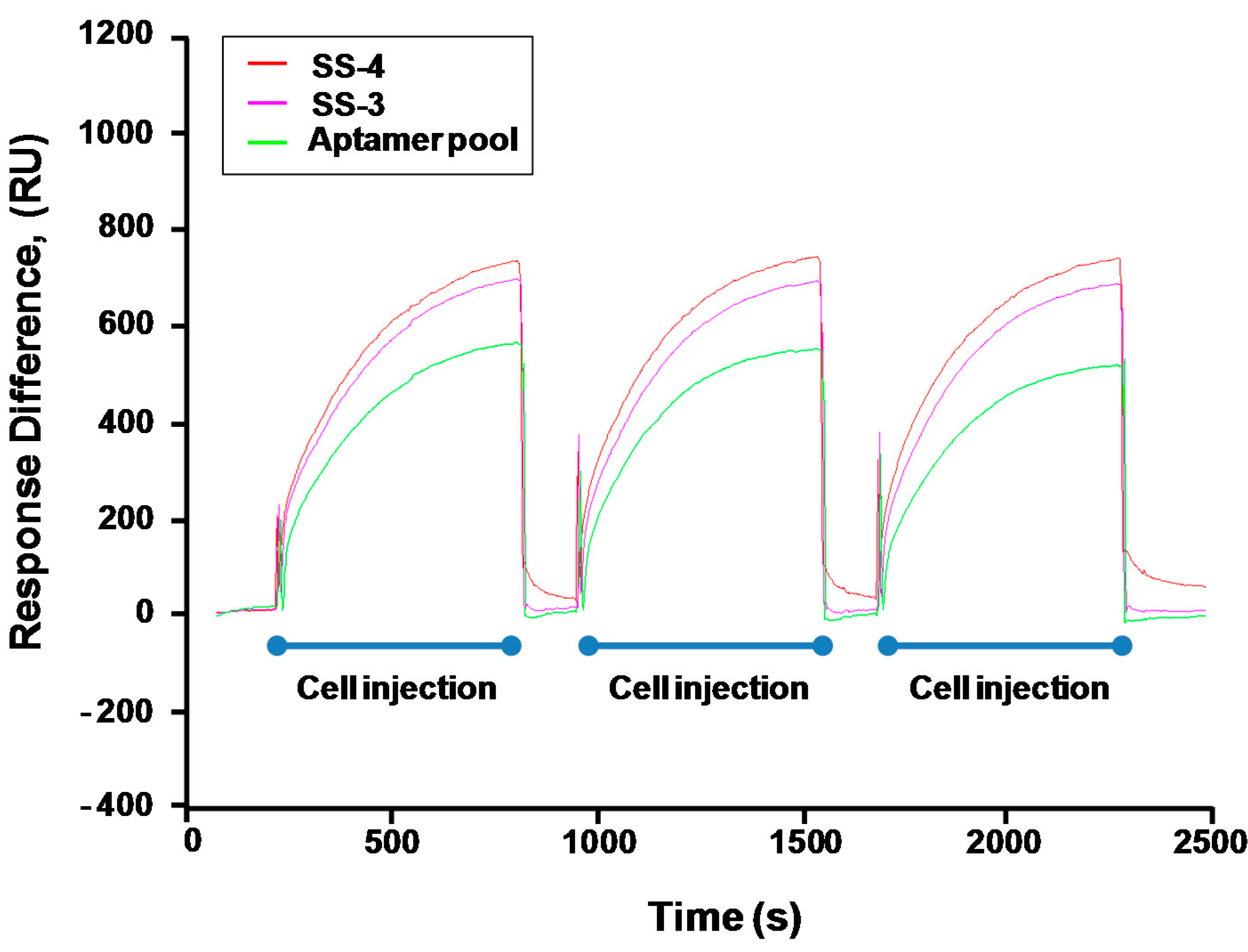
2.3. Binding Test of Aptamer Candidates by Post-SELEX
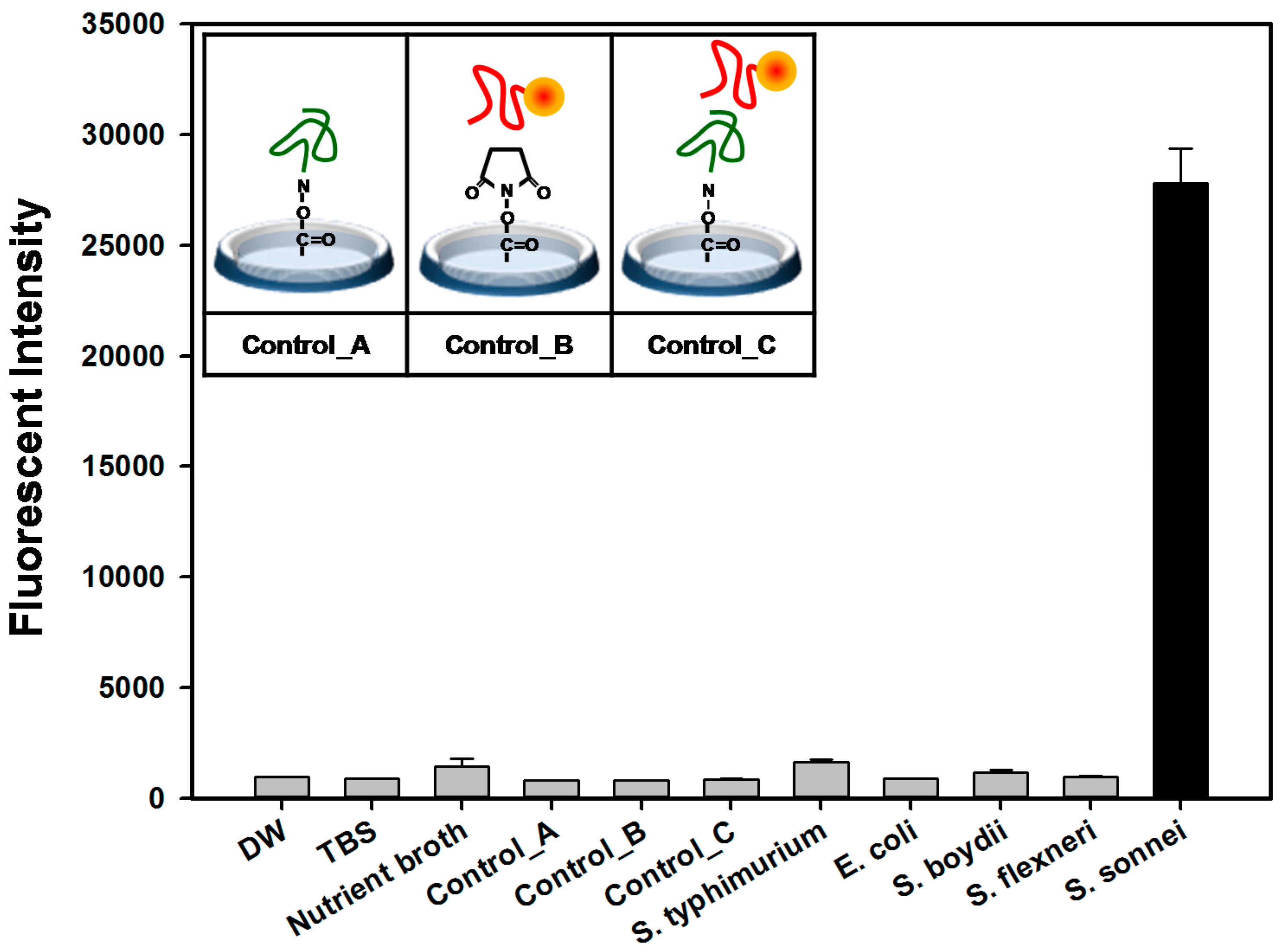
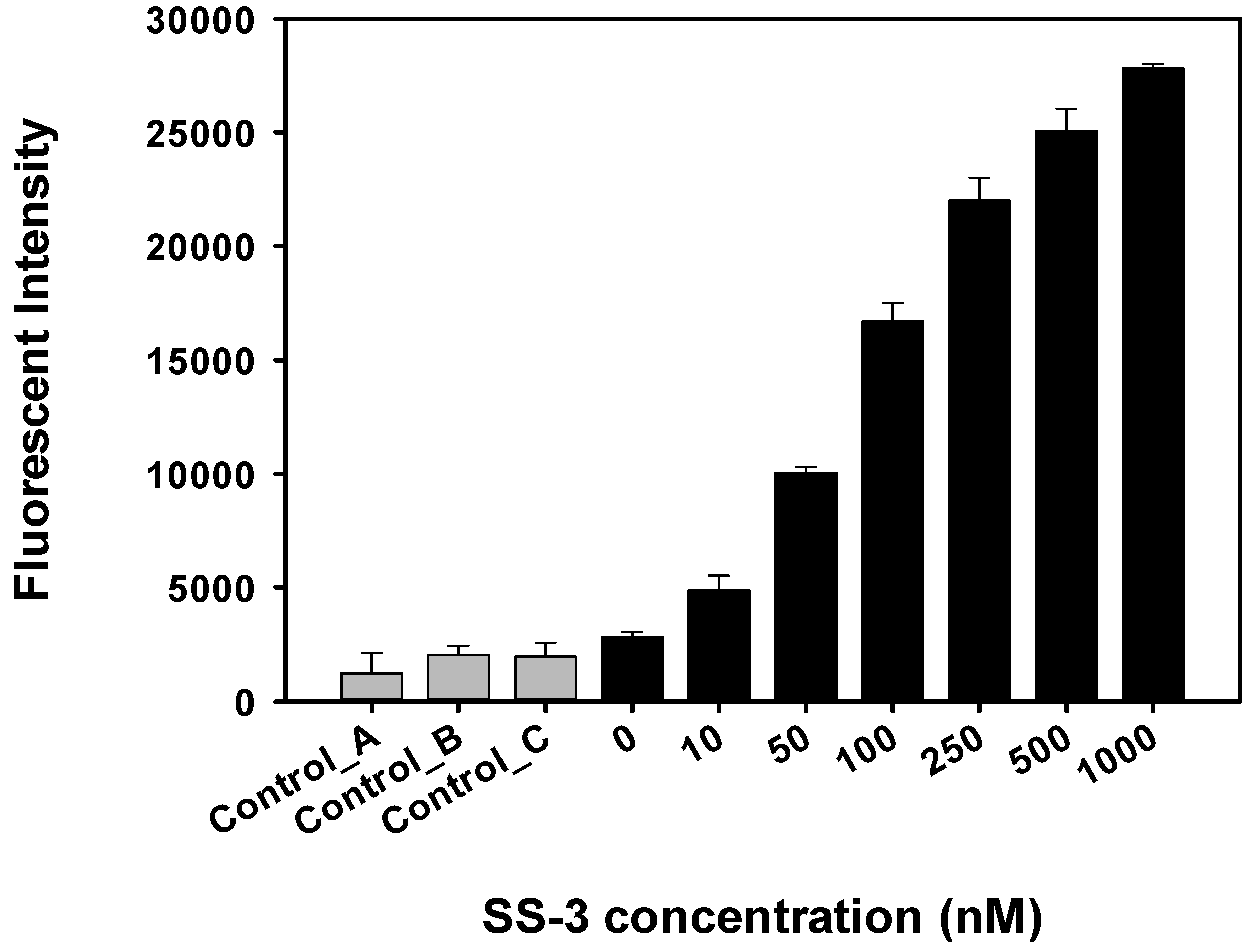
2.4. Detection and Discrimination of S. sonnei by Aptamer-Based Fluorescent Biosensor
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Cell Maintenance
4.2. Whole SELEX Pprocedure
4.3. Real-Time PCR Analysis
4.4. Post-SELEX for the Binding Affinity Test
4.5. Binding Activity of Selected Aptamers
4.6. Sensitivity Detecting of S. sonnei by Aptamer-Based Fluorescent Biosensor Assay
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Campilongo, R.; Di Martino, M.L.; Marcocci, L.; Pietrangeli, P.; Leuzzi, A.; Grossi, M.; Casalino, M.; Nicoletti, M.; Micheli, G.; Colonna, B.; et al. Molecular and functional profiling of the polyamine content in enteroinvasive E. coli: Looking into the gap between commensal E. coli and harmful Shigella. PLoS ONE 2014, 9, e106589. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Oza, S.; Hogan, D.; Perin, J.; Rudan, I.; Lawn, J.E.; Cousens, S.; Mathers, C.; Black, R.E. Global, regional, and national causes of child mortality in 2000–2013, with projections to inform post-2015 priorities: An updated systematic analysis. Lancet 2015, 385, 430–440. [Google Scholar] [CrossRef]
- Kotloff, K.L. Bacterial diarrheal pathogens. Adv. Pediatr. Infect. Dis. 1999, 14, 219–267. [Google Scholar] [PubMed]
- Lew, J.F.; Swerdlow, D.L.; Dance, M.E.; Griffin, P.M.; Bopp, C.A.; Gillenwater, M.J.; Mercatante, T.; Glass, R.I. An outbreak of shigellosis aboard a cruise ship caused by a multiple-antibiotic-resistant strain of Shigella flexneri. Am. J. Epidemiol. 1991, 134, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Gaynor, K.; Park, S.Y.; Kanenaka, R.; Colindres, R.; Mintz, E.; Ram, P.K.; Kitsutani, P.; Nakata, M.; Wedel, S.; Boxrud, D.; et al. International foodborne outbreak of Shigella sonnei infection in airline passengers. Epidemiol. Infect. 2009, 137, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Das, S.K.; Chisti, M.J.; Malek, M.A.; Salam, M.A.; Ahmed, T.; Faruque, A.S.; Mondal, D. Comparison of clinical and laboratory characteristics of intestinal amebiasis with shigellosis among patients visiting a large urban diarrheal disease hospital in Bangladesh. Am. J. Trop. Med. Hyg. 2013, 89, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, K.B.; McCoy, C.B.; Achi, R. Detection of shigella in lettuce by the use of a rapid molecular assay with increased sensitivity. Braz. J. Microbiol. 2010, 41, 993–1000. [Google Scholar] [CrossRef] [PubMed]
- Hien, B.T.; Scheutz, F.; Cam, P.D.; Serichantalergs, O.; Huong, T.T.; Thu, T.M.; Dalsgaard, A. Diarrheagenic Escherichia coli and Shigella strains isolated from children in a hospital case-control study in Hanoi, Vietnam. J. Clin. Microbiol. 2008, 46, 996–1004. [Google Scholar] [CrossRef] [PubMed]
- Mokhtari, C.; Marchadier, E.; Haim-Boukobza, S.; Jeblaoui, A.; Tesse, S.; Savary, J.; Roque-Afonso, A.M. Comparison of real-time RT-PCR assays for hepatitis E virus RNA detection. J. Clin. Virol. 2013, 58, 36–40. [Google Scholar] [CrossRef] [PubMed]
- Sethabutr, O.; Venkatesan, M.; Yam, S.; Pang, L.W.; Smoak, B.L.; Sang, W.K.; Echeverria, P.; Taylor, D.N.; Isenbarger, D.W. Detection of PCR products of the ipaH gene from Shigella and enteroinvasive Escherichia coli by enzyme linked immunosorbent assay. Diagn. Microbiol. Infect. Dis. 2000, 37, 11–16. [Google Scholar] [CrossRef]
- Huang, C.J.; Knoll, W.; Sessitsch, A.; Dostalek, J. SPR bacterial pathogen biosensor: The importance of fluidic conditions and probing depth. Talanta 2014, 122, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Pierce, S.E.; Bell, R.L.; Hellberg, R.S.; Cheng, C.M.; Chen, K.S.; Williams-Hill, D.M.; Martin, W.B.; Allard, M.W. Detection and Identification of Salmonella enterica, Escherichia coli, and Shigella spp. via PCR-electrospray ionization mass spectrometry: Isolate testing and analysis of food samples. Appl. Environ. Microbiol. 2012, 78, 8403–8411. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Feng, F.; Zhao, L.; Wang, C.; Wang, H.; Tian, M.; Qin, J.; Duan, Y.; He, X. Aptamer/thrombin/aptamer-AuNPs sandwich enhanced surface plasmon resonance sensor for the detection of subnanomolar thrombin. Biosens. Bioelectron. 2013, 47, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Mazumdar, D.; Lu, Y. A simple and sensitive “dipstick” test in serum based on lateral flow separation of aptamer-linked nanostructures. Angew. Chem. Int. Ed. 2006, 45, 7955–7959. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Wu, C.; Liu, H.; Zou, Y.; Zhang, X.; Kang, H.; Yang, C.J.; Tan, W. An aptamer cross-linked hydrogel as a colorimetric platform for visual detection. Angew. Chem. Int. Ed. 2010, 49, 1052–1056. [Google Scholar] [CrossRef] [PubMed]
- Sekhon, S.S.; Um, H.J.; Shin, W.R.; Lee, S.H.; Min, J.; Ahn, J.Y.; Kim, Y.H. Aptabody-aptatope interactions in aptablotting Assays. Nanoscale 2017. [Google Scholar] [CrossRef]
- Lee, K.A.; Ahn, J.Y.; Lee, S.H.; Singh Sekhon, S.; Kim, D.G.; Min, J.; Kim, Y.H. Aptamer-based sandwich assay and its clinical outlooks for detecting lipocalin-2 in hepatocellular carcinoma (HCC). Sci. Rep. 2015, 5, 10897. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Ahn, J.Y.; Lee, K.A.; Um, H.J.; Sekhon, S.S.; Sun, P.; Min, J.; Kim, Y.H. Analytical bioconjugates, aptamers, enable specific quantitative detection of Listeria monocytogenes. Biosens. Bioelectron. 2015, 68, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Sekhon, S.S.; Kim, S.G.; Lee, S.H.; Jang, A.; Min, J.; Ahn, J.Y.; Kim, Y.H. Advances in pathogen-associated molecules detection using Aptamer based biosensors. Mol. Cell. Toxicol. 2013, 9, 311–317. [Google Scholar] [CrossRef]
- Sekhon, S.S.; Lee, S.H.; Lee, K.A.; Min, J.; Lee, B.T.; Kim, K.W.; Ahn, J.Y.; Kim, Y.H. Defining the copper binding aptamotif and aptamer integrated recovery platform (AIRP). Nanoscale 2017, 9, 2883–2894. [Google Scholar] [CrossRef] [PubMed]
- Shiao, Y.S.; Chiu, H.H.; Wu, P.H.; Huang, Y.F. Aptamer-Functionalized Gold Nanoparticles As Photoresponsive Nanoplatform for Co-Drug Delivery. ACS Appl. Mater. Interfaces 2014, 6, 21832–21841. [Google Scholar] [CrossRef] [PubMed]
- Shieh, Y.A.; Yang, S.J.; Wei, M.F.; Shieh, M.J. Aptamer-based tumor-targeted drug delivery for photodynamic therapy. ACS Nano 2010, 4, 1433–1442. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Duan, S.; Wu, S.; Huang, Y.; Chen, X.; Wang, Z. Selection, identification, and application of dual DNA aptamers against Shigella sonnei. Anal. Meth. 2015, 7, 3625–3631. [Google Scholar] [CrossRef]
- Ahmed, A.M.; Shimamoto, T. Isolation and molecular characterization of Salmonella enterica, Escherichia coli O157:H7 and Shigella spp. from meat and dairy products in Egypt. Int. J. Food Microbiol. 2014, 168, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Fellner, L.; Bechtel, N.; Witting, M.A.; Simon, S.; Schmitt-Kopplin, P.; Keim, D.; Scherer, S.; Neuhaus, K. Phenotype of htgA (mbiA), a recently evolved orphan gene of Escherichia coli and Shigella, completely overlapping in antisense to yaaW. FEMS Microbiol. Lett. 2014, 350, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, J.G.; Wang, L.; Reeves, P.R. Comparison of O-antigen gene clusters of Escherichia coli (Shigella) sonnei and Plesiomonas shigelloides O17: Sonnei gained its current plasmid-borne O-antigen genes from P. shigelloides in a recent event. Infect. Immun. 2000, 68, 6056–6061. [Google Scholar] [CrossRef] [PubMed]
- Pupo, G.M.; Lan, R.; Reeves, P.R. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl. Acad. Sci. USA 2000, 97, 10567–10572. [Google Scholar] [CrossRef] [PubMed]
- Van De Verg, L.L.; Bendiuk, N.O.; Kotloff, K.; Marsh, M.M.; Ruckert, J.L.; Puryear, J.L.; Taylor, D.N.; Hartman, A.B. Cross-reactivity of Shigella flexneri serotype 2a O antigen antibodies following immunization or infection. Vaccine 1996, 14, 1062–1068. [Google Scholar] [CrossRef]
- Baig, I.A.; Moon, J.Y.; Lee, S.C.; Ryoo, S.W.; Yoon, M.Y. Development of ssDNA aptamers as potent inhibitors of Mycobacterium tuberculosis acetohydroxyacid synthase. Biochim. Biophys. Acta 2015, 1854, 1338–1350. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. In Current Protocols in Bioinformatics; Andreas, D.B., Ed.; Wiley: New York, NY, USA, 2002; Chapter 2, Unit 2.3. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |

| Clone | Selected Sequence | Size (bp) | Secondary Structure | Chemical Modification | KD (nM) |
|---|---|---|---|---|---|
| SS-3 | ATACCAGCTTATTCAATT CCATGGTCCCTCGTGTTTAT TATGTTGTCTGAACTGGCTG AGATTGCACTTACTATCT | 76 |  | Cy5 for detection probe | 39.32 ± 5.02 |
| SS-4 | ATACCAGCTTATTCAATT CCACACATACCAAAAACACA GCACACTTCATCAATTTCACG AGATTGCACTTACTATCT | 77 |  | Amine for capture probe | 15.89 ± 1.77 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, M.-S.; Sekhon, S.S.; Shin, W.-R.; Kim, H.C.; Min, J.; Ahn, J.-Y.; Kim, Y.-H. Detecting and Discriminating Shigella sonnei Using an Aptamer-Based Fluorescent Biosensor Platform. Molecules 2017, 22, 825. https://doi.org/10.3390/molecules22050825
Song M-S, Sekhon SS, Shin W-R, Kim HC, Min J, Ahn J-Y, Kim Y-H. Detecting and Discriminating Shigella sonnei Using an Aptamer-Based Fluorescent Biosensor Platform. Molecules. 2017; 22(5):825. https://doi.org/10.3390/molecules22050825
Chicago/Turabian StyleSong, Myeong-Sub, Simranjeet Singh Sekhon, Woo-Ri Shin, Hyung Cheol Kim, Jiho Min, Ji-Young Ahn, and Yang-Hoon Kim. 2017. "Detecting and Discriminating Shigella sonnei Using an Aptamer-Based Fluorescent Biosensor Platform" Molecules 22, no. 5: 825. https://doi.org/10.3390/molecules22050825






