Arginine: New Insights into Growth Performance and Urinary Metabolomic Profiles of Rats
Abstract
:1. Introduction
2. Results
2.1. Effect of Arginine Supplementation on Growth Performance
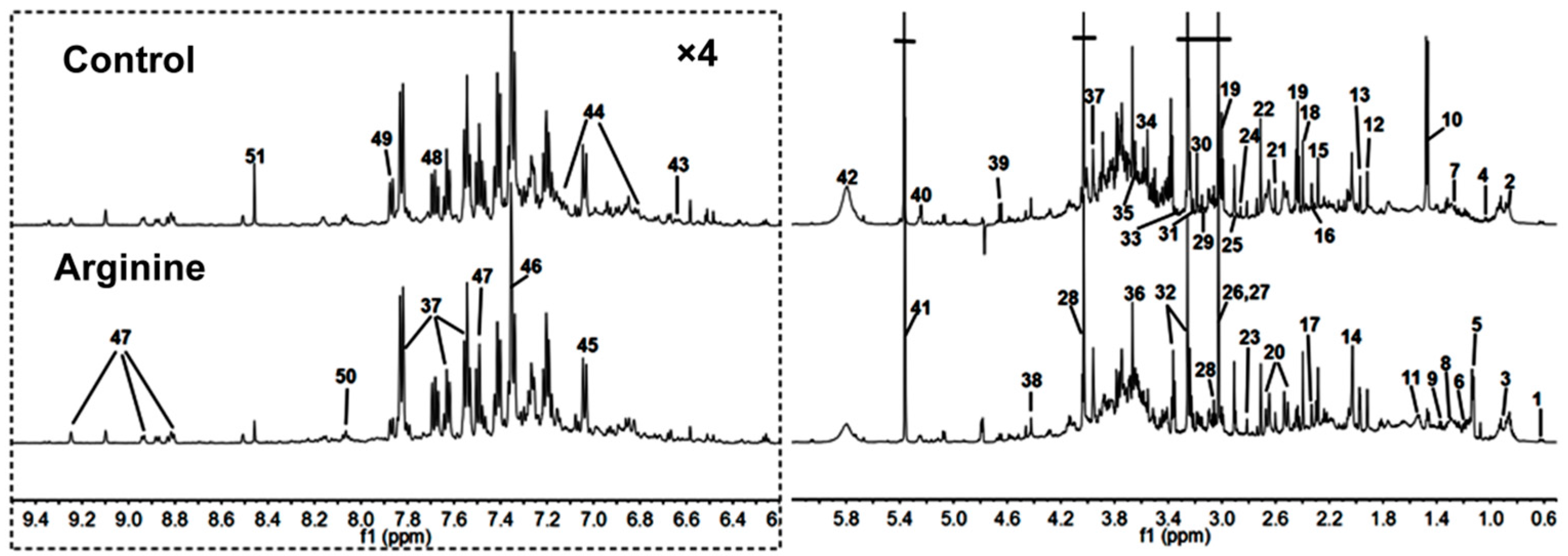
2.2. 1H-NMR Spectra of Urine Samples
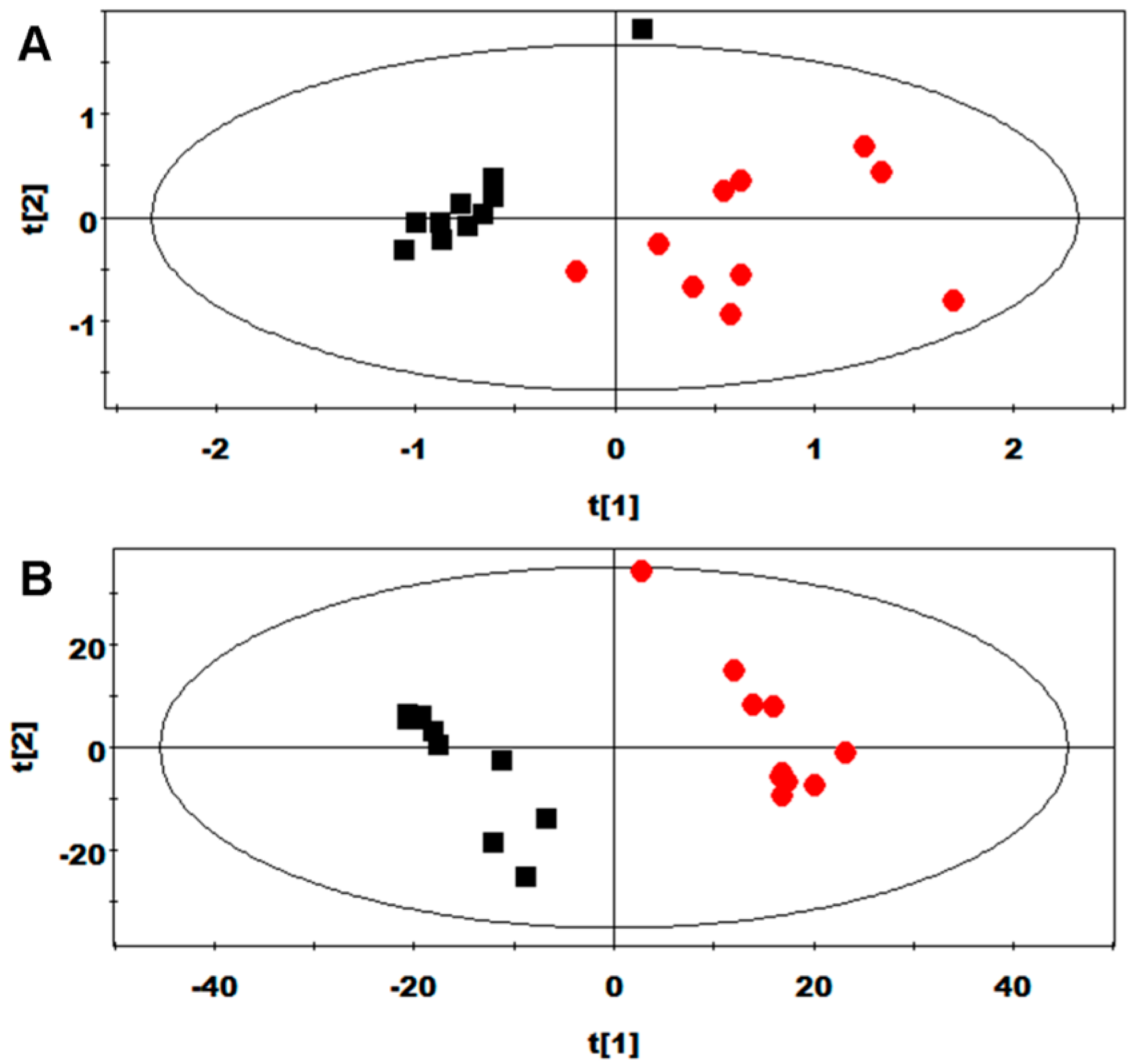
2.3. Multivariate Data Analysis of NMR Data
3. Discussion
4. Materials and Methods
4.1. Animal Experiments and Sample Collection
4.2. Sample Preparation and NMR Spectroscopy
4.3. NMR Spectroscopic Processes and Analyses
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kim, S.W.; Wu, G. Regulatory role for amino acids in mammary gland growth and milk synthesis. Amino Acids 2009, 37, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Bazer, F.W.; Davis, T.A.; Jaeger, L.A.; Johnson, G.A.; Kim, S.W.; Knabe, D.A.; Meininger, C.J.; Spencer, T.E.; Yin, Y.L. Important roles for the arginine family of amino acids in swine nutrition and production. Livest. Sci. 2007, 112, 8–22. [Google Scholar] [CrossRef]
- Hoang, H.H.; Padgham, S.V.; Meininger, C.J. l-Arginine, tetrahydrobiopterin, nitric oxide and diabetes. Curr. Opin. Clin. Nutr. Metab. Care 2013, 16, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Satterfield, M.C.; Dunlap, K.A.; Keisler, D.H.; Bazer, F.W.; Wu, G. Arginine nutrition and fetal brown adipose tissue development in diet-induced obese sheep. Amino Acids 2012, 43, 1593–1603. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.Y.; Knabe, D.A.; Kim, S.W. Arginine nutrition in neonatal pigs. J. Nutr. 2004, 134, 2783S–2790S. [Google Scholar] [PubMed]
- Grimble, G.K. Adverse gastrointestinal effects of arginine and related amino acids. J. Nutr. 2007, 137, 1693S–1701S. [Google Scholar] [PubMed]
- Yao, K.; Yin, Y.; Chu, W.; Liu, Z.; Deng, D.; Li, T.; Huang, R.; Zhang, J.; Tan, B.; Wang, W.; et al. Dietary arginine supplementation increases mTOR signaling activity in skeletal muscle of neonatal pigs. J. Nutr. 2008, 138, 867–872. [Google Scholar] [PubMed]
- Kim, J.; Burghardt, R.C.; Wu, G.; Johnson, G.A.; Spencer, T.E.; Bazer, F.W. Select nutrients in the ovine uterine lumen. VIII. Arginine stimulates proliferation of ovine trophectoderm cells through MTOR-RPS6K-RPS6 signaling cascade and synthesis of nitric oxide and polyamines. Biol. Reprod. 2011, 84, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Bazer, F.W.; Song, G.; Kim, J.; Erikson, D.W.; Johnson, G.A.; Burghardt, R.C.; Gao, H.; Satterfield, M.C.; Spencer, T.E.; Wu, G. Mechanistic mammalian target of rapamycin (MTOR) cell signaling: Effects of select nutrients and secreted phosphoprotein 1 on development of mammalian conceptuses. Mol. Cell. Endocrinol. 2012, 354, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, J.M.; Liu, Y.; Niu, X.; Surendran, S.; Wu, G. Arginine stimulates cdx2-transformed intestinal epithelial cell migration via a mechanism requiring both nitric oxide and phosphorylation of p70S6 kinase. J. Nutr. 2008, 138, 1652–1657. [Google Scholar] [PubMed]
- Tan, B.; Yin, Y.; Kong, X.; Li, P.; Li, X.; Gao, H.; Li, X.; Huang, R.; Wu, G. l-Arginine stimulates proliferation and prevents endotoxin-induced death of intestinal cells. Amino Acids 2010, 38, 1227–1235. [Google Scholar] [CrossRef] [PubMed]
- Jobgen, W.J.; Meininger, C.J.; Jobgen, S.C.; Li, P.; Lee, M.J.; Smith, S.B.; Spencer, T.E.; Fried, S.K.; Wu, G. Dietary l-arginine supplementation reduces white-fat gain and enhances skeletal muscle and brown fat masses in diet-induced obese rats. J. Nutr. 2009, 139, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Hurt, R.T.; Ebbert, J.O.; Schroeder, D.R.; Croghan, I.T.; Bauer, B.A.; McClave, S.A.; Miles, J.M.; McClain, C.J. l-Arginine for the treatment of centrally obese subjects: A pilot study. J. Diet. Suppl. 2014, 11, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.; Li, X.; Yin, Y.; Wu, Z.; Liu, C.; Tekwe, C.D.; Wu, G. Regulatory roles for l-arginine in reducing white adipose tissue. Front. Biosci. 2012, 17, 2237–2246. [Google Scholar] [CrossRef]
- McKnight, J.R.; Satterfield, M.C.; Jobgen, W.S.; Smith, S.B.; Spencer, T.E.; Meininger, C.J.; McNeal, C.J.; Wu, G. Beneficial effects of l-arginine on reducing obesity: Potential mechanisms and important implications for human health. Amino Acids 2010, 39, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.; Yin, Y.; Liu, Z.; Li, X.; Xu, H.; Kong, X.; Huang, R.; Tang, W.; Shinzato, I.; Smith, S.B.; et al. Dietary l-arginine supplementation increases muscle gain and reduces body fat mass in growing-finishing pigs. Amino Acids 2009, 37, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Yin, Y.; Liu, G.; Yu, X.; Li, Y.; Yang, G.; Li, T.; Wu, G. Effect of dietary arginine supplementation on reproductive performance of mice with porcine circovirus type 2 infection. Amino Acids 2012, 42, 2089–2094. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Bazer, F.W.; Satterfield, M.C.; Li, X.; Wang, X.; Johnson, G.A.; Burghardt, R.C.; Dai, Z.; Wang, J.; Wu, Z. Impacts of arginine nutrition on embryonic and fetal development in mammals. Amino Acids 2013, 45, 241–256. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Yin, Y.; Li, D.; Kim, S.W.; Wu, G. Amino acids and immune function. Br. J. Nutr. 2007, 98, 237–252. [Google Scholar] [CrossRef] [PubMed]
- Bronte, V.; Serafini, P.; Mazzoni, A.; Segal, D.M.; Zanovello, P. l-Arginine metabolism in myeloid cells controls T-lymphocyte functions. Trends Immunol. 2003, 24, 301–305. [Google Scholar] [CrossRef]
- Peranzoni, E.; Marigo, I.; Dolcetti, L.; Ugel, S.; Sonda, N.; Taschin, E.; Mantelli, B.; Bronte, V.; Zanovello, P. Role of arginine metabolism in immunity and immunopathology. Immunobiology 2008, 212, 795–812. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Liu, Y.; Xie, X.; Huang, J.; Hou, Y. Effect of l-arginine on intestinal mucosal immune barrier function in weaned pigs after Escherichia coli LPS challenge. Innate Immun. 2012, 19, 242–252. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Morris, S.M., Jr. Aginine metabolism: Nitric oxide and beyond. Biochem. J. 1998, 336. [Google Scholar] [CrossRef]
- Agostinelli, E. Polyamines and transglutaminase: Biological, clinical, and biotechnological perspectives. Amino Acids 2014, 46, 475–485. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Fang, T.; Yan, T.; Jia, G.; Zhao, H.; Chen, X.; Wang, J.; Xue, B. Metabolomic strategy for the detection of metabolic effects of spermine supplementation in weaned rats. J. Agric. Food Chem. 2014, 62, 9035–9042. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Fang, T.; Yan, T.; Jia, G.; Zhao, H.; Chen, X.; Wu, C.; Wang, J. Systemic responses of weaned rats to spermine against oxidative stress revealed by a metabolomic strategy. RSC Adv. 2014, 4, 56766–56778. [Google Scholar] [CrossRef]
- Liu, G.; Yan, T.; Fang, T.; Jia, G.; Chen, X.; Zhao, H.; Wang, J.; Wu, C. Nutrimetabolomic analysis provides new insights into spermine-induced ileum-system alterations for suckling rats. RSC Adv. 2015, 5, 48769–48778. [Google Scholar] [CrossRef]
- Liu, G.M.; Xiao, L.; Fang, T.T.; Cai, Y.M.; Jia, G.; Zhao, H.; Wang, J.; Chen, X.L.; Wu, C.M. Pea fiber and wheat bran fiber show distinct metabolic profiles in rats as investigated by a 1H-NMR-based metabollomic approach. PLoS ONE 2014, 9, e115561. [Google Scholar]
- He, Q.; Kong, X.; Wu, G.; Ren, P.; Tang, H.; Hao, F.; Huang, R.; Li, T.; Tan, B.; Li, P.; et al. Metabolomic analysis of the response of growing pigs to dietary l-arginine supplementation. Amino Acids 2009, 37, 199–208. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Tang, H.; Ren, P.; Kong, X.; Wu, G.; Yin, Y.; Wang, Y. Dietary supplementation with l-arginine partially conteracts serum metabonome induced by weaning stress in piglets. J. Proteome Res. 2011, 10, 5214–5221. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.M.; Xiao, L.; Cao, W.; Fang, T.T.; Jia, G.; Chen, X.L.; Zhao, H.; Wu, C.M.; Wang, J. Changes in the metabolome of rats after exposure to arginine and N-carbamylglutamate in combination with diquat a compound that causes oxidative stress assessed by 1H-NMR spectroscopy. Food Funct. 2016, 7, 964–974. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.M.; Heaney, R.P.; Rafferty, K. Decline in muscle mass with age in woman: A longitudinal study using an indirect measure. Metabolism 2002, 51, 935–939. [Google Scholar] [CrossRef] [PubMed]
- Huynh, N.N.; Chin-Dusting, J. Amino acids, arginase and nitric oxide in vascular health. Clin. Exp. Pharmacol. Physiol. 2006, 33. [Google Scholar] [CrossRef] [PubMed]
- Meijer, A.J.; Verhoeven, A.J. N-acetylglutamate and urea synthesis. Biochem. J. 1984, 56, 559–560. [Google Scholar] [CrossRef]
- Arias-Barrau, E.; Olivera, E.R.; Luengo, J.M.; Fernández, C.; Galán, B.; García, J.L.; Díaz, E.; Miñambres, B. The homogentisate pathway: A central catabolic pathway involved in the degradation of l-phenylalanine, l-tyrosine, and 3-hydroxyphenylacetate in Pseudomonas putida. J. Bacteriol. 2004, 186, 5062–5077. [Google Scholar] [CrossRef] [PubMed]
- Rechner, A.R.; Kuhnle, G.; Hu, H.; Roedig-Penman, A.; van den Braak, M.H.; Moore, K.P.; Rice-Evans, C.A. The metabolism of dietary polyphenols and the relative to circulating levels of conjugated metabolites. Free Radic. Res. 2002, 36, 1229–1241. [Google Scholar] [CrossRef] [PubMed]
- Bohus, E.; Coen, M.; Keun, H.C.; Ebbels, T.M.; Beckonert, O.; Lindon, J.C.; Holmes, E.; Noszál, B.; Nicholson, J.K. Temporal metabonomic modeling of l-arginine-induced exocrine pancreatitis. J. Proteome Res. 2008, 7, 4435–4445. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Foxall, P.J.D. 750MHz 1H-13C-NMR spectroscopy of human blood plasma. Anal. Chem. 1995, 67, 793–811. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.W.M. Metabolite profiling by one- and two-dimensional NMR analysis of complex mixtures. Prog. Nucl. Magn. Reson. Spectrosc. 1996, 28, 161–219. [Google Scholar] [CrossRef]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. HMDB 3.0-The Human Metabolome Database in 2013. Nucleic Acids Res. 2013, 41, D801–D807. [Google Scholar] [CrossRef] [PubMed]
- Trygg, J.; Wold, S. Orthogonal projections to latent structures (O-PLS). J. Chemom. 2002, 16, 119–128. [Google Scholar] [CrossRef]
- Cloarec, O.; Dumas, M.E.; Trygg, J.; Craig, A.; Barton, R.H.; Lindon, J.C.; Nicholson, J.K.; Holmes, E. Evaluation of the orthogonal projection on latent structure model limitations caused by chemical shift variability and improved visualization of biomarker changes in 1H-NMR spectroscopic metabonomic studies. Anal. Chem. 2005, 77, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Lindgren, F.; Hansen, B.; Karcher, W.; SjÖstrÖm, M.; Eriks, L. Model validation by permutation tests: Applications to variable selection. J. Chemom. 1996, 10, 521–532. [Google Scholar] [CrossRef]
- Sample Availability: Samples of arginine are available from the authors.

| Parameters | Control | Arginine | SEM | p Value |
|---|---|---|---|---|
| Mean daily body weight gain (g) | 2.95 | 3.60 | 0.16 | 0.03 |
| Mean daily food intake (g) | 21.95 | 23.91 | 0.59 | 0.14 |
| Food intake/body weight gain ratio | 8.86 | 7.19 | 1.04 | 0.25 |
| Keys | Metabolites | Moieties | δ 1H (ppm) and Multiplicity |
|---|---|---|---|
| 1 | Bile acids | CH3 | 0.64 (m), 0.75 (m) |
| 2 | α-Hydroxy-iso-valerate | δCH3, CH3 | 0.83 (d), 0.97 (d) |
| 3 | α-Hydroxybutyrate | CH3 | 0.89 (t) |
| 4 | Propionate | CH3 | 1.06 (t) |
| 5 | Isobutyrate | CH3 | 1.13 (d) |
| 6 | Ethanol | CH3 | 1.19 (t) |
| 7 | Methylmalonate | CH3, CH | 1.25 (d), 3.75 (m) |
| 8 | α-Hydroxy-n-valerate | CH3, γCH2 | 0.89 (t), 1.31 (m) |
| 9 | Lactate | αCH, βCH3 | 4.14 (q), 1.33 (d) |
| 10 | Alanine | αCH, βCH3 | 3.77 (q), 1.47 (d) |
| 11 | Citrulline | γCH2, βCH2 | 1.56 (m), 1.82 (m) |
| 12 | Acetate | CH3 | 1.92 (s) |
| 13 | Acetamide | CH3 | 1.99 (s) |
| 14 | N-Acetylglutamate | βCH2, γCH2, CH3 | 2.06 (m), 1.87 (m), 2.03 (s) |
| 15 | Acetone | CH3 | 2.24 (s) |
| 16 | Acetoacetate | CH3 | 2.28 (s) |
| 17 | Pyruvate | CH3 | 2.33 (s) |
| 18 | Succinate | CH2 | 2.40 (s) |
| 19 | α-Ketoglutarate | βCH2, γCH2 | 2.45 (t), 3.01 (t) |
| 20 | Citrate | CH2 | 2.54 (d), 2.68 (d) |
| 21 | Methylamine | CH3 | 2.61 (s) |
| 22 | Dimethylamine | CH3 | 2.71 (s) |
| 23 | Methylguanidine | CH3 | 2.81 (s) |
| 24 | Trimethylamine | CH3 | 2.88 (s) |
| 25 | Dimethylglycine | CH3 | 2.93 (s) |
| 26 | Creatine | CH3, CH2 | 3.04 (s), 3.93 (s) |
| 27 | Creatinine | CH3, CH2 | 3.04 (s), 4.05 (s) |
| 28 | Ornithine | CH2 | 3.06 (t) |
| 29 | Ethanolamine | CH2 | 3.11 (t) |
| 30 | Malonate | CH2 | 3.15 (s) |
| 31 | Choline | OCH2, NCH2, N(CH3)3 | 4.07 (t), 3.53 (t), 3.21 (s) |
| 32 | Taurine | -CH2-S, -CH2-NH2 | 3.27 (t), 3.43 (t) |
| 33 | TMAO a | CH3 | 3.27 (s) |
| 34 | Glycine | CH2 | 3.57 (s) |
| 35 | Sarcosine | CH2 | 3.6 (s) |
| 36 | Phenylacetyglycine | 2,6-CH, 3,5-CH, 7-CH, 10-CH | 7.30 (t), 7.36 (m), 7.42 (m), 3.67 (s) |
| 37 | Hippurate | CH2, 3,5-CH, 4-CH, 2,6-CH | 3.97 (d), 7.55 (t), 7.63 (t), 7.84 (d) |
| 38 | N-Methylnicotinamide | CH3, 5-CH, 4-CH, 6-CH, CH2 | 4.42 (s), 8.21 (d), 8.87 (d), 8.93 (d), 9.24 (s) |
| 39 | β-Glucose | 1-CH, 2-CH, 3-CH, 4-CH, 5-CH, 6-CH | 4.47 (d), 3.25 (dd), 3.49 (t), 3.41 (dd), 3.46 (m), 3.73 (dd), 3.90 (dd) |
| 40 | α-Glucose | 1-CH, 2-CH, 3-CH, 4-CH, 5-CH, 6-CH | 5.24 (d), 3.54 (dd), 3.71 (dd), 3.42 (dd), 3.84 (m), 3.78 (m) |
| 41 | Allantoin | CH | 5.39 (s) |
| 42 | Urea | NH2 | 5.82 (s) |
| 43 | Homogentisate | 6-CH, 5-CH | 6.67 (d), 6.82 (d), |
| 44 | p-Hydroxyphenylacetate | 6-CH, 2-CH, 3,5-CH | 3.6 (s), 6.85 (d), 7.15 (d) |
| 45 | m-Hydroxyphenylacetate | 6-CH, 4-CH, 3-CH | 6.92 (m), 7.04 (d), 7.26 (t) |
| 46 | Indoxyl sulfate | 4-CH, 5-CH, 6-CH, 7-CH, CH | 7.51 (m), 7.22 (m), 7.28 (m), 7.71 (m), 7.37 (s) |
| 47 | Nicotinamide | 2-CH, 4-CH, 5-CH, 6-CH | 8.94 (d), 8.61 (dd), 8.25 (m), 7.5 (dd) |
| 48 | 4-Aminohippurate | CH2, CH | 7.6 (d), 6.8 (d), 3.9 (d) |
| 49 | Benzoate | 2,6-CH, 3,5-CH, 4-CH | 7.87 (d),7.49 (dd), 7.56 (t) |
| 50 | Trigonelline | 2-CH, 4-CH, 6-CH, 5-CH, CH3 | 9.09 (s), 8.85 (m), 8.81 (dd), 8.07 (m), 4.44 (s) |
| 51 | Formate | CH | 8.46 (s) |
| Metabolite | OPLS-DA Coefficient (r) a | p Value b |
|---|---|---|
| B (vs. A) | B (vs. A) | |
| 4-Aminohippurate (48) | 0.798 | <0.05 |
| Acetamide (13) | −0.661 | <0.05 |
| Acetate (12) | 0.760 | <0.05 |
| β-Glucose (39) | −0.603 | <0.05 |
| Citrulline (11) | −0.889 | <0.05 |
| Creatine (26) | 0.794 | <0.05 |
| Creatinine (27) | 0.915 | <0.05 |
| Ethanol (6) | −0.932 | <0.05 |
| Ethanolamine (29) | 0.618 | <0.05 |
| Formate (51) | 0.660 | <0.05 |
| Glycine (34) | −0.832 | <0.05 |
| Hippurate (37) | 0.632 | <0.05 |
| Homogentisate (43) | 0.654 | <0.05 |
| Indoxyl sulfate (46) | 0.742 | <0.05 |
| Isobutyrate (5) | −0.871 | <0.05 |
| Lactate (9) | −0.715 | <0.05 |
| Malonate (30) | −0.716 | <0.05 |
| Methymalonate (7) | −0.953 | <0.05 |
| N-Acetylglutamate (14) | −0.855 | <0.05 |
| N-Methylnicotinamide (38) | −0.642 | <0.05 |
| Phenylacetyglycine (36) | 0.650 | <0.05 |
| Propionate (4) | −0.825 | <0.05 |
| α-Hydroxy-n-valerate (8) | −0.748 | <0.05 |
| α-Ketoglutarate (19) | 0.665 | <0.05 |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, G.; Wu, X.; Jia, G.; Chen, X.; Zhao, H.; Wang, J.; Wu, C.; Cai, J. Arginine: New Insights into Growth Performance and Urinary Metabolomic Profiles of Rats. Molecules 2016, 21, 1142. https://doi.org/10.3390/molecules21091142
Liu G, Wu X, Jia G, Chen X, Zhao H, Wang J, Wu C, Cai J. Arginine: New Insights into Growth Performance and Urinary Metabolomic Profiles of Rats. Molecules. 2016; 21(9):1142. https://doi.org/10.3390/molecules21091142
Chicago/Turabian StyleLiu, Guangmang, Xianjian Wu, Gang Jia, Xiaoling Chen, Hua Zhao, Jing Wang, Caimei Wu, and Jingyi Cai. 2016. "Arginine: New Insights into Growth Performance and Urinary Metabolomic Profiles of Rats" Molecules 21, no. 9: 1142. https://doi.org/10.3390/molecules21091142





