Interactions of Bovine Serum Albumin with Anti-Cancer Compounds Using a ProteOn XPR36 Array Biosensor and Molecular Docking
Abstract
:1. Introduction
2. Results
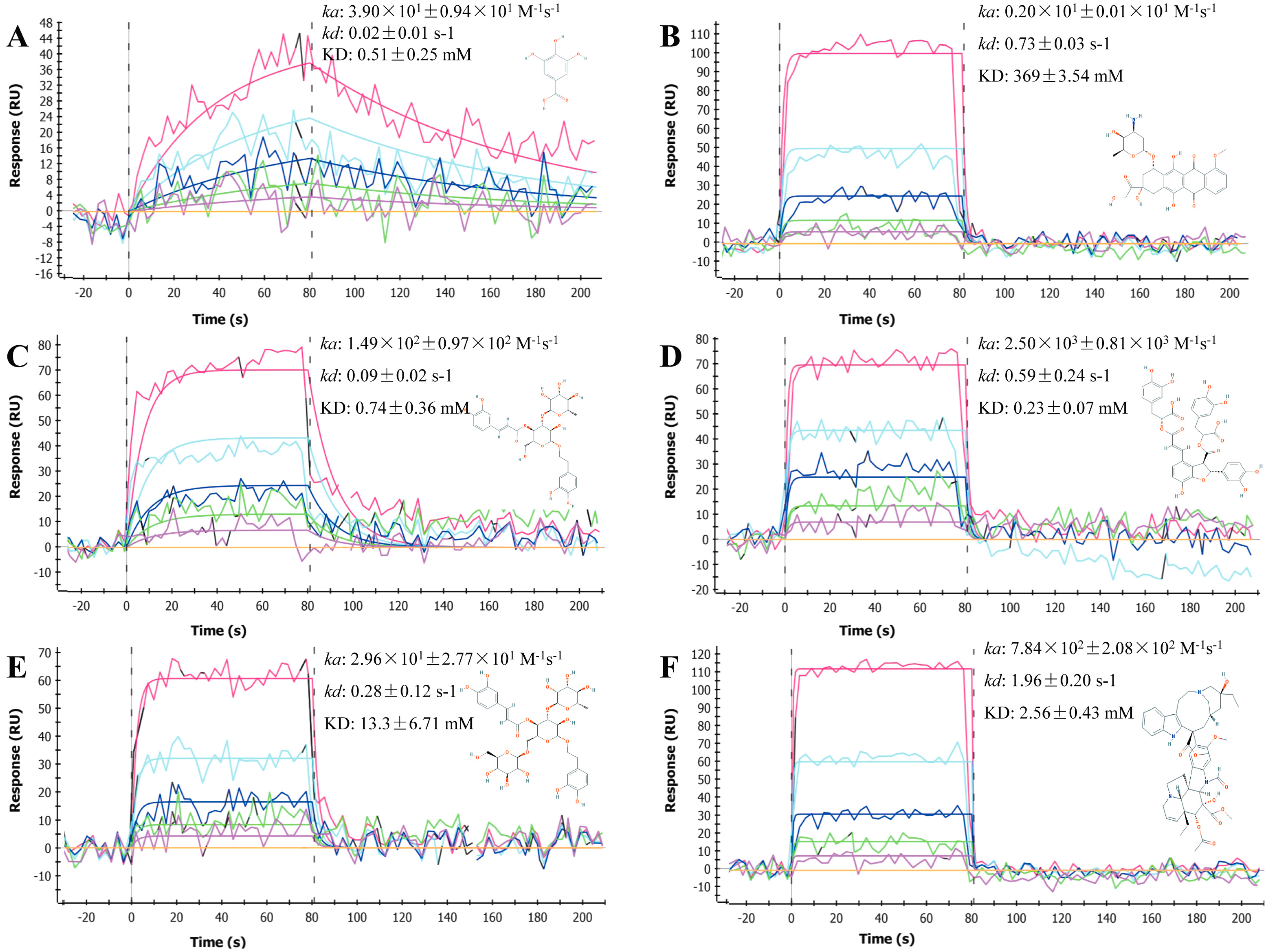
2.1. Sensorgrams of Anti-Cancer Compounds
2.2. Determination and Comparison of Kinetic Binding Constants
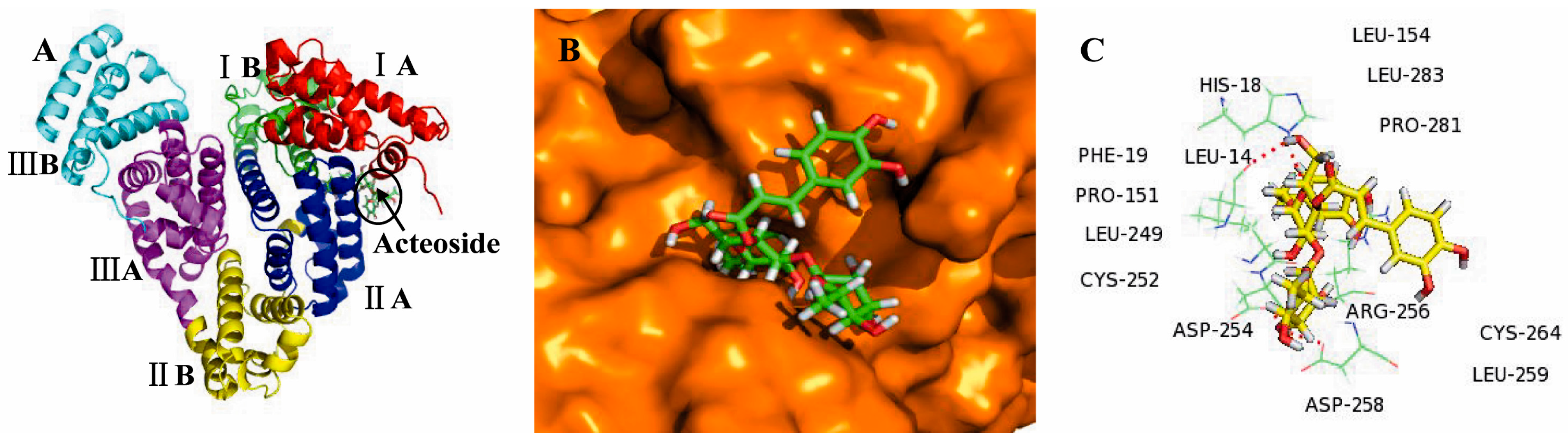
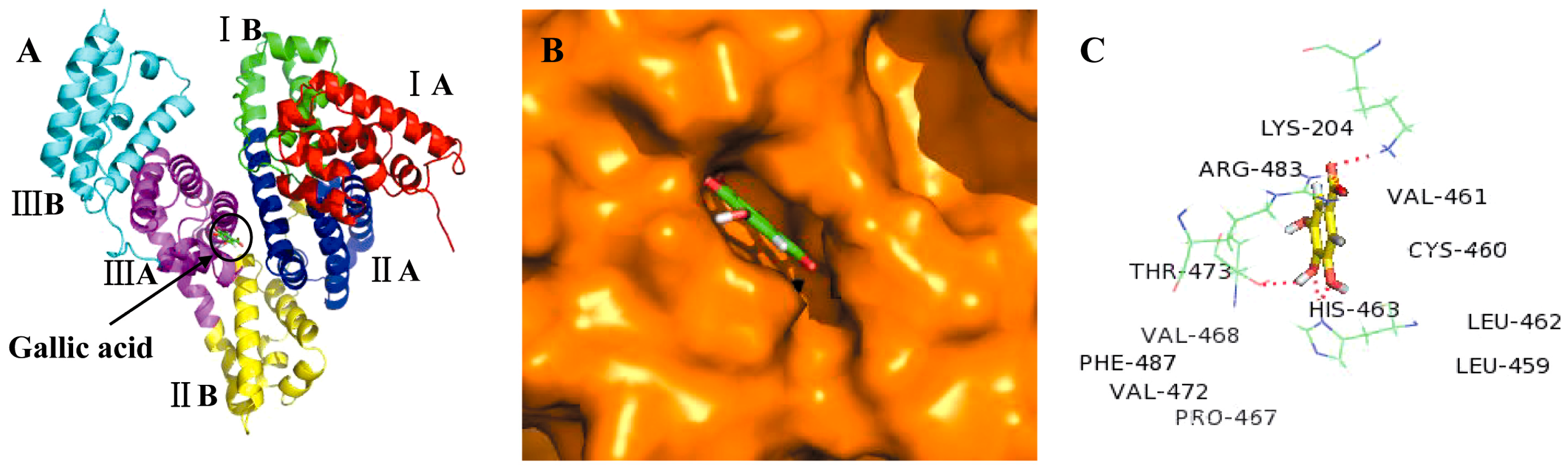
2.3. Molecular Docking Studies
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Initialization of GLH Sensor Chip
4.3. Preconditioning of GLH Sensor Chip
4.4. Immobilization of BSA
4.5. Injection of Analytes
4.6. Data Processing and Analysis
4.7. Molecular Docking
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| SPR | surface plasmon resonance |
| ka | association rate constant |
| kd | dissociation rate constant |
| KD | equilibrium dissociation constant |
| SA | Serum albumin |
| HSA | human serum albumin |
| BSA | bovine serum albumin |
| EDC | N-ethyl-N′-(3-dimethylaminopropyl)carbodiimide |
| sulfo-NHS | sulfo-N-hydroxysuccinimide |
| RU | Resonance Units |
References
- Qin, P.; Liu, R.; Pan, X.; Fang, X.; Mou, Y. Impact of carbon chain length on binding of perfluoroalkyl acids to bovine serum albumin determined by spectroscopic methods. J. Agric. Food Chem. 2010, 58, 5561–5567. [Google Scholar] [CrossRef] [PubMed]
- Sugio, S.; Kashima, A.; Mochizuki, S.; Noda, M.; Kobayashi, K. Crystal structure of human serum albumin at 2.5 Å resolution. Protein Eng. 1999, 12, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Han, X.-L.; Tian, F.-F.; Ge, Y.-S.; Jiang, F.-L.; Lai, L.; Li, D.W.; Yu, Q.L.; Wang, J.; Lin, C.; Liu, Y. Spectroscopic, structural and thermodynamic properties of chlorpyrifos bound to serum albumin: A comparative study between BSA and HSA. J. Photochem. Photobiol. B Biol. 2012, 109, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Atanu Singha, R.; Debi Ranjan, T.; Angshuman, C.; Swagata, D. A spectroscopic study of the interaction of the antioxidant naringin with bovine serum albumin. J. Biophys. Chem. 2010, 1, 141–152. [Google Scholar]
- Liang, H.; Xin, B.; Wang, X.; Yuan, Y.; Zhou, Y.; Shen, P. Equilibrium dialysis study on the interaction between Cu (II) and HSA or BSA. Sci. Bull. 1998, 43, 404–409. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, Z.; Fu, Z.; Xiong, Y.; Zhang, X. A flow-injection ultrafiltration sampling chemiluminescence system for on-line determination of drug–protein interaction. Anal. Bioanal. Chem. 2003, 377, 660–665. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.W.; Dong, C.Q.; Huang, X.Y.; Ren, J.C. Using capillary electrophoresis mobility shift assay to study the interaction of CdTe quantum dots with bovine serum albumin. Chin. Chem. Lett. 2008, 19, 707–710. [Google Scholar] [CrossRef]
- Van der Vlies, C. Binding of a diphenhydramine analogue to BSA: An NMR investigation. Biochem. Pharmacol. 1970, 19, 859–864. [Google Scholar] [CrossRef]
- Mir, M.U.H.; Maurya, J.K.; Ali, S.; Ubaid-ullah, S.; Khan, A.B.; Patel, R. Molecular interaction of cationic gemini surfactant with bovine serum albumin: A spectroscopic and molecular docking study. Process Biochem. 2014, 49, 623–630. [Google Scholar] [CrossRef]
- Zhang, X.; Li, L.; Xu, Z.; Liang, Z.; Su, J.; Huang, J.; Li, B. Investigation of the interaction of naringin palmitate with bovine serum albumin: Spectroscopic analysis and molecular docking. PLoS ONE 2013, 8, E59106. [Google Scholar] [CrossRef] [PubMed]
- Skrt, M.; Benedik, E.; Podlipnik, Č.; Ulrih, N.P. Interactions of different polyphenols with bovine serum albumin using fluorescence quenching and molecular docking. Food Chem. 2012, 135, 2418–2424. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Zhang, Y. Spectroscopic investigation on the interaction of salidroside with bovine serum albumin. J. Mol. Struct. 2008, 889, 20–27. [Google Scholar] [CrossRef]
- Gelamo, E.; Silva, C.; Imasato, H.; Tabak, M. Interaction of bovine (BSA) and human (HSA) serum albumins with ionic surfactants: Spectroscopy and modelling. Biochim. Biophys. Acta 2002, 1594, 84–99. [Google Scholar] [CrossRef]
- Myszka, D.G. Analysis of small-molecule interactions using Biacore S51 technology. Anal. Biochem. 2004, 329, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Long, S.; Myszka, D. Affinity-Based Optical Biosensors. Handbook of Affinity Chromatography; Hage, D.S., Cazes, J., Eds.; CRC Press: Boca Raton, FL, USA, 2005; pp. 685–698. [Google Scholar]
- Bravman, T.; Bronner, V.; Lavie, K.; Notcovich, A.; Papalia, G.A.; Myszka, D.G. Exploring “one-shot” kinetics and small molecule analysis using the ProteOn XPR36 array biosensor. Anal. Biochem. 2006, 358, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Rich, R.L.; Myszka, D.G. Survey of the year 2004 commercial optical biosensor literature. J. Mol. Recognit. 2005, 18, 431–478. [Google Scholar] [CrossRef] [PubMed]
- Reichmann, D.; Rahat, O.; Albeck, S.; Meged, R.; Dym, O.; Schreiber, G. The modular architecture of protein–protein binding interfaces. Proc. Natl. Acad. Sci. USA 2005, 102, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Ao, J.; Gao, L.; Yuan, T.; Jiang, G. Interaction mechanisms between organic UV filters and bovine serum albumin as determined by comprehensive spectroscopy exploration and molecular docking. Chemosphere 2015, 119, 590–600. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.-H.; Wang, J.; Zhu, Y.-Y.; Chen, J. Characterization of interaction between isoliquiritigenin and bovine serum albumin: Spectroscopic and molecular docking methods. J. Lumin. 2014, 145, 643–650. [Google Scholar] [CrossRef]
- Hu, X.; Lin, S.; Yu, D.; Qiu, S.; Zhang, X.; Mei, R. A preliminary study: The anti-proliferation effect of salidroside on different human cancer cell lines. Cell Biol. Toxicol. 2010, 26, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Luo, C.; Liu, J. Hydroxytyrosol induces apoptosis in human colon cancer cells through ROS generation. Food Funct. 2014, 5, 1909–1914. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, J.; Ma, H.; Chen, X.; Liu, T.; Jiao, Z.; He, W.; Wang, F.; Liu, X.; Zeng, X. Protective role of autophagy in matrine‑induced gastric cancer cell death. Int. J. Oncol. 2013, 42, 1417–1426. [Google Scholar] [CrossRef] [PubMed]
- Villegas, I.; Sánchez-Fidalgo, S.; Alarcón de la Lastra, C. New mechanisms and therapeutic potential of curcumin for colorectal cancer. Mol. Nutr. Food Res. 2008, 52, 1040–1061. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-W.; Kim, H.J.; Lee, Y.S.; Park, H.-J.; Choi, J.-W.; Ha, J.; Kyung-Tae, L. Acteoside inhibits human promyelocytic HL-60 leukemia cell proliferation via inducing cell cycle arrest at G0/G1 phase and differentiation into monocyte. Carcinogenesis 2007, 28, 1928–1936. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Zhu, W.; Zhang, Y.; Shu, Y.; Liu, P. Paeoniflorin modulates multicompound resistance of a human gastric cancer cell line via the inhibition of NF-κB activation. Mol. Med. Rep. 2012, 5, 351–356. [Google Scholar] [PubMed]
- Lee, Y.; Jin, Y.; Lim, W.; Ji, S.; Choi, S.; Jang, S.; Lee, S. A ginsenoside-Rh1, a component of ginseng saponin, activates estrogen receptor in human breast carcinoma MCF-7 cells. J. Steroid Biochem. Mol. Biol. 2003, 84, 463–468. [Google Scholar] [CrossRef]
- Zhao, Y.; Guo, Y.; Gu, X. Salvianolic Acid B, a potential chemopreventive agent, for head and neck squamous cell cancer. J. Oncol. 2011, 2010. [Google Scholar] [CrossRef] [PubMed]
- You, B.R.; Park, W.H. Gallic acid-induced lung cancer cell death is related to glutathione depletion as well as reactive oxygen species increase. Toxicol. In Vitro 2010, 24, 1356–1362. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Yu, D.; Wu, N.; Wang, H.; Niu, J.; Wang, Y.; Zou, Z. Echinacoside Induces Apoptosis in Human SW480 Colorectal Cancer Cells by Induction of Oxidative DNA Damages. Int. J. Mol. Sci. 2015, 16, 14655–14668. [Google Scholar] [CrossRef] [PubMed]
- Bujacz, A. Structures of bovine, equine and leporine serum albumin. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 1278–1289. [Google Scholar] [CrossRef] [PubMed]
- Paul, B.K.; Samanta, A.; Guchhait, N. Exploring hydrophobic subdomain IIA of the protein bovine serum albumin in the native, intermediate, unfolded, and refolded states by a small fluorescence molecular reporter. J. Phys. Chem. B 2010, 114, 6183–6196. [Google Scholar] [CrossRef] [PubMed]
- Frostell-Karlsson, Å.; Remaeus, A.; Roos, H.; Andersson, K.; Borg, P.; Hämäläinen, M.; Karlsson, R. Biosensor analysis of the interaction between immobilized human serum albumin and drug compounds for prediction of human serum albumin binding levels. J. Med. Chem. 2000, 43, 1986–1992. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, B.; Tanious, F.A.; Wilson, W.D. Biosensor-surface plasmon resonance: Quantitative analysis of small molecule–nucleic acid interactions. Methods 2007, 42, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Abdi, K.; Nafisi, S.; Manouchehri, F.; Bonsaii, M.; Khalaj, A. Interaction of 5-Fluorouracil and its derivatives with bovine serum albumin. J. Photochem. Photobiol. B Biol. 2012, 107, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Xu, X.; Liu, M.; Li, L.; Sun, D. Study on the interaction of matrine with bovine serum albumin. J. Solut. Chem. 2010, 39, 77–85. [Google Scholar] [CrossRef]
- Barik, A.; Mishra, B.; Kunwar, A.; Priyadarsini, K.I. Interaction of curcumin with human serum albumin: Thermodynamic properties, fluorescence energy transfer and denaturation effects. Chem. Phys. Lett. 2007, 436, 239–243. [Google Scholar] [CrossRef]
- Qiu, J.; Shao, S.; You, J.-Z. Studies on the Interaction between Oxaliplatin and Bovine Serum Albumin by Fluorescence Spectrometry. J. Instrum. Anal. 2007, 26, 51–54. [Google Scholar]
- Sample Availability: Samples of the compounds fluorouracil, hydroxytyrosol, gallic acid, matrine, salidroside, curcumin, oxaliplatin, paeoniflorin, doxorubicin, acteoside, ginsenoside Rh1, salvianolic acid B, echinacoside, and vincristine are available from the authors.

| Sample | Compound | Molecular Mass (Da) |
|---|---|---|
| A | Fluorouracil | 130.08 |
| B | Hydroxytyrosol | 138.16 |
| C | Gallic acid | 170.12 |
| D | Matrine | 248.37 |
| E | Salidroside | 300.31 |
| F | Curcumin | 368.39 |
| G | Oxaliplatin | 397.29 |
| H | Paeoniflorin | 480.45 |
| I | Doxorubicin | 543.52 |
| J | Acteoside | 624.59 |
| K | Ginsenoside Rh1 | 638.87 |
| L | Salvianolic acid B | 718.62 |
| M | Echinacoside | 786.72 |
| N | Vincristine | 824.96 |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Cai, Q.-Y.; Cai, Z.-X.; Fang, Y.; Zheng, C.-S.; Wang, L.-L.; Lin, S.; Chen, D.-X.; Peng, J. Interactions of Bovine Serum Albumin with Anti-Cancer Compounds Using a ProteOn XPR36 Array Biosensor and Molecular Docking. Molecules 2016, 21, 1706. https://doi.org/10.3390/molecules21121706
Zhang L, Cai Q-Y, Cai Z-X, Fang Y, Zheng C-S, Wang L-L, Lin S, Chen D-X, Peng J. Interactions of Bovine Serum Albumin with Anti-Cancer Compounds Using a ProteOn XPR36 Array Biosensor and Molecular Docking. Molecules. 2016; 21(12):1706. https://doi.org/10.3390/molecules21121706
Chicago/Turabian StyleZhang, Ling, Qiao-Yan Cai, Zhi-Xiong Cai, Yi Fang, Chun-Song Zheng, Li-Li Wang, Shan Lin, Da-Xin Chen, and Jun Peng. 2016. "Interactions of Bovine Serum Albumin with Anti-Cancer Compounds Using a ProteOn XPR36 Array Biosensor and Molecular Docking" Molecules 21, no. 12: 1706. https://doi.org/10.3390/molecules21121706






