Computational Study of Symmetric Methylation on Histone Arginine Catalyzed by Protein Arginine Methyltransferase PRMT5 through QM/MM MD and Free Energy Simulations
Abstract
:1. Introduction
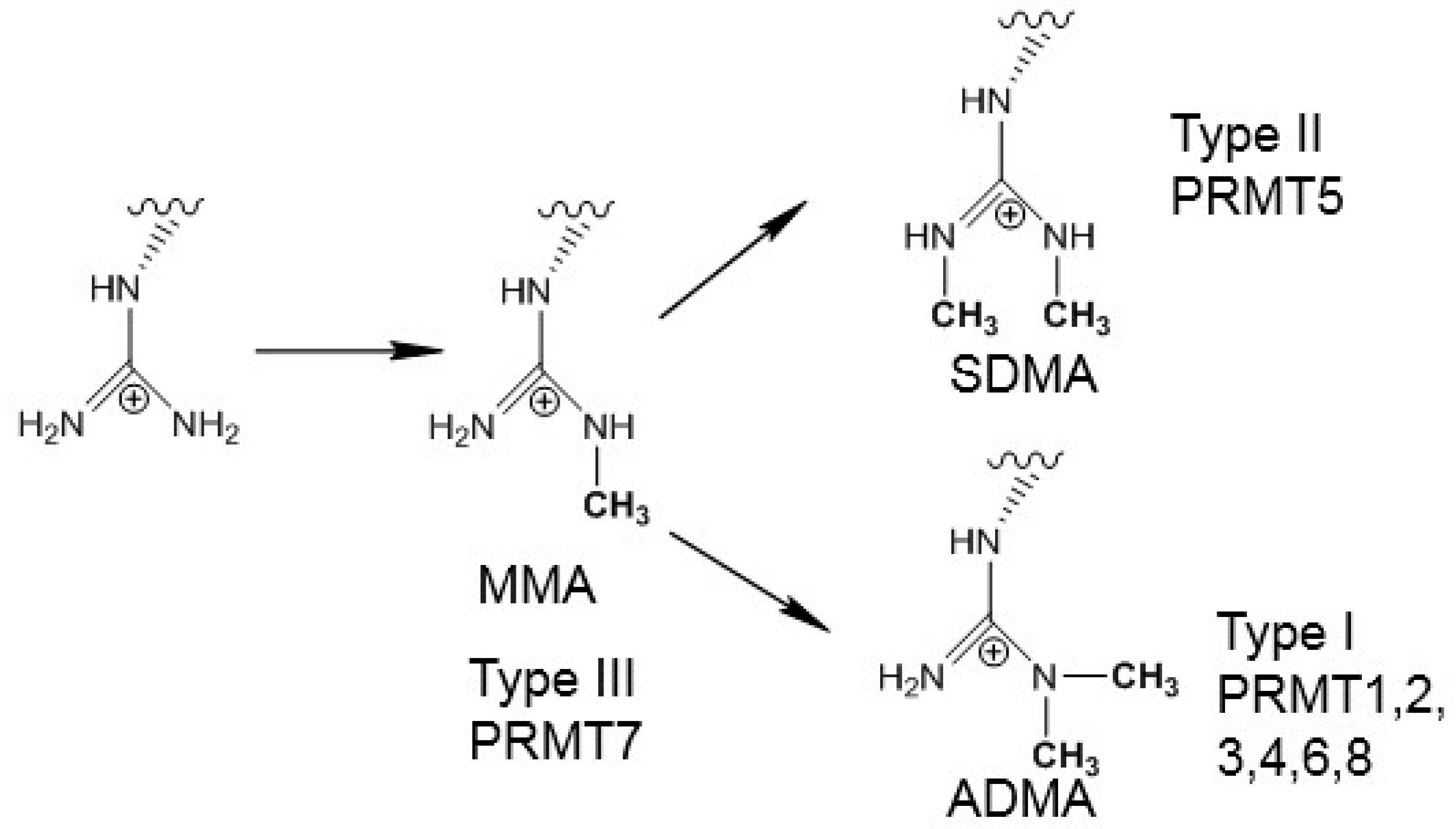
2. Results and Discussion
2.1. Comparison of the Active Site Structures of Type II PRMT5 and Type I PRMT3
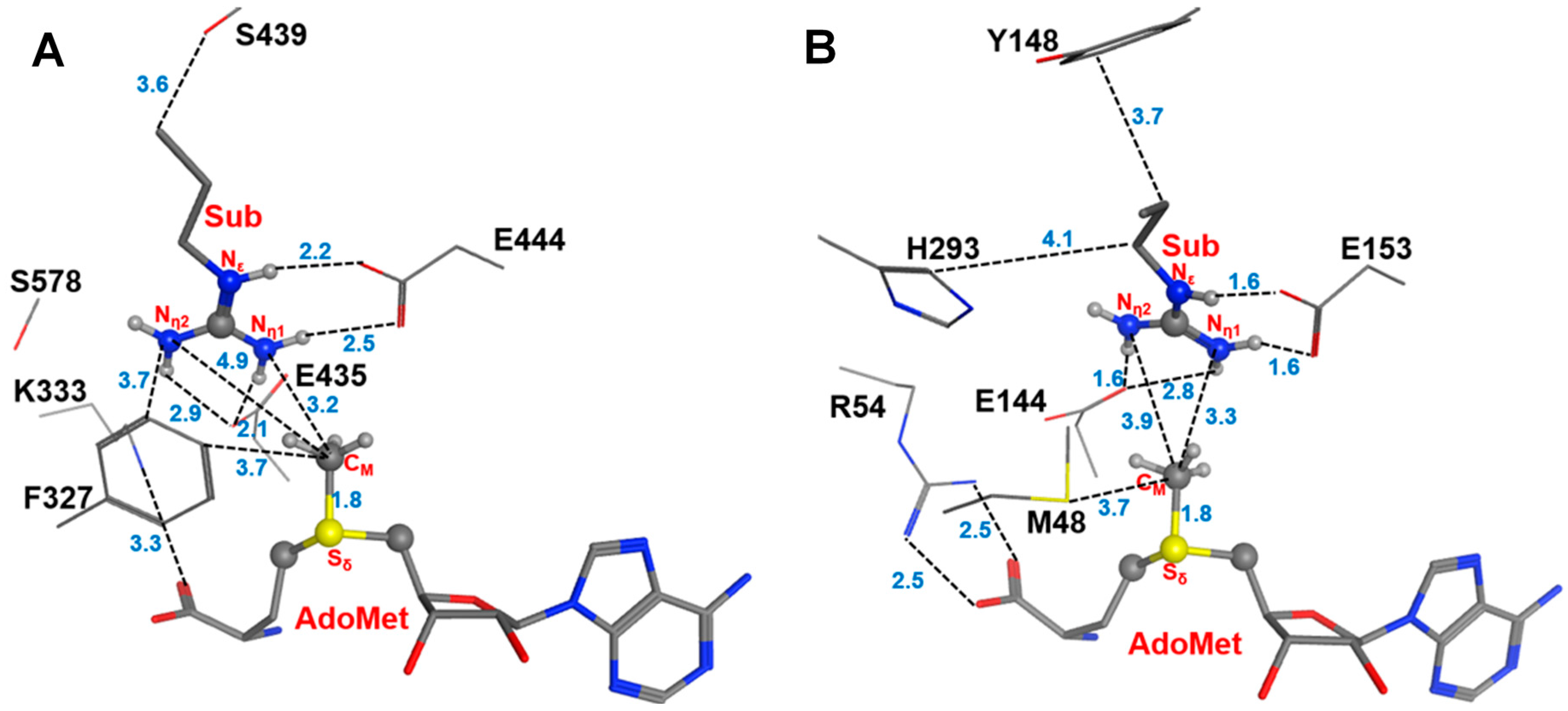
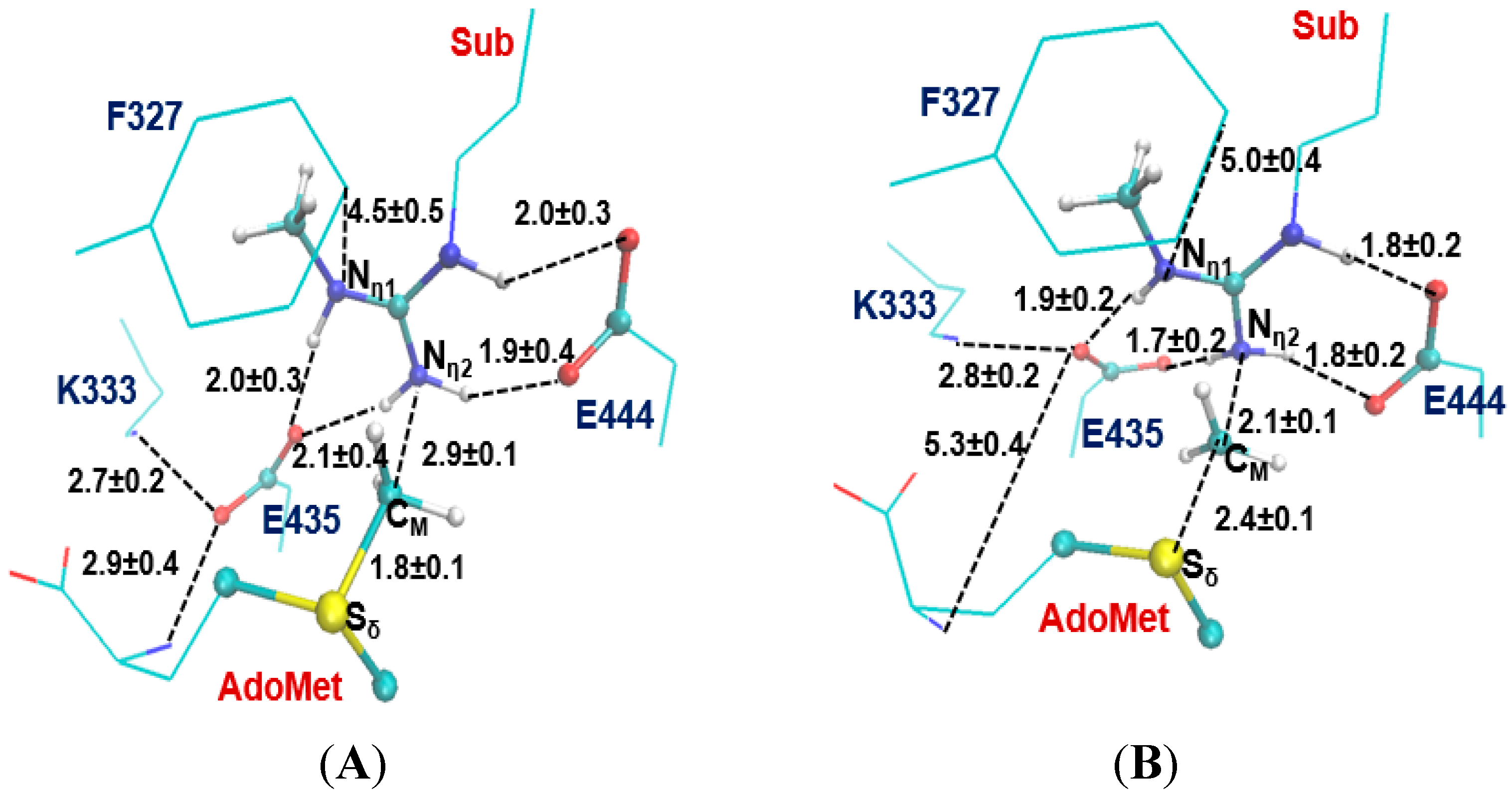
2.2. The First Methylation Catalyzed by PRMT5

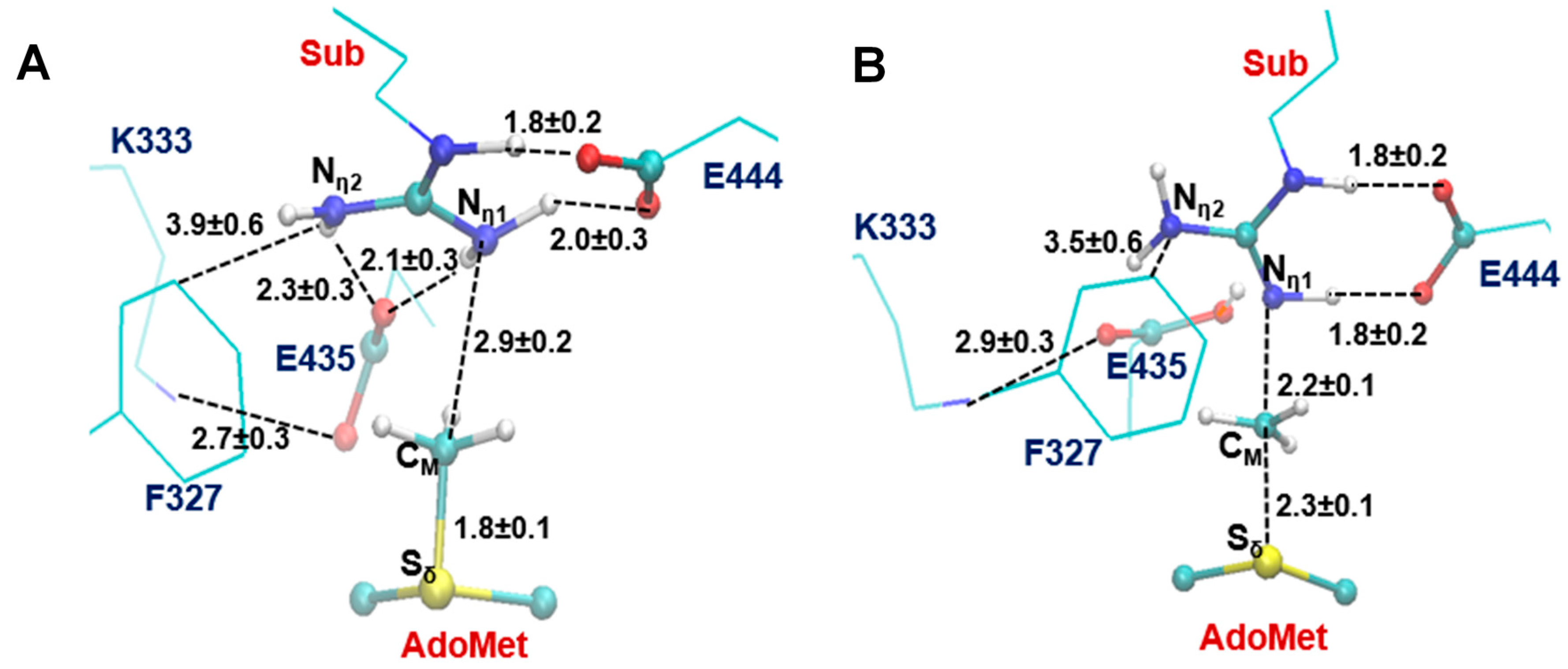
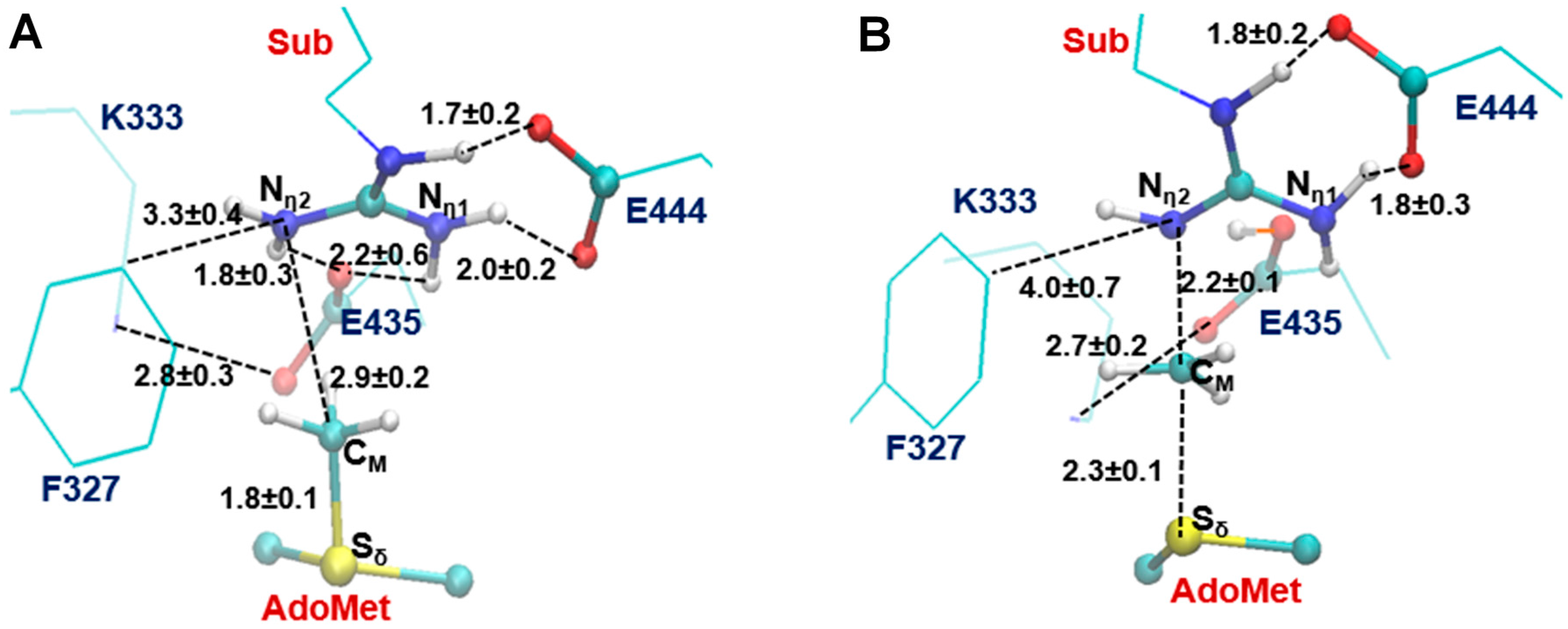
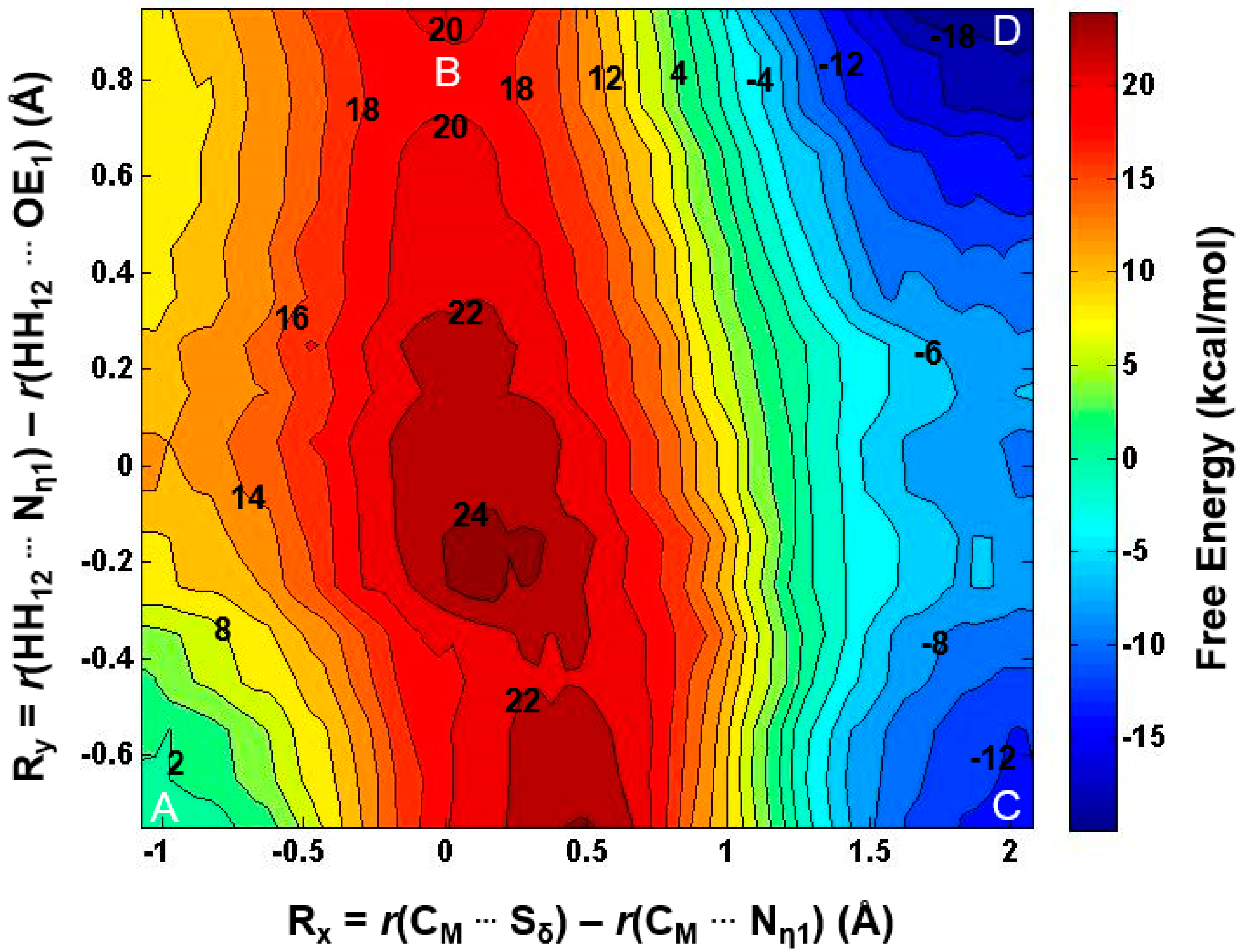
2.3. The Second Methylation Catalyzed by PRMT5
 | |||||
| To Nη1 | To Nη2 | To Nη1 | To Nη2 | To Nη1 | To Nη2 |
| 32.3 kcal/mol | 36 kcal/mol | 31.3 kcal/mol | 20.1 kcal/mol | 43.9 kcal/mol | 32.5 kcal/mol |
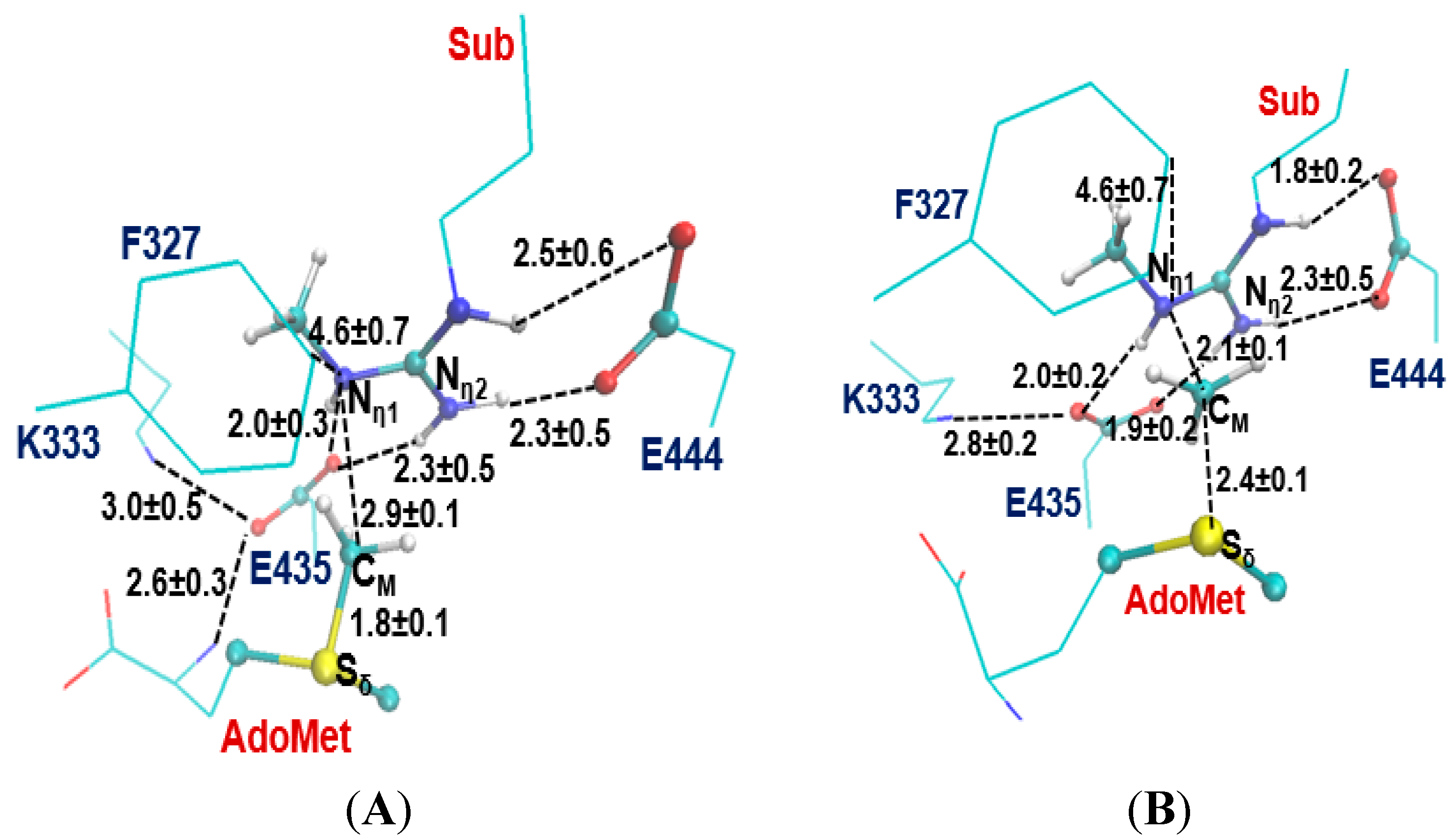

3. Experimental Section
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bedford, M.T.; Clarke, S.G. Protein arginine methylation in mammals: Who, what, and why. Mol. Cell 2009, 33, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Di Lorenzo, A.; Bedford, M.T. Histone arginine methylation. FEBS Lett. 2011, 585, 2024–2031. [Google Scholar] [CrossRef] [PubMed]
- Biggar, K.K.; Li, S.S. Non-histone protein methylation as a regulator of cellular signalling and function. Nat. Rev. Mol. Cell Biol. 2015, 16, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Xu, R.M.; Thompson, P.R. Substrate specificity, processivity, and kinetic mechanism of protein arginine methyltransferase 5. Biochemistry 2013, 52, 5430–5440. [Google Scholar] [CrossRef] [PubMed]
- Wysocka, J.; Allis, C.D.; Coonrod, S. Histone arginine methylation and its dynamic regulation. Front. Biosci. 2006, 11, 344–355. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, S.E.; Petrillo, E.; Beckwith, E.J.; Zhang, X.; Rugnone, M.L.; Hernando, C.E.; Cuevas, J.C.; Godoy Herz, M.A.; Depetris-Chauvin, A.; Simpson, C.G.; et al. A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 2010, 468, 112–116. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Song, H.R.; Lutz, K.; Kerstetter, R.A.; Michael, T.P.; McClung, C.R. Type II protein arginine methyltransferase 5 (prmt5) is required for circadian period determination in arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 21211–21216. [Google Scholar] [CrossRef] [PubMed]
- Meister, G.; Eggert, C.; Buhler, D.; Brahms, H.; Kambach, C.; Fischer, U. Methylation of sm proteins by a complex containing prmt5 and the putative u snrnp assembly factor picln. Curr. Biol. 2001, 11, 1990–1994. [Google Scholar] [CrossRef]
- Deng, X.; Gu, L.; Liu, C.; Lu, T.; Lu, F.; Lu, Z.; Cui, P.; Pei, Y.; Wang, B.; Hu, S.; et al. Arginine methylation mediated by the arabidopsis homolog of prmt5 is essential for proper pre-mrna splicing. Proc. Natl. Acad. Sci. USA 2010, 107, 19114–19119. [Google Scholar] [CrossRef] [PubMed]
- Nishida, K.M.; Okada, T.N.; Kawamura, T.; Mituyama, T.; Kawamura, Y.; Inagaki, S.; Huang, H.; Chen, D.; Kodama, T.; Siomi, H.; et al. Functional involvement of tudor and dprmt5 in the pirna processing pathway in drosophila germlines. EMBO J. 2009, 28, 3820–3831. [Google Scholar] [CrossRef] [PubMed]
- Gonsalvez, G.B.; Rajendra, T.K.; Tian, L.; Matera, A.G. The sm-protein methyltransferase, dart5, is essential for germ-cell specification and maintenance. Curr. Biol. 2006, 16, 1077–1089. [Google Scholar] [CrossRef] [PubMed]
- Jansson, M.; Durant, S.T.; Cho, E.C.; Sheahan, S.; Edelmann, M.; Kessler, B.; la Thangue, N.B. Arginine methylation regulates the p53 response. Nat. Cell Biol. 2008, 10, 1431–1439. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Sun, J.; Sun, X.; Shen, Q.; Gao, Z.; Yang, C. Caenorhabditis elegans protein arginine methyltransferase prmt-5 negatively regulates DNA damage-induced apoptosis. PLoS Genet. 2009, 5, e1000514. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wang, M.; Lv, Z.; Yang, N.; Liu, Y.; Bao, S.; Gong, W.; Xu, R.M. Structural insights into protein arginine symmetric dimethylation by prmt5. Proc. Natl. Acad. Sci. USA 2011, 108, 20538–20543. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, Y.; Ma, Q.; Zhang, Z.; Xue, Y.; Bao, S.; Chong, K. Skb1-mediated symmetric dimethylation of histone h4r3 controls flowering time in arabidopsis. EMBO J. 2007, 26, 1934–1941. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Rank, G.; Tan, Y.T.; Li, H.; Moritz, R.L.; Simpson, R.J.; Cerruti, L.; Curtis, D.J.; Patel, D.J.; Allis, C.D.; et al. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat. Struct. Mol. Biol. 2009, 16, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Huang, Z.Q.; Xia, L.; Feng, Q.; Erdjument-Bromage, H.; Strahl, B.D.; Briggs, S.D.; Allis, C.D.; Wong, J.; Tempst, P.; et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 2001, 293, 853–857. [Google Scholar] [CrossRef] [PubMed]
- Strahl, B.D.; Briggs, S.D.; Brame, C.J.; Caldwell, J.A.; Koh, S.S.; Ma, H.; Cook, R.G.; Shabanowitz, J.; Hunt, D.F.; Stallcup, M.R.; et al. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator prmt1. Curr. Biol. 2001, 11, 996–1000. [Google Scholar] [CrossRef]
- Zhang, X.; Cheng, X. Structure of the predominant protein arginine methyltransferase prmt1 and analysis of its binding to substrate peptides. Structure 2003, 11, 509–520. [Google Scholar] [CrossRef]
- Siarheyeva, A.; Senisterra, G.; Allali-Hassani, A.; Dong, A.; Dobrovetsky, E.; Wasney, G.A.; Chau, I.; Marcellus, R.; Hajian, T.; Liu, F.; et al. An allosteric inhibitor of protein arginine methyltransferase 3. Structure 2012, 20, 1425–1435. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhu, Y.; Chen, J.; Li, X.; Peng, J.; Chen, J.; Zou, Y.; Zhang, Z.; Jin, H.; Yang, P.; et al. Crystal structure of arginine methyltransferase 6 from trypanosoma brucei. PLoS ONE 2014, 9, e87267. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Frazier, M.; Lu, F.; Cao, X.; Redinbo, M.R. Crystal structure of the plant epigenetic protein arginine methyltransferase 10. J. Mol. Biol. 2011, 414, 106–122. [Google Scholar] [CrossRef] [PubMed]
- Antonysamy, S.; Bonday, Z.; Campbell, R.M.; Doyle, B.; Druzina, Z.; Gheyi, T.; Han, B.; Jungheim, L.N.; Qian, Y.; Rauch, C.; et al. Crystal structure of the human PRMT5:MEP50 complex. Proc. Natl. Acad. Sci. USA 2012, 109, 17960–17965. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhu, Y.; Caceres, T.B.; Liu, L.; Peng, J.; Wang, J.; Chen, J.; Chen, X.; Zhang, Z.; Zuo, X.; et al. Structural determinants for the strict monomethylation activity by trypanosoma brucei protein arginine methyltransferase 7. Structure 2014, 22, 756–768. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Li, X.; Liang, Z.; Zhu, K.; Lu, J.; Kong, X.; Ouyang, S.; Li, L.; Zheng, Y.G.; Luo, C. Theoretical insights into catalytic mechanism of protein arginine methyltransferase 1. PLoS ONE 2013, 8, e72424. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.; Li, G.; Guo, H. QM/MM md and free energy simulations of the methylation reactions catalyzed by protein arginine methyltransferase prmt3. Can. J. Chem. 2013, 91, 605–612. [Google Scholar] [CrossRef]
- Guo, H.B.; Guo, H. Mechanism of histone methylation catalyzed by protein lysine methyltransferase set7/9 and origin of product specificity. Proc. Natl. Acad. Sci. USA 2007, 104, 8797–8802. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Chu, Y.Z.; Guo, H.B.; Smith, J.C.; Guo, H. Energy triplets for writing epigenetic marks: Insights from QM/MM free-energy simulations of protein lysine methyltransferases. Chemistry 2009, 15, 12596–12599. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.; Yao, J.; Guo, H. QM/MM MD and free energy simulations of G9a-like protein (glp) and its mutants: Understanding the factors that determine the product specificity. PLoS ONE 2012, 7, e37674. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.; Xu, Q.; Guo, H. Understanding energetic origins of product specificity of set8 from qm/mm free energy simulations: What causes the stop of methyl addition during histone lysine methylation? J. Chem. Theory Comput. 2010, 6, 1380–1389. [Google Scholar] [CrossRef]
- Yao, J.; Xu, Q.; Chen, F.; Guo, H. QM/MM free energy simulations of salicylic acid methyltransferase: Effects of stabilization of ts-like structures on substrate specificity. J. Phys. Chem. B 2010, 115, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; Guo, H. Quantum mechanical/molecular mechanical study of catalytic mechanism and role of key residues in methylation reactions catalyzed by dimethylxanthine methyltransferase in caffeine biosynthesis. J. Chem. Inf. Model. 2014, 54, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.D.; Collins, R.E.; Zhang, X. Structural and sequence motifs of protein (histone) methylation enzymes. Annu. Rev. Biophys. Biomol. Struct. 2005, 34, 267–294. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Fuhrmann, J.; Thompson, P.R. Protein arginine methyltransferase 5 catalyzes substrate dimethylation in a distributive fashion. Biochemistry 2014, 53, 7884–7892. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.R.; Bruccoleri, R.E.; Olafson, B.D.; States, D.J.; Swaminathan, S.; Karplus, M. CHARMM: A program for macro-molecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983, 4, 187–217. [Google Scholar] [CrossRef]
- Jorgensen, W.; Chandrasekhar, J.; Madura, J.; Impey, R.; Klein, M. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Brooks, C.L.; Brunger, A.; Karplus, M. Active site dynamics in protein molecules: A stochastic boundary molecular-dynamics approach. Biopolymers 1985, 24, 843–865. [Google Scholar] [CrossRef] [PubMed]
- MacKerell, A.D.; Bashford, D.; Bellott, M.; Dunbrack, R.L.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef] [PubMed]
- Elstner, M.; Porezag, D.; Jungnickel, G.; Elsner, J.; Haugk, M.; Frauenheim, T.; Suhai, S.; Seifert, G. Self-consistent-charge density functional tight-binding method for simulations of complex materials properties. Phys. Rev. B 1998, 58, 7260–7268. [Google Scholar] [CrossRef]
- Cui, Q.; Elstner, M.; Kaxiras, E.; Frauenheim, T.; Karplus, M. A QM/MM Implementation of the Self-Consistent Charge Density Functional Tight Binding (SCC-DFTB) Method. J. Phys. Chem. B 2001, 105, 569–585. [Google Scholar] [CrossRef]
- Field, M.J.; Bash, P.A.; Karplus, M. A combined quantum mechanical and molecular mechanical potential for molecular dynamics simulations. J. Comput. Chem. 1990, 11, 700–733. [Google Scholar] [CrossRef]
- Torrie, G.M.; Valleau, J.P. Monte-Carlo free energy estimates using non-Boltzmann sampling. Application to the subcritical Lennard Jones fluid. Chem. Phys. Lett. 1974, 28, 578–581. [Google Scholar] [CrossRef]
- Kumar, S.; Bouzida, D.; Swendsen, R.H.; Kollman, P.A.; Rosenberg, J.M. The weighted histogram analysis method for free energy calculations on biomolecules. I. The method. J. Comput. Chem. 1992, 13, 1011–1021. [Google Scholar] [CrossRef]
- Sample Availability: Not apply.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yue, Y.; Chu, Y.; Guo, H. Computational Study of Symmetric Methylation on Histone Arginine Catalyzed by Protein Arginine Methyltransferase PRMT5 through QM/MM MD and Free Energy Simulations. Molecules 2015, 20, 10032-10046. https://doi.org/10.3390/molecules200610032
Yue Y, Chu Y, Guo H. Computational Study of Symmetric Methylation on Histone Arginine Catalyzed by Protein Arginine Methyltransferase PRMT5 through QM/MM MD and Free Energy Simulations. Molecules. 2015; 20(6):10032-10046. https://doi.org/10.3390/molecules200610032
Chicago/Turabian StyleYue, Yufei, Yuzhuo Chu, and Hong Guo. 2015. "Computational Study of Symmetric Methylation on Histone Arginine Catalyzed by Protein Arginine Methyltransferase PRMT5 through QM/MM MD and Free Energy Simulations" Molecules 20, no. 6: 10032-10046. https://doi.org/10.3390/molecules200610032
APA StyleYue, Y., Chu, Y., & Guo, H. (2015). Computational Study of Symmetric Methylation on Histone Arginine Catalyzed by Protein Arginine Methyltransferase PRMT5 through QM/MM MD and Free Energy Simulations. Molecules, 20(6), 10032-10046. https://doi.org/10.3390/molecules200610032





