Tryptophan as a Probe to Study the Anticancer Mechanism of Action and Specificity of α-Helical Anticancer Peptides
Abstract
:1. Introduction
2. Results
2.1. Peptide Design
| Peptides | Sequence a |
|---|---|
| P(PNW) | Ac-K-W-K-S-F-L-K-T-F-K-S-L-K-K-T-V-L-H-T-L-L-K-A-I-S-S-amide |
| PMW | Ac-K-I-K-S-F-L-K-T-F-K-S-W-K-K-T-V-L-H-T-L-L-K-A-I-S-S-amide |
| PCW | Ac-K-I-K-S-F-L-K-T-F-K-S-L-K-K-T-V-L-H-T-L-L-K-A-W-S-S-amide |
2.2. Peptide Secondary Structure
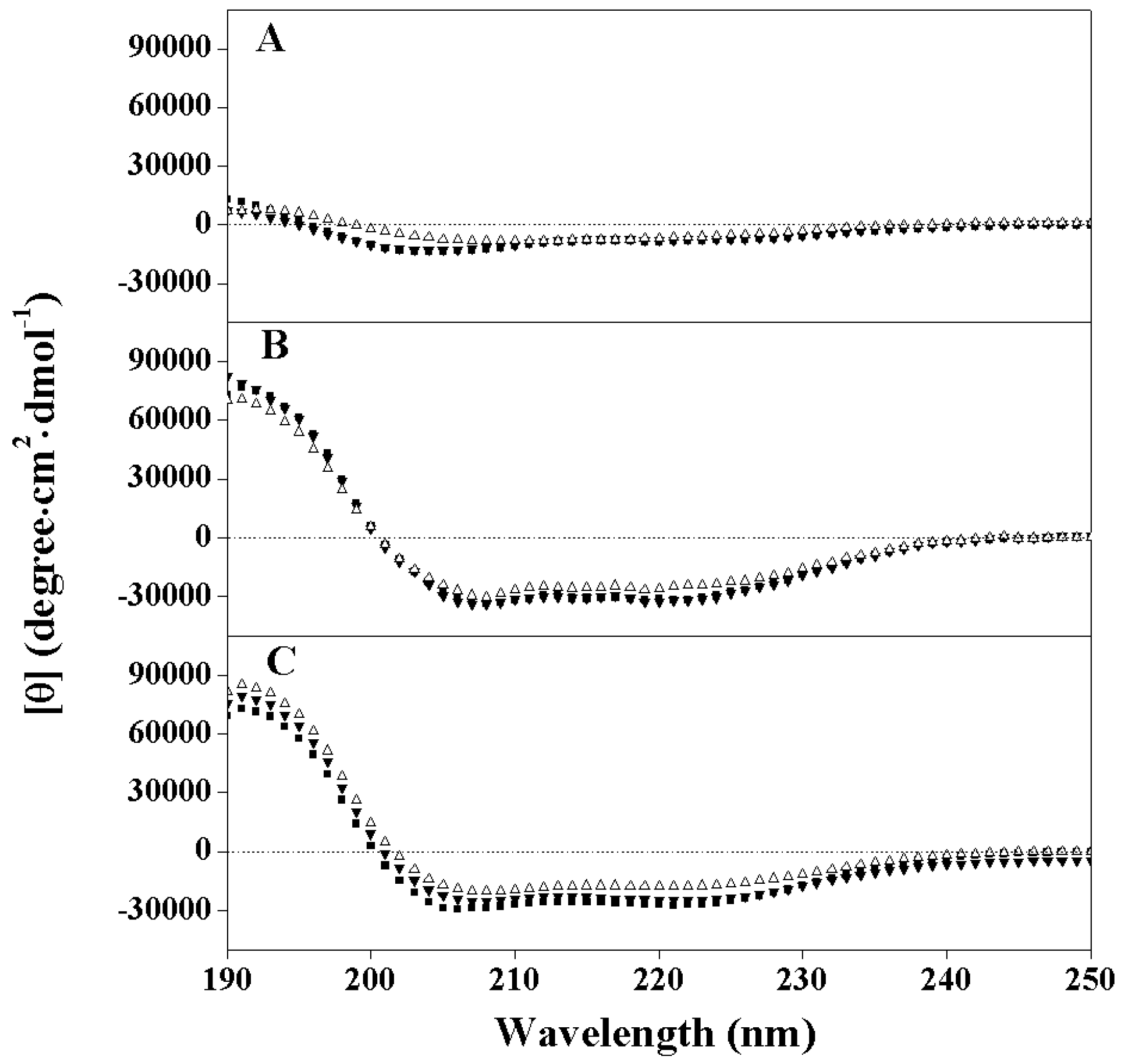
2.3. Hydrophobicity and Peptide Self-Association
| Peptides | tR a | Benign b | 50%TFE c | SDS in Buffer d | PA (min) e | |||
|---|---|---|---|---|---|---|---|---|
| [θ]222 | % Helix f | [θ]222 | % Helix | [θ]222 | % Helix | |||
| PNW | 46.8 | −7800 | 24 | −31700 | 99 | −27,300 | 86 | 5.7 |
| PMW | 46.5 | −7400 | 23 | −31900 | 100 | −25,050 | 79 | 5.3 |
| PCW | 45.6 | −5900 | 18 | −24300 | 76 | −17,800 | 56 | 4.5 |

2.4. Anticancer Activity and Hemolytic Activity
| Peptides | IC50 (μM) a | MHC (μM) b |
|---|---|---|
| PNW | 2.35 ± 0.28 | 10.41 ± 0.02 |
| PMW | 2.47 ± 0.18 | 10.41 ± 0.01 |
| PCW | 2.33 ± 0.12 | 10.41 ± 0.01 |
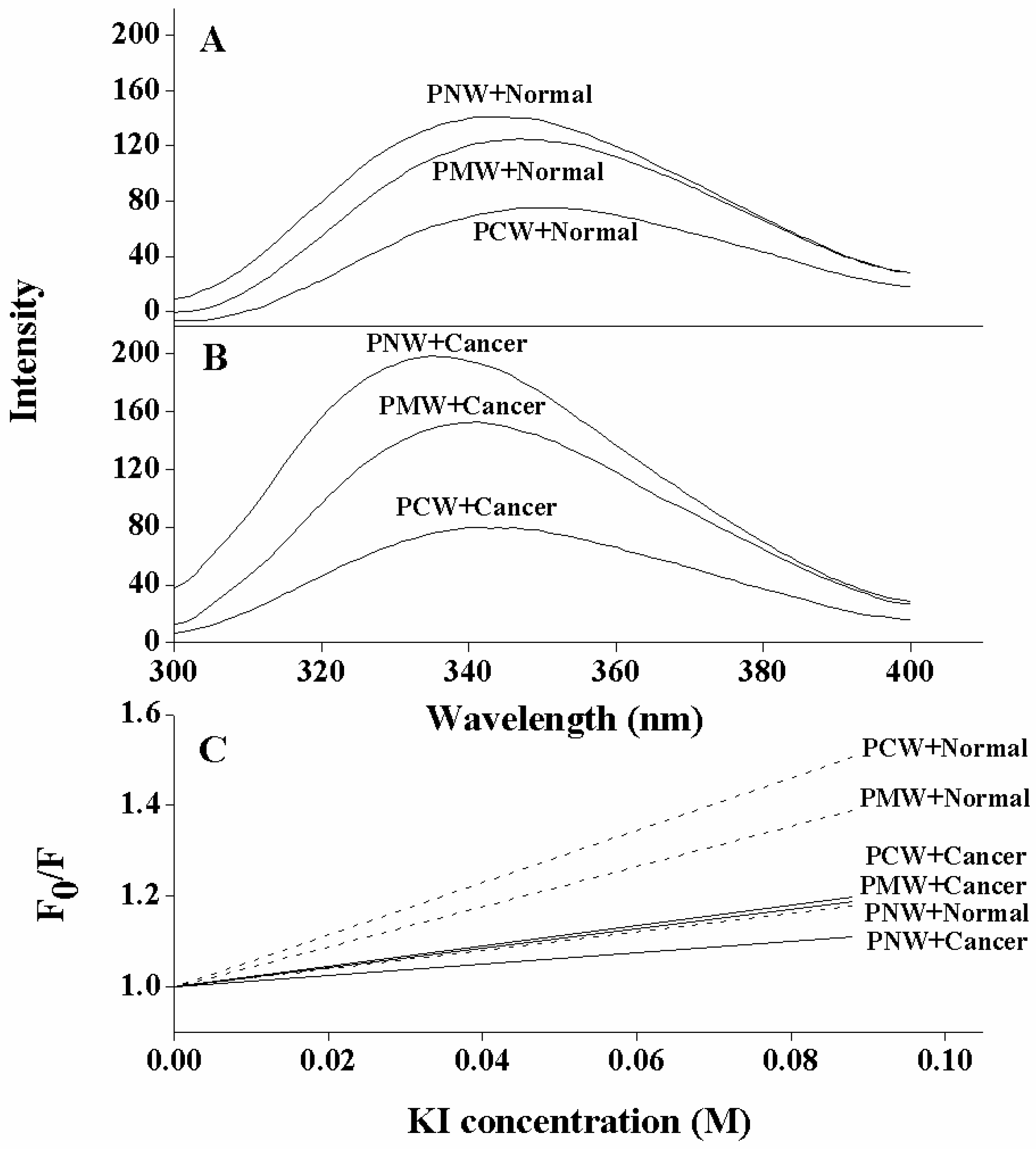
2.5. Tryptophan Fluorescence and Quenching Experiments
| Peptides | Membranes a | Intensity change b | Blue shift (nm) c |
|---|---|---|---|
| PNW | cancer LUVs | 81 | 14 |
| normal LUVs | 23 | 5 | |
| PMW | cancer LUVs | 38 | 9 |
| normal LUVs | 11 | 3 | |
| PCW | cancer LUVs | 2 | 6 |
| normal LUVs | 0 | 0 |
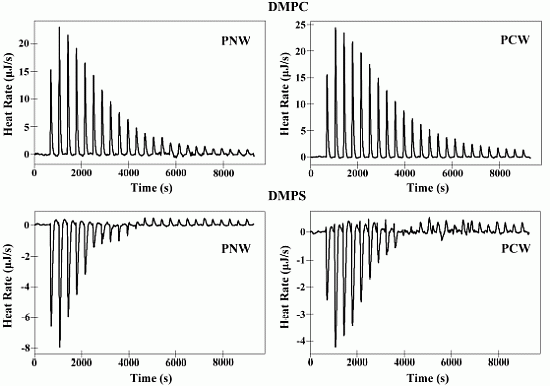
2.6. Isothermal Titration Calorimetry (ITC)
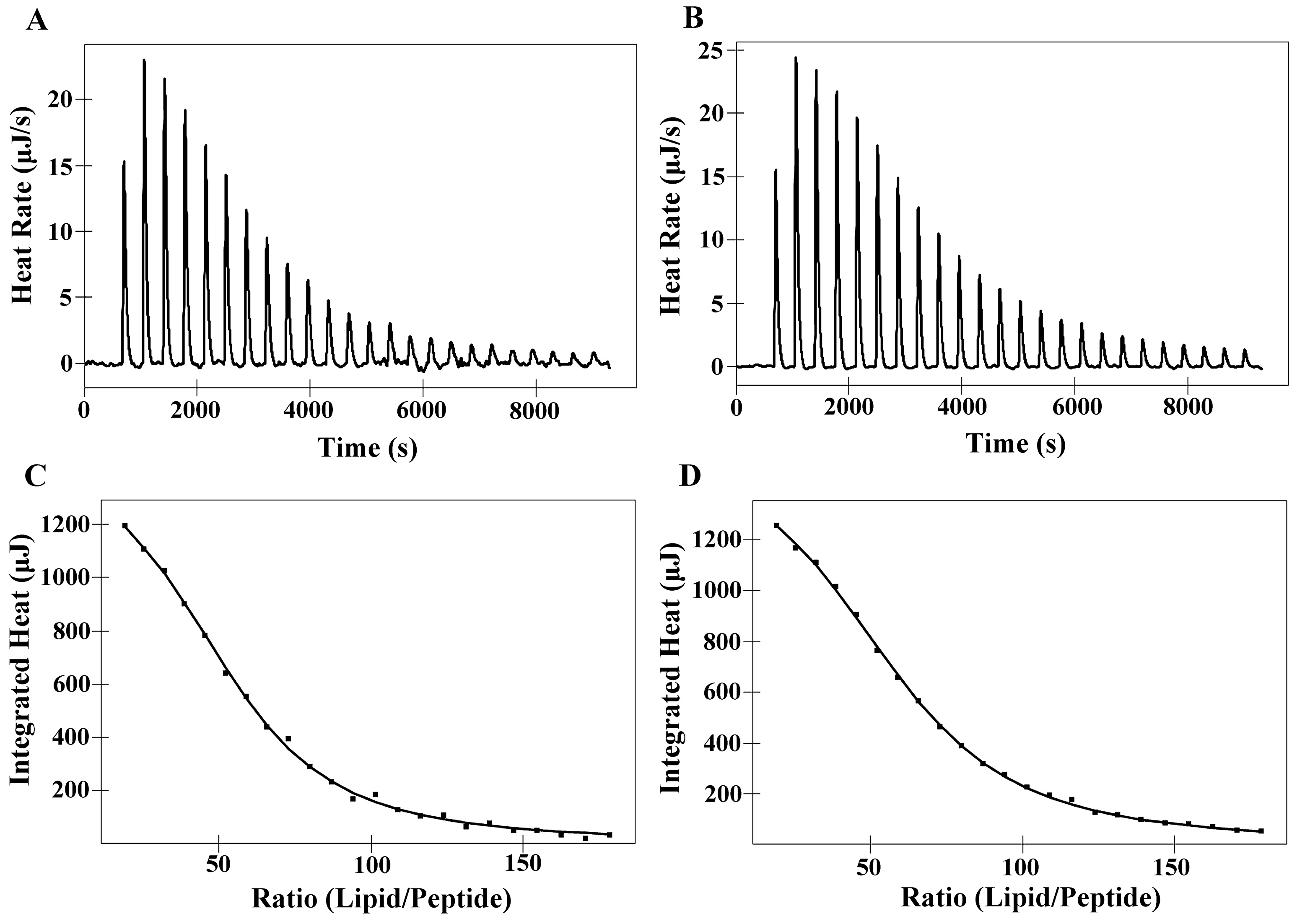
| Membranes | Peptides | Ka a [M]−1× 103 | ∆H b (kJ M−1) | ∆G c (kJ M−1) | T d ∆S (kJ M−1) | ∆S e(J M−1K−1) | N f |
|---|---|---|---|---|---|---|---|
| DMPC | PNW | 5.00 | 10.36 | −21.12 | 31.48 | 105.60 | 52.51 |
| PCW | 4.15 | 10.93 | −20.64 | 31.57 | 105.90 | 58.60 | |
| DMPS | PNW | 20.69 | −4.74 | −24.64 | 19.90 | 66.72 | 16.91 |
| PCW | 17.70 | −3.20 | −24.24 | 21.04 | 70.58 | 18.96 |
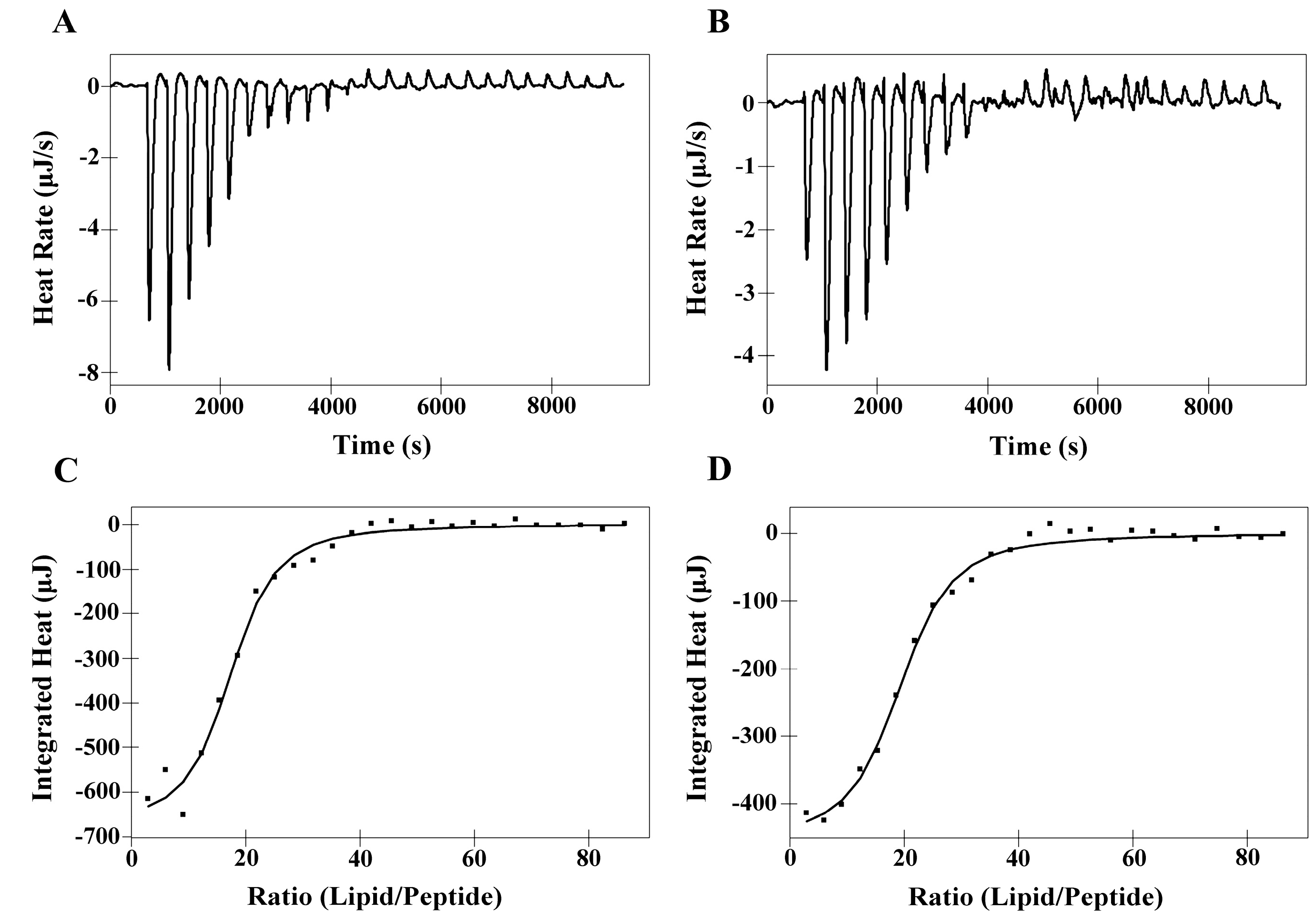
2.7. Differential Scanning Calorimetry Experiment (DSC)

3. Discussion
4. Experimental
4.1. Reagents
4.2. Peptide Synthesis and Purification
4.3. Analytical RP-HPLC and Temperature Profiling of Peptides
4.4. Circular Dichroism Spectroscopy
4.5. Measurement of Anticancer Activity
4.6. Measurement of Hemolytic Activity
4.7. Preparation of MLVs and LUVs
4.8. Tryptophan Fluorescence and Quenching Experiments
4.9. Isothermal Titration Experiment (ITC)
4.10. Differential Scanning Calorimetry (DSC)
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Smith, L.L.; Brown, K.; Carthew, P.; Lim, C.K.; Martin, E.A.; Styles, J.; White, I.N. Chemoprevention of breast cancer by tamoxifen: Risks and opportunities. Crit. Rev. Toxicol. 2000, 30, 571–594. [Google Scholar] [CrossRef] [PubMed]
- Borst, P.; Evers, R.; Kool, M.; Wijnholds, J. A family of drug transporters: The multidrug resistance-associated proteins. J. Natl. Cancer Inst. 2000, 92, 1295–1302. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Riedl, S.; Zweytick, D.; Lohner, K. Membrane-active host defense peptides—challenges and perspectives for the development of novel anticancer drugs. Chem. Phys. Lipids 2011, 164, 766–781. [Google Scholar] [PubMed]
- Jenssen, H.; Hamill, P.; Hancock, R.E. Peptide antimicrobial agents. Clin. Microbiol. Rev. 2006, 19, 491–511. [Google Scholar] [CrossRef] [PubMed]
- Hoskin, D.W.; Ramamoorthy, A. Studies on anticancer activities of antimicrobial peptides. Biochim. Biophys. Acta 2008, 1778, 357–375. [Google Scholar]
- Papo, N.; Shai, Y. Host defense peptides as new weapons in cancer treatment. Cell Mol. Life Sci. 2005, 62, 784–790. [Google Scholar] [CrossRef] [PubMed]
- Yoon, W.H.; Park, H.D.; Lim, K.; Hwang, B.D. Effect of O-glycosylated mucin on invasion and metastasis of HM7 human colon cancer cells. Biochem. Biophys. Res. Commun. 1996, 222, 694–699. [Google Scholar] [CrossRef] [PubMed]
- Papo, N.; Shai, Y. New lytic peptides based on the d,l-amphipathic helix motif preferentially kill tumor cells. Biochemistry 2003, 42, 9346–9354. [Google Scholar] [CrossRef] [PubMed]
- Zachowski, A. Phospholipids in animal eukaryotic membranes: Transverse asymmetry and movement. Biochem. J. 1993, 294, 1–14. [Google Scholar] [PubMed]
- Jiang, Z.; Vasil, A.I.; Hale, J.D.; Hancock, R.E.; Vasil, M.L.; Hodges, R.S. Effects of net charge and the number of positively charged residues on the biological activity of amphipathic alpha-helical cationic antimicrobial peptides. Biopolymers 2008, 90, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.B.; He, L.Y.; Jiang, H.Y.; Chen, Y.X. Role of helicity on the anticancer mechanism of action of cationic-helical peptides. Int. J. Mol. Sci. 2012, 13, 6849–6862. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.B.; Wang, X.F.; Wang, H.Y.; Liu, Y.; Chen, Y. Studies on mechanism of action of anticancer peptides by modulation of hydrophobicity within a defined structural framework. Mol. Cancer Ther. 2011, 10, 416–426. [Google Scholar] [CrossRef] [PubMed]
- Schiopu, I.; Mereuta, L.; Apetrei, A.; Park, Y.; Hahm, K.S.; Luchian, T. The role of tryptophan spatial arrangement for antimicrobial-derived, membrane-active peptides adsorption and activity. Mol. Biosyst. 2012, 8, 2860–2863. [Google Scholar] [CrossRef] [PubMed]
- Rekdal, O.; Haug, B.E.; Kalaaji, M.; Hunter, H.N.; Lindin, I.; Israelsson, I.; Solstad, T.; Yang, N.; Brandl, M.; Mantzilas, D.; et al. Relative spatial positions of tryptophan and cationic residues in helical membrane-active peptides determine their cytotoxicity. J. Biol. Chem. 2012, 287, 233–244. [Google Scholar] [CrossRef] [PubMed]
- De Jesus, A.J.; Allen, T.W. The role of tryptophan side chains in membrane protein anchoring and hydrophobic mismatch. Biochim. Biophys. Acta 2013, 1828, 864–876. [Google Scholar]
- Ladokhin, A.S. Evaluation of lipid exposure of tryptophan residues in membrane peptides and proteins. Anal. Biochem. 1999, 276, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Mant, C.T.; Farmer, S.W.; Hancock, R.E.; Vasil, M.L.; Hodges, R.S. Rational design of alpha-helical antimicrobial peptides with enhanced activities and specificity/therapeutic index. J. Biol. Chem. 2005, 280, 12316–12329. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Guarnieri, M.T.; Vasil, A.I.; Vasil, M.L.; Mant, C.T.; Hodges, R.S. Role of peptide hydrophobicity in the mechanism of action of alpha-helical antimicrobial peptides. Antimicrob. Agents Chemother. 2007, 51, 1398–1406. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Benz, R.; Hancock, R.E. Influence of proline residues on the antibacterial and synergistic activities of alpha-helical peptides. Biochemistry 1999, 38, 8102–8111. [Google Scholar] [CrossRef] [PubMed]
- Mant, C.T.; Chen, Y.; Hodges, R.S. Temperature profiling of polypeptides in reversed-phase liquid chromatography. I. Monitoring of dimerization and unfolding of amphipathic alpha-helical peptides. J. Chromatogr. A 2003, 1009, 29–43. [Google Scholar]
- Chen, Y.; Mant, C.T.; Hodges, R.S. Determination of stereochemistry stability coefficients of amino acid side-chains in an amphipathic alpha-helix. J. Pept. Res. 2002, 59, 18–33. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.E.; Mant, C.T.; Hodges, R.S. Effect of preferred binding domains on peptide retention behavior in reversed-phase chromatography: Amphipathic alpha-helices. Pept. Res. 1990, 3, 8–20. [Google Scholar] [PubMed]
- Mant, C.T.; Tripet, B.; Hodges, R.S. Temperature profiling of polypeptides in reversed-phase liquid chromatography. II. Monitoring of folding and stability of two-stranded alpha-helical coiled-coils. J. Chromatogr. A 2003, 1009, 45–59. [Google Scholar]
- Killian, J.A.; Keller, R.C.; Struyve, M.; de Kroon, A.I.; Tommassen, J.; de Kruijff, B. Tryptophan fluorescence study on the interaction of the signal peptide of the Escherichia coli outer membrane protein PhoE with model membranes. Biochemistry 1990, 29, 8131–8137. [Google Scholar] [CrossRef] [PubMed]
- Seelig, J. Thermodynamics of lipid-peptide interactions. Biochim. Biophys. Acta 2004, 1666, 40–50. [Google Scholar]
- Seto, G.W.; Marwaha, S.; Kobewka, D.M.; Lewis, R.N.; Separovic, F.; McElhaney, R.N. Interactions of the Australian tree frog antimicrobial peptides aurein 1.2, citropin 1.1 and maculatin 1.1 with lipid model membranes: Differential scanning calorimetric and Fourier transform infrared spectroscopic studies. Biochim. Biophys. Acta 2007, 1768, 2787–2800. [Google Scholar]
- Nguyen, L.T.; Haney, E.F.; Vogel, H.J. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 2011, 29, 464–472. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Vasil, A.I.; Rehaume, L.; Mant, C.T.; Burns, J.L.; Vasil, M.L.; Hancock, R.E.; Hodges, R.S. Comparison of biophysical and biologic properties of alpha-helical enantiomeric antimicrobial peptides. Chem. Biol. Drug Des. 2006, 67, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Ehrenstein, G.; Lecar, H. Electrically gated ionic channels in lipid bilayers. Q. Rev. Biophys. 1977, 10, 1–34. [Google Scholar] [CrossRef]
- Shai, Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by alpha-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta 1999, 1462, 55–70. [Google Scholar]
- Ross, P.D.; Subramanian, S. Thermodynamics of protein association reactions: Forces contributing to stability. Biochemistry 1981, 20, 3096–3102. [Google Scholar] [CrossRef] [PubMed]
- Andrushchenko, V.V.; Vogel, H.J.; Prenner, E.J. Interactions of tryptophan-rich cathelicidin antimicrobial peptides with model membranes studied by differential scanning calorimetry. Biochim. Biophys. Acta 2007, 1768, 2447–2458. [Google Scholar]
- Fiske, C.H.; Subbarow, Y. The colorimetric determination of phosphorus. J. Biol. Chem. 1925, 66, 375–400. [Google Scholar]
- Eftink, M.R.; Ghiron, C.A. Exposure of tryptophanyl residues in proteins. Quantitative determination by fluorescence quenching studies. Biochemistry 1976, 15, 672–680. [Google Scholar]
- Sample Availability: Samples of the compounds are not available from the authors.
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Li, G.; Huang, Y.; Feng, Q.; Chen, Y. Tryptophan as a Probe to Study the Anticancer Mechanism of Action and Specificity of α-Helical Anticancer Peptides. Molecules 2014, 19, 12224-12241. https://doi.org/10.3390/molecules190812224
Li G, Huang Y, Feng Q, Chen Y. Tryptophan as a Probe to Study the Anticancer Mechanism of Action and Specificity of α-Helical Anticancer Peptides. Molecules. 2014; 19(8):12224-12241. https://doi.org/10.3390/molecules190812224
Chicago/Turabian StyleLi, Guirong, Yibing Huang, Qi Feng, and Yuxin Chen. 2014. "Tryptophan as a Probe to Study the Anticancer Mechanism of Action and Specificity of α-Helical Anticancer Peptides" Molecules 19, no. 8: 12224-12241. https://doi.org/10.3390/molecules190812224