An Alternative Approach to Investigate Biofilm in Medical Devices: A Feasibility Study
Abstract
:1. Introduction
2. Materials and Methods
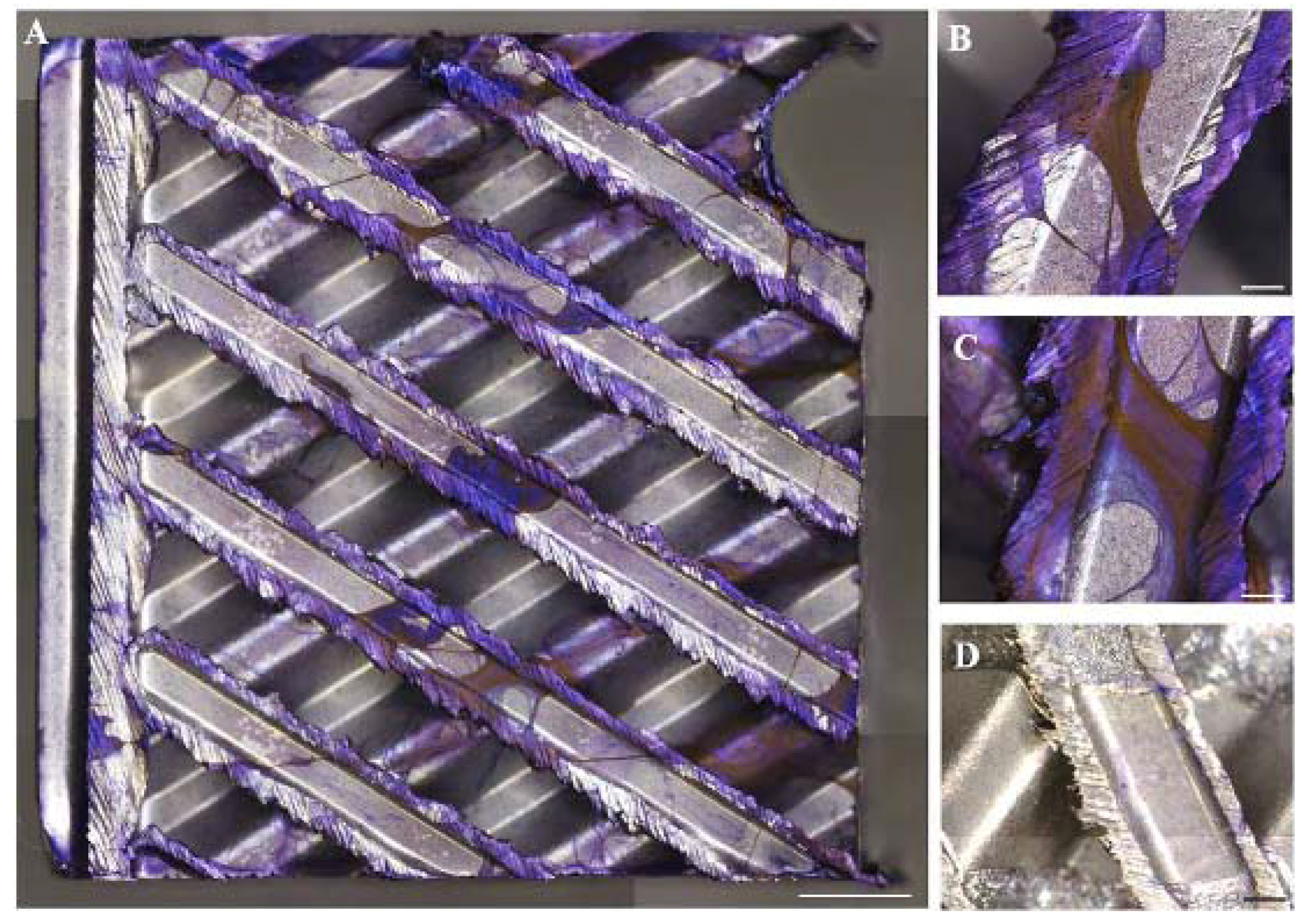
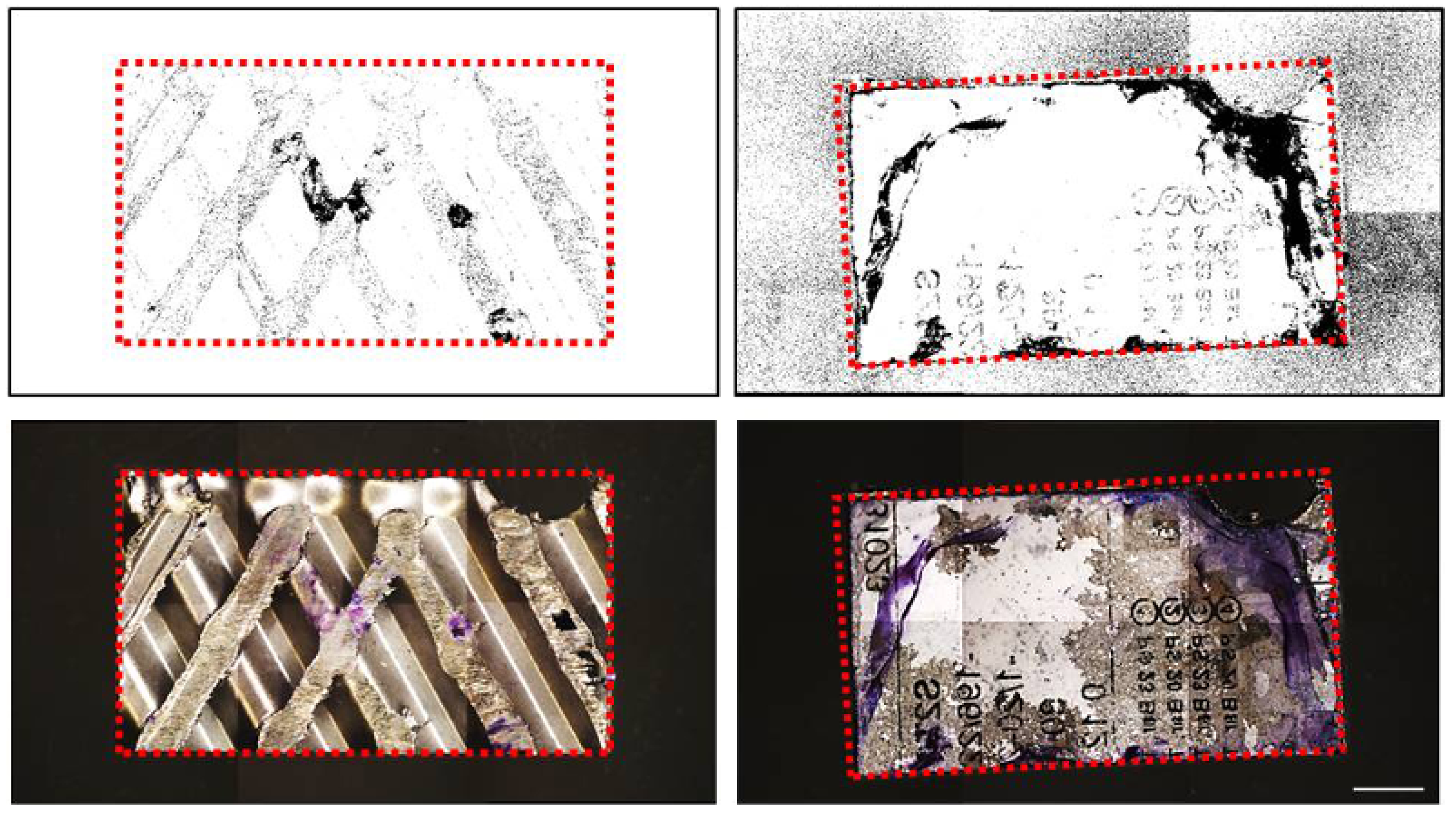
3. Results
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Vinh, D.C.; Embil, J.M. Device-related infections: A review. J. Long Term Eff. Med. Implants 2005, 15, 467–488. [Google Scholar] [CrossRef] [PubMed]
- Seth, A.K.; Geringer, M.R.; Hong, S.J.; Leung, K.P.; Mustoe, T.A.; Galiano, R.D. In vivo modeling of biofilm-infected wounds: A review. J. Surg. Res. 2012, 178, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Van Kleef, E.; Robotham, J.V.; Jit, M.; Deeny, S.R.; Edmunds, W.J. Modelling the transmission of healthcare associated infections: A systematic review. BMC Infect. Dis. 2013, 13, 294. [Google Scholar] [CrossRef] [PubMed]
- Dijkshoorn, L.; Nemec, A.; Seifert, H. An increasing threat in hospitals: Multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 2007, 5, 939–951. [Google Scholar] [CrossRef] [PubMed]
- Bryers, J.D. Medical Biofilms. Biotechnol. Bioeng. 2008, 100, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Reid, G. Biofilms in infectious disease and on medical devices. Int. J. Antimicrob. Agents 1999, 11, 223–226. [Google Scholar] [CrossRef]
- Ferri, M.; Ranucci, E.; Romagnoli, P.; Giaccone, V. Antimicrobial resistance: A global emerging threat to public health systems. Crit. Rev. Food Sci. Nutr. 2017, 57, 2857–2876. [Google Scholar] [CrossRef] [PubMed]
- Sultana, S.T.; Babauta, J.T.; Beyenal, H. Electrochemical biofilm control: A review. Biofouling 2015, 31, 745–758. [Google Scholar] [CrossRef] [PubMed]
- Percival, S.L.; Thomas, J.G.; Williams, D.W. The World of Microbiology and Biofilmology. In Microbiology of Wounds; CRC Press: London, UK, 2010; pp. 1–58. [Google Scholar]
- Lawrence, J.R.; Korber, D.R.; Hoyle, B.D.; Costerton, J.W.; Caldwell, D.E. Optical sectioning of microbial biofilm. J. Bacteriol. 1991, 173, 6558–6567. [Google Scholar] [CrossRef] [PubMed]
- Vu, B.; Chen, M.; Crawford, R.J.; Ivanova, E.P. Bacterial Extracellular Polysaccharides Involved in Biofilm Formation. Molecules 2009, 14, 2535–2554. [Google Scholar] [CrossRef] [PubMed]
- Pantanella, F.; Valenti, P.; Natalizi, T.; Passeri, D.; Berlutti, F. Analytical techniques to study microbial biofilm on abiotic surfaces: Pros and cons of the main techniques currently in use. Ann. Ig. 2013, 25, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Donlan, R.M. Biofilms and Device-Associated Infections. Emerg. Infect. Dis. 2001, 7, 277–281. [Google Scholar] [CrossRef] [PubMed]
- Vertes, A.; Hitchins, V.; Phillips, K.S. Analytical challenges of microbial biofilms on medical devices. Anal. Chem. 2012, 1, 3858–3866. [Google Scholar] [CrossRef] [PubMed]
- IEC (International Electrotechnical Commission). IEC 60601-2-16:2012 Medical Electrical Equipment—Part 2–16: Particular Requirements for Basic Safety and Essential Performance of Haemodialysis, Haemodiafiltration and Haemofiltration Equipment; IEC: Geneva, Switzerland, 2017. [Google Scholar]
- Arnold, J.W. Colorimetric assay for biofilms in wet processing conditions. J. Ind. Microbiol. Biotechnol. 2008, 35, 1475–1480. [Google Scholar] [CrossRef] [PubMed]
- Pittsa, B.; Hamilton, M.A.; Zelverc, N.; Stewart, P.S. A microtiter-plate screening method for biofilm disinfection and removal. Microbiol. Methods 2003, 54, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nature Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Alberink, I.; Ruifrok, A. Repeatability and reproducibility of earprint acquisition. J. Forensic Sci. 2008, 53, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Helps, S.C.; Thornton, E.; Kleinig, T.J.; Manavis, J.; Vink, R. Automatic nonsubjective estimation of antigen content visualized by immunohistochemistry using color deconvolution. Appl. Immunohistochem. Mol. Morphol. 2012, 20, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Marion-Ferey, K.; Pasmore, M.; Stoodley, P.; Wilson, S.; Husson, G.P.; Costerton, J.W. Biofilm removal from silicone tubing: An assessment of the efficacy of dialysis machine decontamination procedures using an in vitro model. J. Hosp. Infect. 2003, 53, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Bakke, R.; Olson, P.Q. Biofilm thickness measurement by light microscopy. J. Microbiol. Methods 1986, 5, 93–98. [Google Scholar] [CrossRef]
- Holt, J.G.; Krieg, N.R.; Sneath, P.H.A.; Staley, J.T.; Williams, S.T. Bergey’s Manual of Determinative Bacteriology, 9th ed.; Lippincott Williams & Wilkins: London, UK, 1994; p. 11. [Google Scholar]

| Contaminated Sample | Stained Area (Pixel2) | mm2 Stained Area | % CV Positive Area | Average (%) |
|---|---|---|---|---|
| #1 | 68,469 | 79,801 | 11.2 | 10.388125 |
| #2 | 26,411 | 30,782 | 4.15 | |
| #3 | 84,321 | 98,276 | 16.13 | |
| #4 | 59,147 | 68,936 | 11.21 | |
| #5 | 211,735 | 246,777 | 16.11 | |
| #6 | 57,876 | 67,455 | 12.135 | |
| #7 | 43,758 | 51 | 6.76 | |
| #8 | 35,147 | 40,964 | 5.41 | |
| Uncontaminated Sample | Stained Area (Pixel2) | mm2 Stained Area | % CV Positive Area | Average (%) |
| #9 | 120 | 3.93 | 0.55 | 0.39125 |
| #10 | 200 | 3.2 | 0.45 | |
| #11 | 136 | 3.27 | 0.54 | |
| #12 | 150 | 2.79 | 0.45 | |
| #13 | 220 | 4.7 | 0.32 | |
| #14 | 164 | 3.74 | 0.24 | |
| #15 | 136 | 1.67 | 0.22 | |
| #16 | 250 | 2.67 | 0.36 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petrachi, T.; Resca, E.; Piccinno, M.S.; Biagi, F.; Strusi, V.; Dominici, M.; Veronesi, E. An Alternative Approach to Investigate Biofilm in Medical Devices: A Feasibility Study. Int. J. Environ. Res. Public Health 2017, 14, 1587. https://doi.org/10.3390/ijerph14121587
Petrachi T, Resca E, Piccinno MS, Biagi F, Strusi V, Dominici M, Veronesi E. An Alternative Approach to Investigate Biofilm in Medical Devices: A Feasibility Study. International Journal of Environmental Research and Public Health. 2017; 14(12):1587. https://doi.org/10.3390/ijerph14121587
Chicago/Turabian StylePetrachi, Tiziana, Elisa Resca, Maria Serena Piccinno, Francesco Biagi, Valentina Strusi, Massimo Dominici, and Elena Veronesi. 2017. "An Alternative Approach to Investigate Biofilm in Medical Devices: A Feasibility Study" International Journal of Environmental Research and Public Health 14, no. 12: 1587. https://doi.org/10.3390/ijerph14121587





