- Article
Hirschioporus abietinus Laccase: Cloning, Heterologous Expression, Characterization and Solvent Tolerance Evaluation
- Ingrida Radveikienė,
- Marius Dagys and
- Vida Časaitė
- + 3 authors
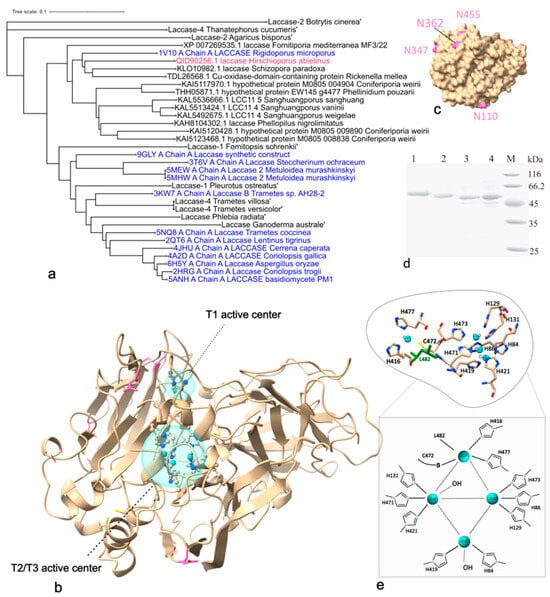
Laccases are versatile biocatalysts with broad industrial relevance. Their heterologous expression enables efficient production, purification, and functional optimization. The white-rot fungus Hirschioporus abietinus produces an effective extracellular laccase (Lac2), inspiring the identification and cloning of its encoding gene. To enable high and stable enzyme production, the gene was expressed in Pichia pastoris and the cultivation conditions for the selected variant were optimized to enhance the yield of recombinant laccase. The Lac2 was then purified and its biochemical properties characterized. The high-redox potential laccase Lac2 exhibited strong tolerance to common metal ions and maintained catalytic activity in the presence of a range of organic solvents. Overall, the results suggest that Lac2 possesses properties compatible with small-scale production and effective use in biosensor systems.
28 January 2026