Supramolecular Nanostructures for Vaccines
Abstract
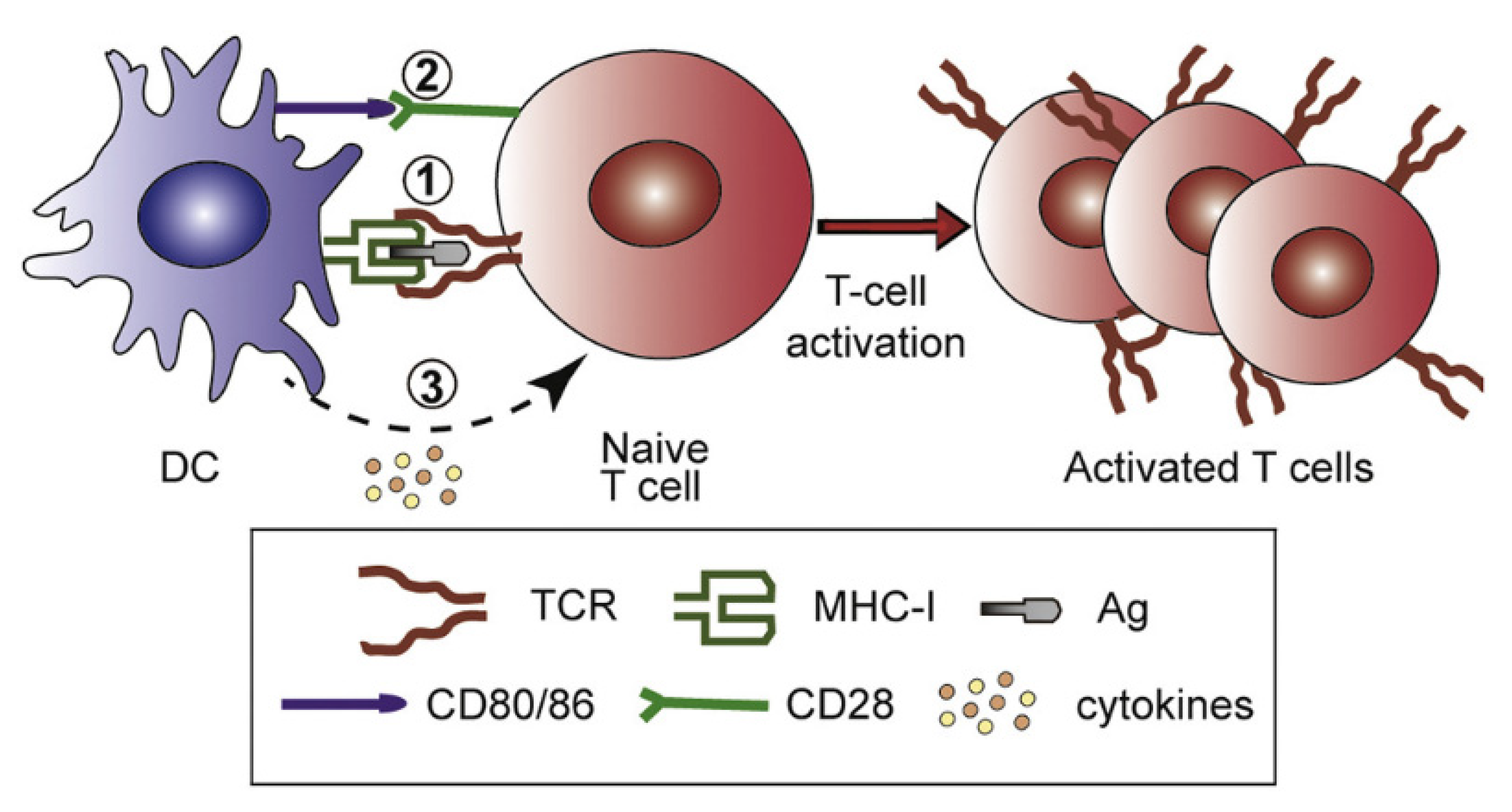
:1. The Importance of Nanoadjuvants for Vaccines
2. Molecular and Supramolecular Assemblies for Vaccines and Beyond
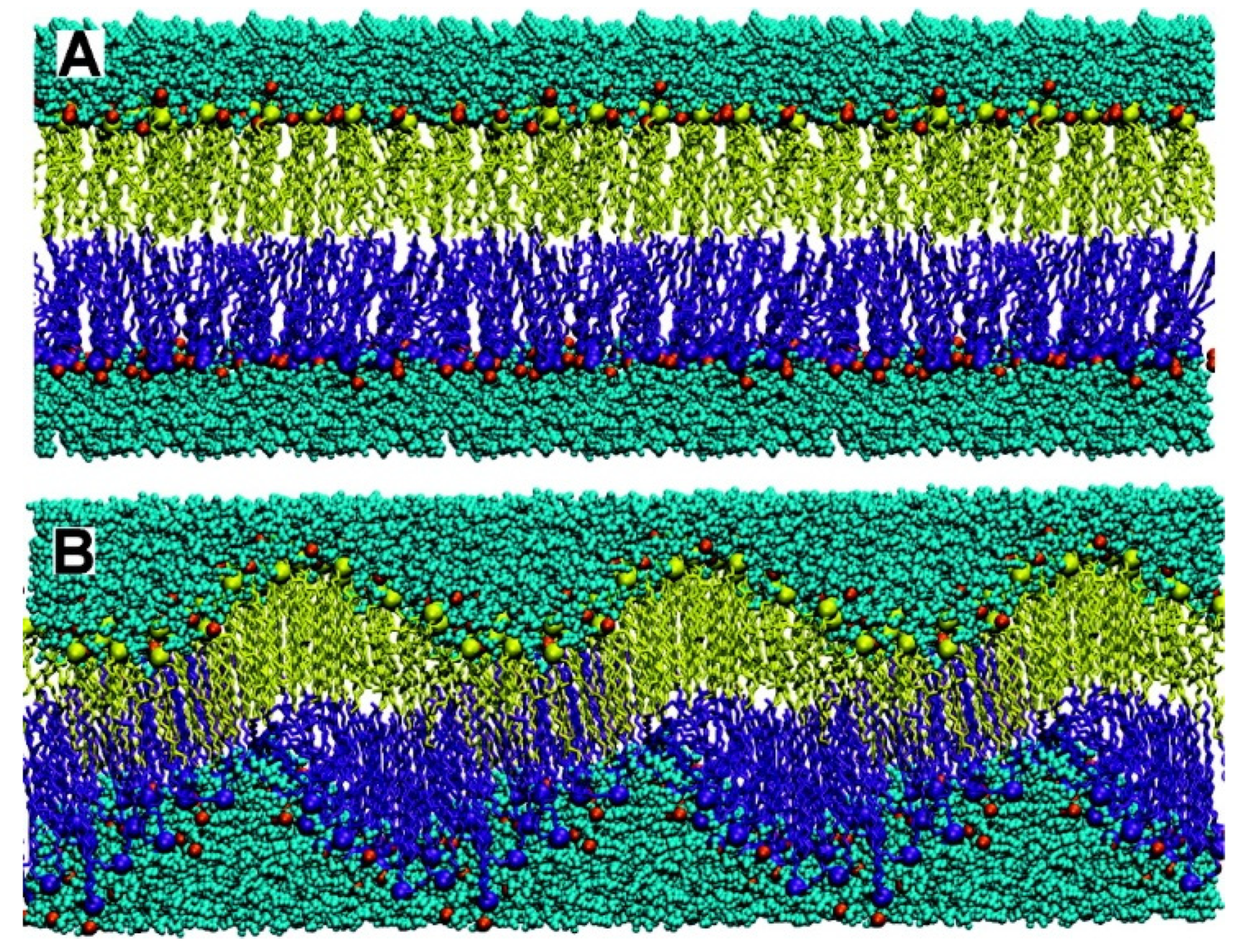
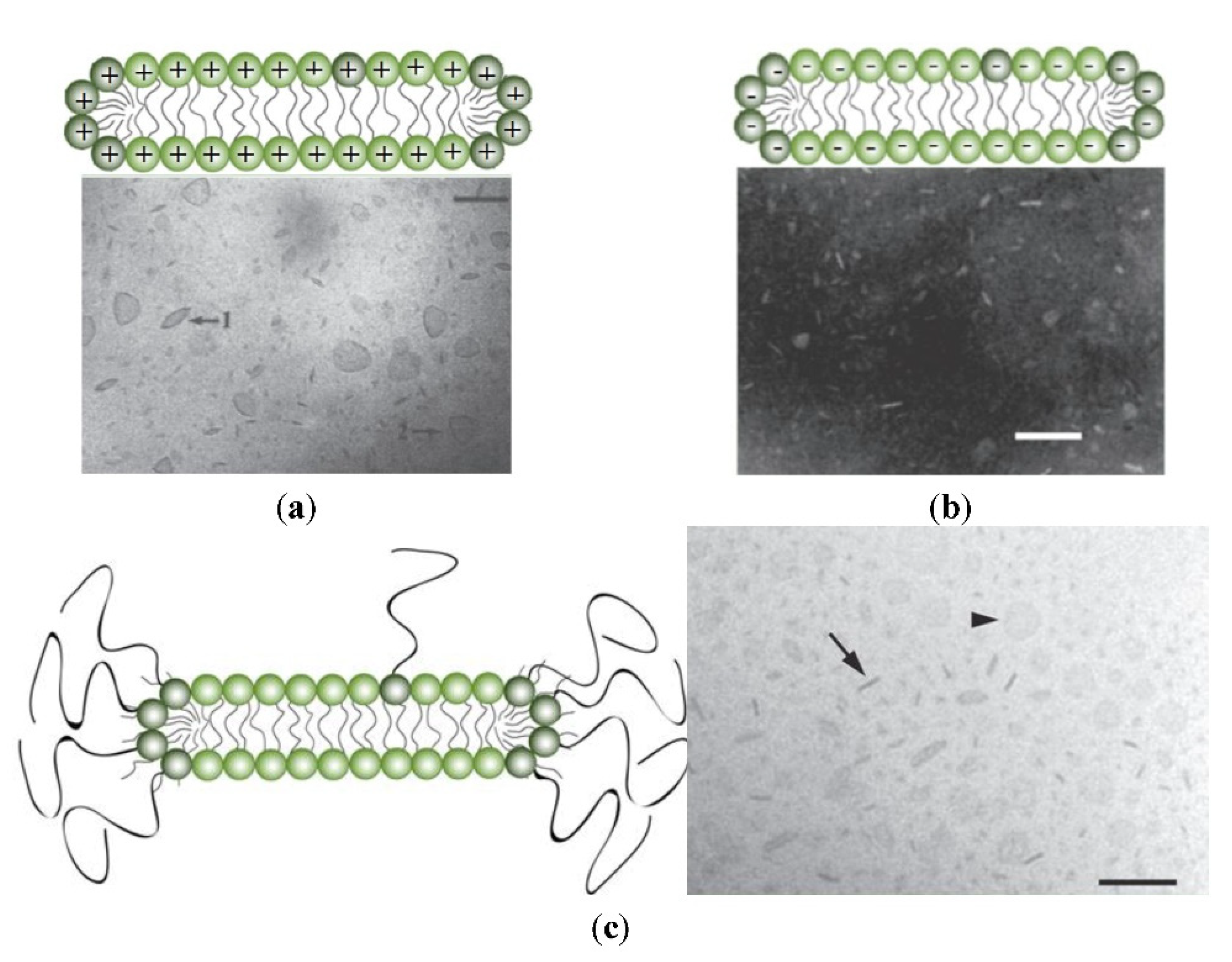
3. Vesicles, Liposomes, Lipid Nanoparticles, Disks, and Niosomes
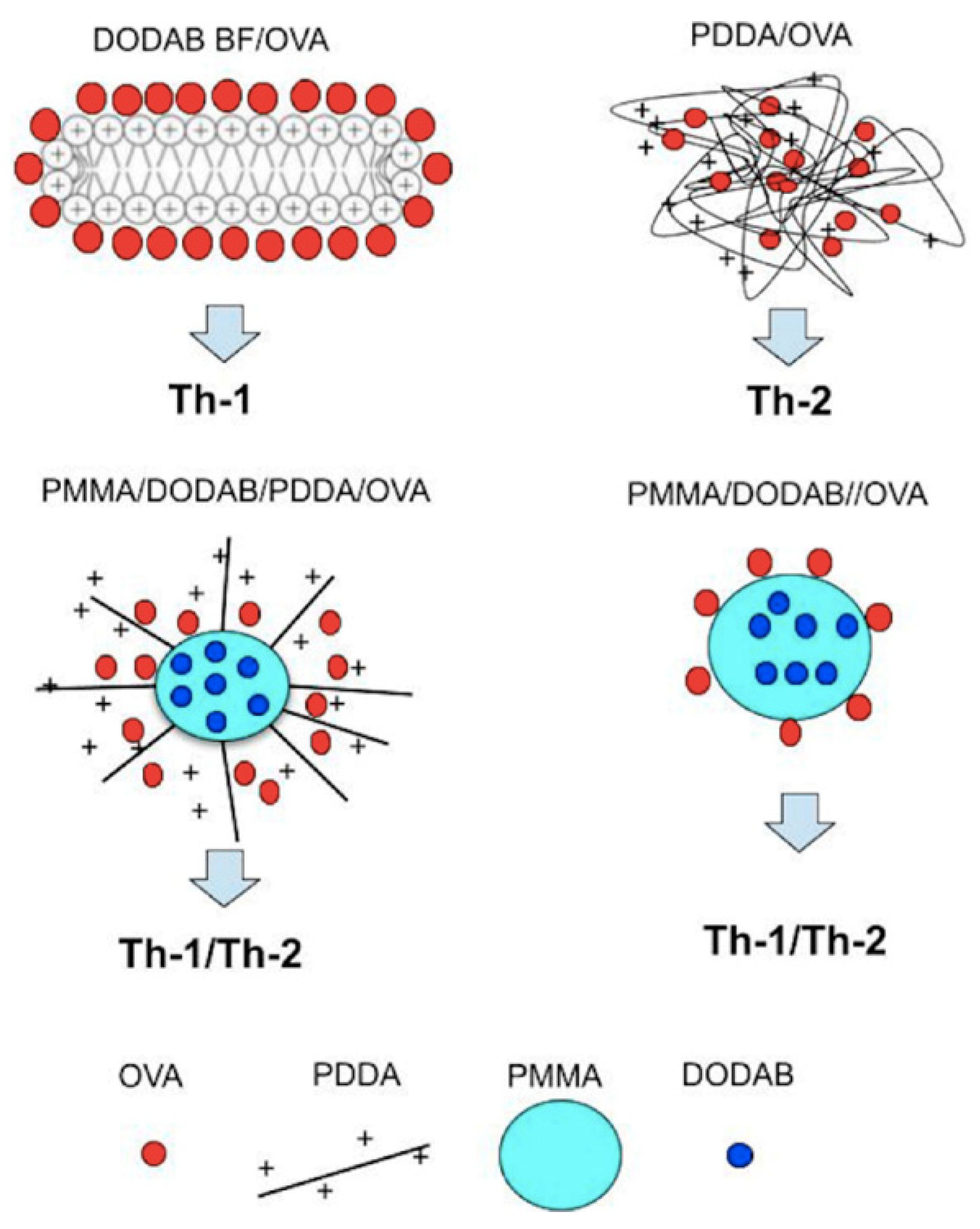
4. Nanoparticles
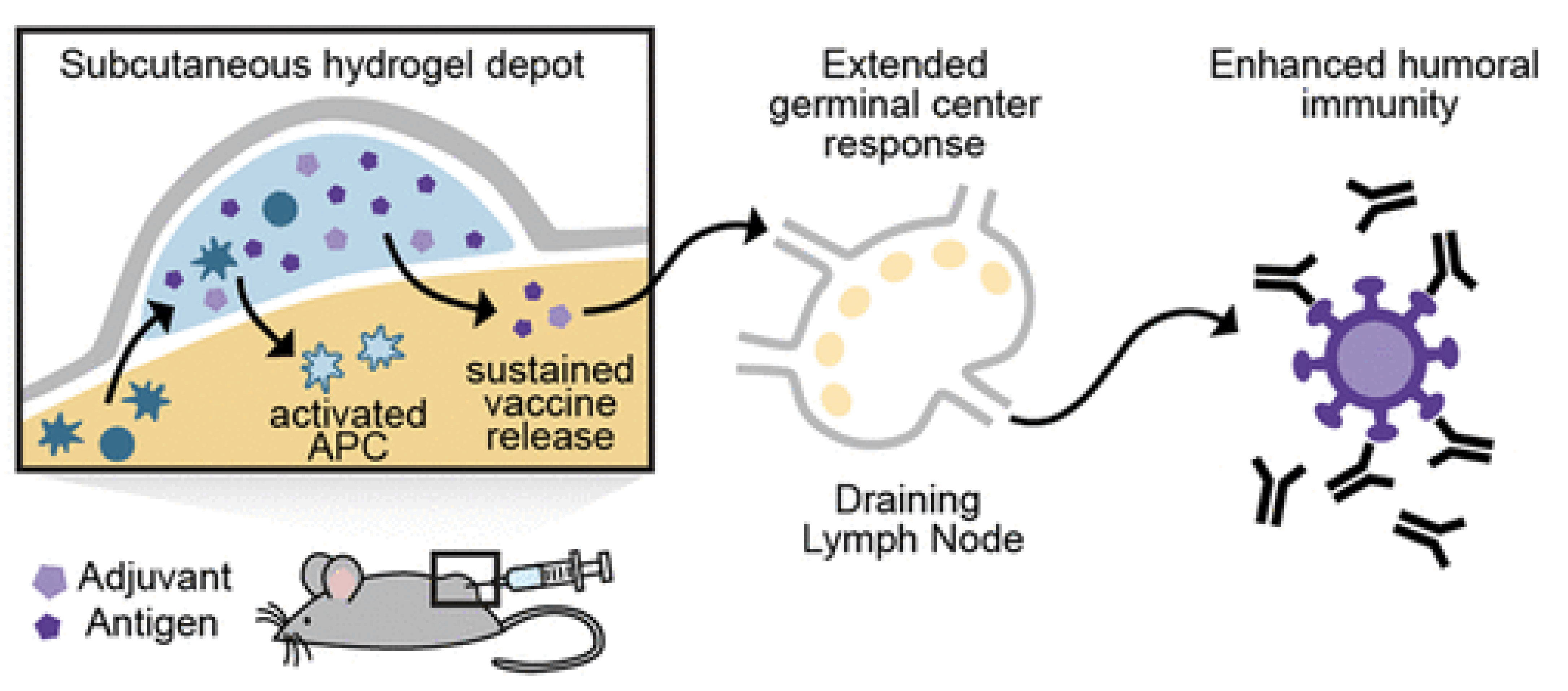
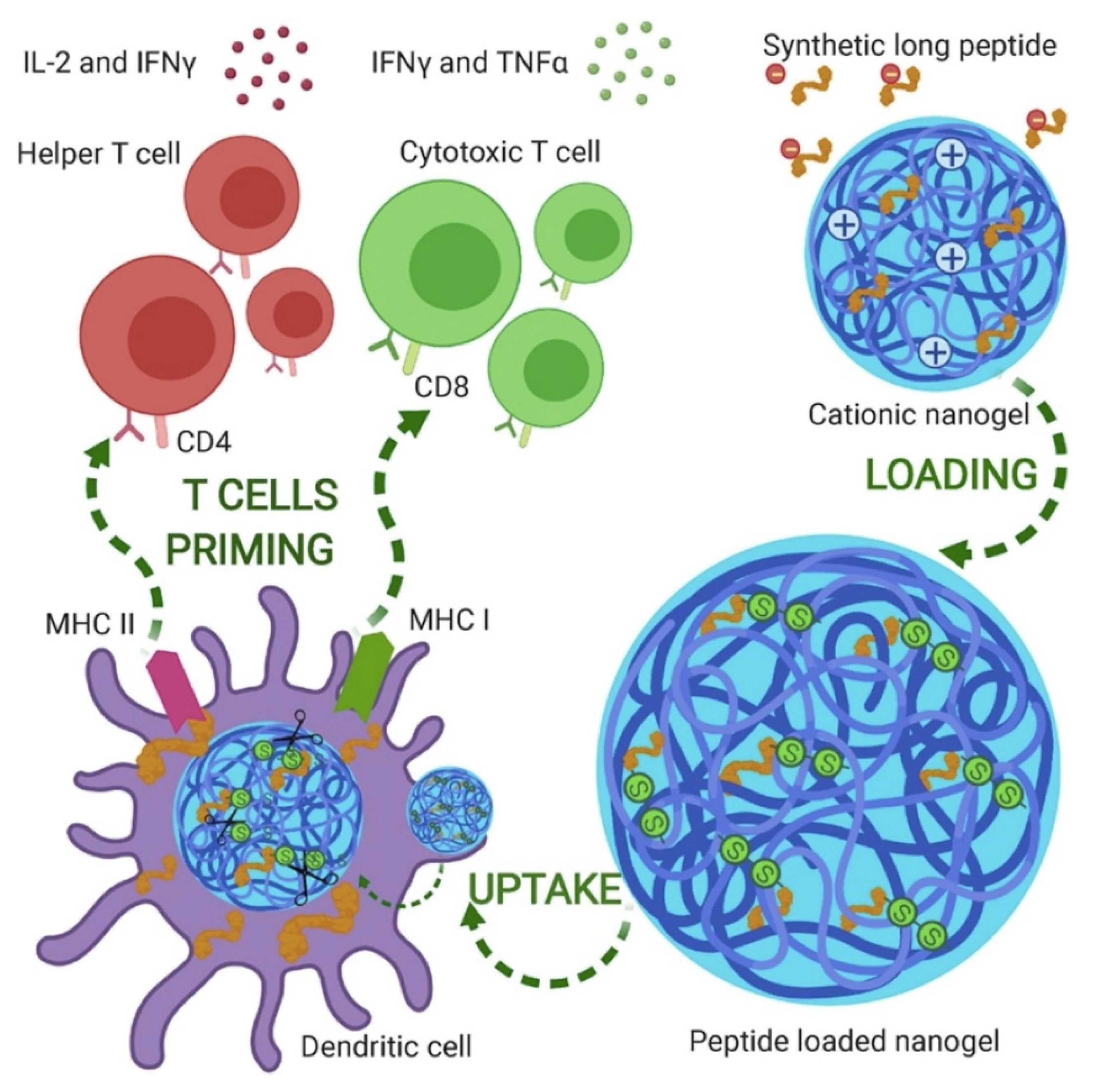
5. Hydrogels
6. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Seyfoori, A.; Shokrollahi Barough, M.; Mokarram, P.; Ahmadi, M.; Mehrbod, P.; Sheidary, A.; Madrakian, T.; Kiumarsi, M.; Walsh, T.; McAlinden, K.D.; et al. Emerging Advances of Nanotechnology in Drug and Vaccine Delivery against Viral Associated Respiratory Infectious Diseases (VARID). Int. J. Mol. Sci. 2021, 22, 6937. [Google Scholar] [CrossRef] [PubMed]
- Perrie, Y.; Mohammed, A.R.; Kirby, D.J.; McNeil, S.E.; Bramwell, V.W. Vaccine Adjuvant Systems: Enhancing the Efficacy of Sub-Unit Protein Antigens. Int. J. Pharm. 2008, 364, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Carmona-Ribeiro, A.M. Nanomaterials Based on Lipids for Vaccine Development. In Micro and Nanotechnology in Vaccine Development; Skwarczynski, M., Toth, I., Eds.; Elsevier: Oxford, UK, 2017; pp. 241–257. ISBN 978-0-323-39981-4. [Google Scholar]
- Carmona-Ribeiro, A.M.; Pérez-Betancourt, Y. Cationic Nanostructures for Vaccines Design. Biomimetics 2020, 5, 32. [Google Scholar] [CrossRef]
- Pérez-Betancourt, Y.; Távora, B.D.C.L.F.; Faquim-Mauro, E.L.; Carmona-Ribeiro, A.M. Biocompatible Lipid Polymer Cationic Nanoparticles for Antigen Presentation. Polymers 2021, 13, 185. [Google Scholar] [CrossRef]
- Foged, C.; Hansen, J.; Agger, E.M. License to Kill: Formulation Requirements for Optimal Priming of CD8(+) CTL Responses with Particulate Vaccine Delivery Systems. Eur. J. Pharm. Sci. Off. J. Eur. Fed. Pharm. Sci. 2012, 45, 482–491. [Google Scholar] [CrossRef] [PubMed]
- Coffman, R.L.; Sher, A.; Seder, R.A. Vaccine Adjuvants: Putting Innate Immunity to Work. Immunity 2010, 33, 492–503. [Google Scholar] [CrossRef] [Green Version]
- Abbas, A.K.; Lichtman, A.H.; Pillai, S. Cellular and Molecular Immunology, 9th ed.; Elsevier Ltd.: Philadelphia, PA, USA, 2018; ISBN 9780323479783. [Google Scholar]
- Manolova, V.; Flace, A.; Bauer, M.; Schwarz, K.; Saudan, P.; Bachmann, M.F. Nanoparticles Target Distinct Dendritic Cell Populations According to Their Size. Eur. J. Immunol. 2008, 38, 1404–1413. [Google Scholar] [CrossRef]
- Fifis, T.; Gamvrellis, A.; Crimeen-Irwin, B.; Pietersz, G.A.; Li, J.; Mottram, P.L.; McKenzie, I.F.C.; Plebanski, M. Size-Dependent Immunogenicity: Therapeutic and Protective Properties of Nano-Vaccines against Tumors. J. Immunol. 2004, 173, 3148–3154. [Google Scholar] [CrossRef] [Green Version]
- Xiang, S.D.; Scholzen, A.; Minigo, G.; David, C.; Apostolopoulos, V.; Mottram, P.L.; Plebanski, M. Pathogen Recognition and Development of Particulate Vaccines: Does Size Matter? Methods 2006, 40, 1–9. [Google Scholar] [CrossRef]
- Carmona-Ribeiro, A.M. Cationic nanostructures for vaccines. In Immune Response Activation; Duc, G.H.T., Ed.; IntechOpen: Rijeka, Croatia, 2014; pp. 1–45. ISBN 978-953-51-1374-4. [Google Scholar]
- Mao, L.; Chen, Z.; Wang, Y.; Chen, C. Design and Application of Nanoparticles as Vaccine Adjuvants against Human Corona Virus Infection. J. Inorg. Biochem. 2021, 219, 111454. [Google Scholar] [CrossRef]
- Tsuruta, L.R.; Quintilio, W.; Costa, M.H.; Carmona-Ribeiro, A.M. Interactions between Cationic Liposomes and an Antigenic Protein: The Physical Chemistry of the Immunoadjuvant Action. J. Lipid Res. 1997, 38, 2003–2011. [Google Scholar] [CrossRef]
- Carmona-Ribeiro, A.M. Synthetic Amphiphile Vesicles. Chem Soc. Rev. 1992, 21, 209–214. [Google Scholar] [CrossRef]
- Carmona-Ribeiro, A.M. Lipid Bilayer Fragments and Disks in Drug Delivery. Curr. Med. Chem. 2006, 13, 1359–1370. [Google Scholar] [CrossRef] [PubMed]
- Lincopan, N.; Espíndola, N.M.; Vaz, A.J.; da Costa, M.H.B.; Faquim-Mauro, E.; Carmona-Ribeiro, A.M. Novel Immunoadjuvants Based on Cationic Lipid: Preparation, Characterization and Activity in Vivo. Vaccine 2009, 27, 5760–5771. [Google Scholar] [CrossRef] [PubMed]
- Rozenfeld, J.H.K.; Silva, S.R.; Raneia, P.A.; Faquim-Mauro, E.; Carmona-Ribeiro, A.M. Stable Assemblies of Cationic Bilayer Fragments and CpG Oligonucleotide with Enhanced Immunoadjuvant Activity in Vivo. J. Control. Release Off. J. Control. Release Soc. 2012, 160, 367–373. [Google Scholar] [CrossRef]
- de Almeida, A.F.; De Gaspari, E. Dioctadecyldimethylammonium Bromide (DODAB-BF) as a New Adjuvant for Maternal-Fetal Immunization in Mice against Neisseria Meningitidis: Evaluation of Humoral Response. Pathog. Dis. 2018, 76, ftx128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersson, M.; Hammarstroem, L.; Edwards, K. Effect of Bilayer Phase Transitions on Vesicle Structure, and Its Influence on the Kinetics of Viologen Reduction. J. Phys. Chem. 1995, 99, 14531–14538. [Google Scholar] [CrossRef]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the MRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef]
- Lincopan, N.; Espindola, N.M.; Vaz, A.J.; Carmona-Ribeiro, A.M. Cationic Supported Lipid Bilayers for Antigen Presentation. Int. J. Pharm. 2007, 340, 216–222. [Google Scholar] [CrossRef]
- Naves, A.F.; Palombo, R.R.; Carrasco, L.D.M.; Carmona-Ribeiro, A.M. Antimicrobial Particles from Emulsion Polymerization of Methyl Methacrylate in the Presence of Quaternary Ammonium Surfactants. Langmuir ACS J. Surf. Colloids 2013, 29, 9677–9684. [Google Scholar] [CrossRef]
- Sanches, L.M.; Petri, D.F.S.; de Melo Carrasco, L.D.; Carmona-Ribeiro, A.M. The Antimicrobial Activity of Free and Immobilized Poly (Diallyldimethylammonium) Chloride in Nanoparticles of Poly (Methylmethacrylate). J. Nanobiotechnol. 2015, 13, 58. [Google Scholar] [CrossRef] [Green Version]
- Xavier, G.R.S.; Carmona-Ribeiro, A.M. Cationic Biomimetic Particles of Polystyrene/Cationic Bilayer/Gramicidin for Optimal Bactericidal Activity. Nanomaterials 2017, 7, 422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathiazzi, B.I.; Carmona-Ribeiro, A.M. Hybrid Nanoparticles of Poly (Methyl Methacrylate) and Antimicrobial Quaternary Ammonium Surfactants. Pharmaceutics 2020, 12, 340. [Google Scholar] [CrossRef]
- Ribeiro, A.M.; Chaimovich, H. Preparation and Characterization of Large Dioctadecyldimethylammonium Chloride Liposomes and Comparison with Small Sonicated Vesicles. Biochim. Biophys. Acta 1983, 733, 172–179. [Google Scholar] [CrossRef]
- Carmona-Ribeiro, A.M.; Chaimovich, H. Salt-Induced Aggregation and Fusion of Dioctadecyldimethylammonium Chloride and Sodium Dihexadecylphosphate Vesicles. Biophys. J. 1986, 50, 621–628. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, L.A.; Carmona-Ribeiro, A.M. Interactions between Cationic Vesicles and Serum Proteins. Langmuir 1998, 14, 6077–6081. [Google Scholar] [CrossRef]
- Lincopan, N.; Carmona-Ribeiro, A.M. Protein Assembly onto Cationic Supported Bilayers. J. Nanosci. Nanotechnol. 2009, 9, 3578–3586. [Google Scholar] [CrossRef]
- Carvalho, C.A.; Olivares-Ortega, C.; Soto-Arriaza, M.A.; Carmona-Ribeiro, A.M. Interaction of Gramicidin with DPPC/DODAB Bilayer Fragments. Biochim. Biophys. Acta 2012, 1818, 3064–3071. [Google Scholar] [CrossRef] [PubMed]
- Ragioto, D.A.M.T.; Carrasco, L.D.M.; Carmona-Ribeiro, A.M. Novel Gramicidin Formulations in Cationic Lipid as Broad-Spectrum Microbicidal Agents. Int. J. Nanomed. 2014, 9, 3183–3192. [Google Scholar] [CrossRef] [Green Version]
- Rozenfeld, J.H.K.; Oliveira, T.R.; Lamy, M.T.; Carmona-Ribeiro, A.M. Interaction of Cationic Bilayer Fragments with a Model Oligonucleotide. Biochim. Biophys. Acta 2011, 1808, 649–655. [Google Scholar] [CrossRef]
- Kikuchi, I.S.; Viviani, W.; Carmona-Ribeiro, A.M. Nucleotide Insertion in Cationic Bilayers. J. Phys. Chem. A 1999, 103, 8050–8055. [Google Scholar] [CrossRef]
- Nantes, I.L.; Correia, F.M.; Faljoni-Alario, A.; Kawanami, A.E.; Ishiki, H.M.; Amaral, A.T.; Carmona-Ribeiro, A.M. Nucleotide Conformational Change Induced by Cationic Bilayers. Arch. Biochem. Biophys. 2003, 416, 25–30. [Google Scholar] [CrossRef]
- Rosa, H.; Petri, D.F.S.; Carmona-Ribeiro, A.M. Interactions between Bacteriophage DNA and Cationic Biomimetic Particles. J. Phys. Chem. B 2008, 112, 16422–16430. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, I.S.; Carmona-Ribeiro, A.M. Interactions between DNA and Synthetic Cationic Liposomes. J. Phys. Chem. B 2000, 104, 2829–2835. [Google Scholar] [CrossRef]
- Carmona-Ribeiro, A.M.; Midmore, B.R. Synthetic Bilayer Adsorption onto Polystyrene Microspheres. Langmuir 1992, 8, 801–806. [Google Scholar] [CrossRef]
- Carmona-Ribeiro, A.M.; de Moraes Lessa, M. Interactions between Bilayer Membranes and Latex. Colloids Surf. Physicochem. Eng. Asp. 1999, 153, 355–361. [Google Scholar] [CrossRef]
- Pereira, E.M.A.; Vieira, D.B.; Carmona-Ribeiro, A.M. Cationic Bilayers on Polymeric Particles: Effect of Low NaCl Concentration on Surface Coverage. J. Phys. Chem. B 2004, 108, 11490–11495. [Google Scholar] [CrossRef]
- Rapuano, R.; Carmona-Ribeiro, A.M. Supported Bilayers on Silica. J. Colloid Interface Sci. 2000, 226, 299–307. [Google Scholar] [CrossRef]
- Moura, S.P.; Carmona-Ribeiro, A.M. Cationic Bilayer Fragments on Silica at Low Ionic Strength: Competitive Adsorption and Colloid Stability. Langmuir 2003, 19, 6664–6667. [Google Scholar] [CrossRef]
- Lincopan, N.; Santana, M.R.; Faquim-Mauro, E.; da Costa, M.H.B.; Carmona-Ribeiro, A.M. Silica-Based Cationic Bilayers as Immunoadjuvants. BMC Biotechnol. 2009, 9, 5. [Google Scholar] [CrossRef] [Green Version]
- Ribeiro, R.T.; Braga, V.H.A.; Carmona-Ribeiro, A.M. Biomimetic Cationic Nanoparticles Based on Silica: Optimizing Bilayer Deposition from Lipid Films. Biomimetics 2017, 2, 20. [Google Scholar] [CrossRef] [PubMed]
- Pereira, E.M.A.; Kosaka, P.M.; Rosa, H.; Vieira, D.B.; Kawano, Y.; Petri, D.F.S.; Carmona-Ribeiro, A.M. Hybrid Materials from Intermolecular Associations between Cationic Lipid and Polymers. J. Phys. Chem. B 2008, 112, 9301–9310. [Google Scholar] [CrossRef]
- Pérez-Betancourt, Y.; Araujo, P.M.; Távora, B.D.C.L.F.; Pereira, D.R.; Faquim-Mauro, E.L.; Carmona-Ribeiro, A.M. Cationic and Biocompatible Polymer/Lipid Nanoparticles as Immunoadjuvants. Pharmaceutics 2021, 13, 1859. [Google Scholar] [CrossRef]
- Mbow, M.L.; De Gregorio, E.; Valiante, N.M.; Rappuoli, R. New Adjuvants for Human Vaccines. Curr. Opin. Immunol. 2010, 22, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Giudice, G.D.; Rappuoli, R.; Didierlaurent, A.M. Correlates of Adjuvanticity: A Review on Adjuvants in Licensed Vaccines. Semin. Immunol. 2018, 39, 14–21. [Google Scholar] [CrossRef]
- Leroux-Roels, G. Unmet Needs in Modern Vaccinology: Adjuvants to Improve the Immune Response. Vaccine 2010, 28 (Suppl. 3), C25–C36. [Google Scholar] [CrossRef] [PubMed]
- Villa, L.L. HPV Prophylactic Vaccination: The First Years and What to Expect from Now. Cancer Lett. 2011, 305, 106–112. [Google Scholar] [CrossRef]
- Cupovic, J.; Ring, S.S.; Onder, L.; Colston, J.M.; Lütge, M.; Cheng, H.-W.; De Martin, A.; Provine, N.M.; Flatz, L.; Oxenius, A.; et al. Adenovirus Vector Vaccination Reprograms Pulmonary Fibroblastic Niches to Support Protective Inflating Memory CD8+ T Cells. Nat. Immunol. 2021, 22, 1042–1051. [Google Scholar] [CrossRef]
- Del Médico Zajac, M.P.; Molinari, P.; Gravisaco, M.J.; Maizon, D.O.; Morón, G.; Gherardi, M.M.; Calamante, G. MVAΔ008 Viral Vector Encoding the Model Protein OVA Induces Improved Immune Response against the Heterologous Antigen and Equal Levels of Protection in a Mice Tumor Model than the Conventional MVA. Mol. Immunol. 2021, 139, 115–122. [Google Scholar] [CrossRef]
- Pinto, P.B.A.; Assis, M.L.; Vallochi, A.L.; Pacheco, A.R.; Lima, L.M.; Quaresma, K.R.L.; Pereira, B.A.S.; Costa, S.M.; Alves, A.M.B. T Cell Responses Induced by DNA Vaccines Based on the DENV2 E and NS1 Proteins in Mice: Importance in Protection and Immunodominant Epitope Identification. Front. Immunol. 2019, 10, 1522. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.A. A Comparison of Plasmid DNA and MRNA as Vaccine Technologies. Vaccines 2019, 7, 37. [Google Scholar] [CrossRef] [Green Version]
- Hobernik, D.; Bros, M. DNA Vaccines—How Far From Clinical Use? Int. J. Mol. Sci. 2018, 19, 3605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, S.; Yang, K.; Li, R.; Zhang, L. MRNA Vaccine Era-Mechanisms, Drug Platform and Clinical Prospection. Int. J. Mol. Sci. 2020, 21, 6582. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. MRNA Vaccines—A New Era in Vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef] [Green Version]
- Qiao, D.; Liu, L.; Chen, Y.; Xue, C.; Gao, Q.; Mao, H.-Q.; Leong, K.W.; Chen, Y. Potency of a Scalable Nanoparticulate Subunit Vaccine. Nano Lett. 2018, 18, 3007–3016. [Google Scholar] [CrossRef]
- Silva, A.L.; Soema, P.C.; Slütter, B.; Ossendorp, F.; Jiskoot, W. PLGA Particulate Delivery Systems for Subunit Vaccines: Linking Particle Properties to Immunogenicity. Hum. Vaccines Immunother. 2016, 12, 1056–1069. [Google Scholar] [CrossRef] [PubMed]
- Francis, M.J. Recent Advances in Vaccine Technologies. Vet. Clin. N. Am. Small Anim. Pract. 2018, 48, 231–241. [Google Scholar] [CrossRef]
- Silveira, M.M.; Moreira, G.M.S.G.; Mendonça, M. DNA Vaccines against COVID-19: Perspectives and Challenges. Life Sci. 2021, 267, 118919. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 MRNA Covid-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Pulendran, B.; Arunachalam, P.S.; O’Hagan, D.T. Emerging Concepts in the Science of Vaccine Adjuvants. Nat. Rev. Drug Discov. 2021, 20, 454–475. [Google Scholar] [CrossRef] [PubMed]
- Reed, S.G.; Orr, M.T.; Fox, C.B. Key Roles of Adjuvants in Modern Vaccines. Nat. Med. 2013, 19, 1597–1608. [Google Scholar] [CrossRef]
- Bastola, R.; Noh, G.; Keum, T.; Bashyal, S.; Seo, J.-E.; Choi, J.; Oh, Y.; Cho, Y.; Lee, S. Vaccine Adjuvants: Smart Components to Boost the Immune System. Arch. Pharm. Res. 2017, 40, 1238–1248. [Google Scholar] [CrossRef]
- Mutwiri, G.; Gerdts, V.; van Drunen Littel-van den Hurk, S.; Auray, G.; Eng, N.; Garlapati, S.; Babiuk, L.A.; Potter, A. Combination Adjuvants: The next Generation of Adjuvants? Expert Rev. Vaccines 2011, 10, 95–107. [Google Scholar] [CrossRef]
- Carmona-Ribeiro, A.M. Biomimetic Nanomaterials from the Assembly of Polymers, Lipids, and Surfactants. In Surfactants and Detergents; Dutta, A.K., Ed.; IntechOpen: Rijeka, Croatia, 2019; pp. 1–16. ISBN 978-1-78984-661-4. [Google Scholar]
- Carmona-Ribeiro, A.M. Biomimetic Nanoparticles: Preparation, Characterization and Biomedical Applications. Int. J. Nanomed. 2010, 5, 249–259. [Google Scholar] [CrossRef] [Green Version]
- Carmona-Ribeiro, A.M. Biomimetic Lipid Polymer Nanoparticles for Drug Delivery. In Nanoparticles in Biology and Medicine: Methods and Protocols; Ferrari, E., Soloviev, M., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2020; pp. 45–60. ISBN 978-1-07-160319-2. [Google Scholar]
- Carmona-Ribeiro, A.M.; Barbassa, L.; de Melo, L.D. Antimicrobial Biomimetics. In Biomimetic Based Applications; Cavrak, M., Ed.; InTech: Rijeka, Croatia, 2011; pp. 227–284. ISBN 978-953-307-195-4. [Google Scholar]
- Carmona-Ribeiro, A.M. Self-Assembled Antimicrobial Nanomaterials. Int. J. Environ. Res. Public Health 2018, 15, 1408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Israelachvili, J.N.; Mitchell, D.J.; Ninham, B.W. Theory of Self-Assembly of Lipid Bilayers and Vesicles. Biochim. Biophys. Acta BBA-Biomembr. 1977, 470, 185–201. [Google Scholar] [CrossRef]
- Israelachvili, J.N. Intermolecular and Surface Forces; Academic Press: Cambridge, MA, USA, 2015; ISBN 978-0-08-092363-5. [Google Scholar]
- Carmona-Ribeiro, A.M. Biomimetic Systems in Nanomedicine. In Handbook of Nanobiomedical Research: Fundamentals, Applications and Recent Developments; Torchilin, W., Ed.; World Scientific Publishing Co. Pte. Ltd.: Singapore, 2014; Volume 4, pp. 401–456. ISBN 978-981-4520-64-5. [Google Scholar]
- Carmona-Ribeiro, A.M. The versatile dioctadecyldimethylammonium bromide. In Application and Characterization of Surfactants; Najjar, R., Ed.; IntechOpen: Rijeka, Croatia, 2017; pp. 157–182. ISBN 978-953-51-3326-1. [Google Scholar]
- Carmona-Ribeiro, A.M.; Yoshida, L.S.; Chaimovich, H. Salt Effects on the Stability of Dioctadecyldimethylammonium Chloride and Sodium Dihexadecyl Phosphate Vesicles. J. Phys. Chem. 1985, 89, 2928–2933. [Google Scholar] [CrossRef]
- Claesson, P.; Carmona-Ribeiro, A.M.; Kurihara, K. Dihexadecyl Phosphate Monolayers: Intralayer and Interlayer Interactions. J. Phys. Chem. 1989, 93, 917–922. [Google Scholar] [CrossRef]
- Carmona-Ribeiro, A.M. Interactions between Charged Spheric Vesicles. J. Phys. Chem. 1993, 97, 11843–11846. [Google Scholar] [CrossRef]
- Tsuruta, L.R.; Lessa, M.M.; Carmona-Ribeiro, A.M. Interactions between Dioctadecyldimethylammonium Chloride or Bromide Bilayers in Water. Langmuir 1995, 11, 2938–2943. [Google Scholar] [CrossRef]
- Jamróz, D.; Kepczynski, M.; Nowakowska, M. Molecular Structure of the Dioctadecyldimethylammonium Bromide (DODAB) Bilayer. Langmuir 2010, 26, 15076–15079. [Google Scholar] [CrossRef]
- Feitosa, E.; Adati, R.D.; Hansson, P.; Malmsten, M. Thermal and Structural Behavior of Dioctadecyldimethylammonium Bromide Dispersions Studied by Differential Scanning Calorimetry and X-Ray Scattering. PLoS ONE 2012, 7, e44702. [Google Scholar] [CrossRef] [PubMed]
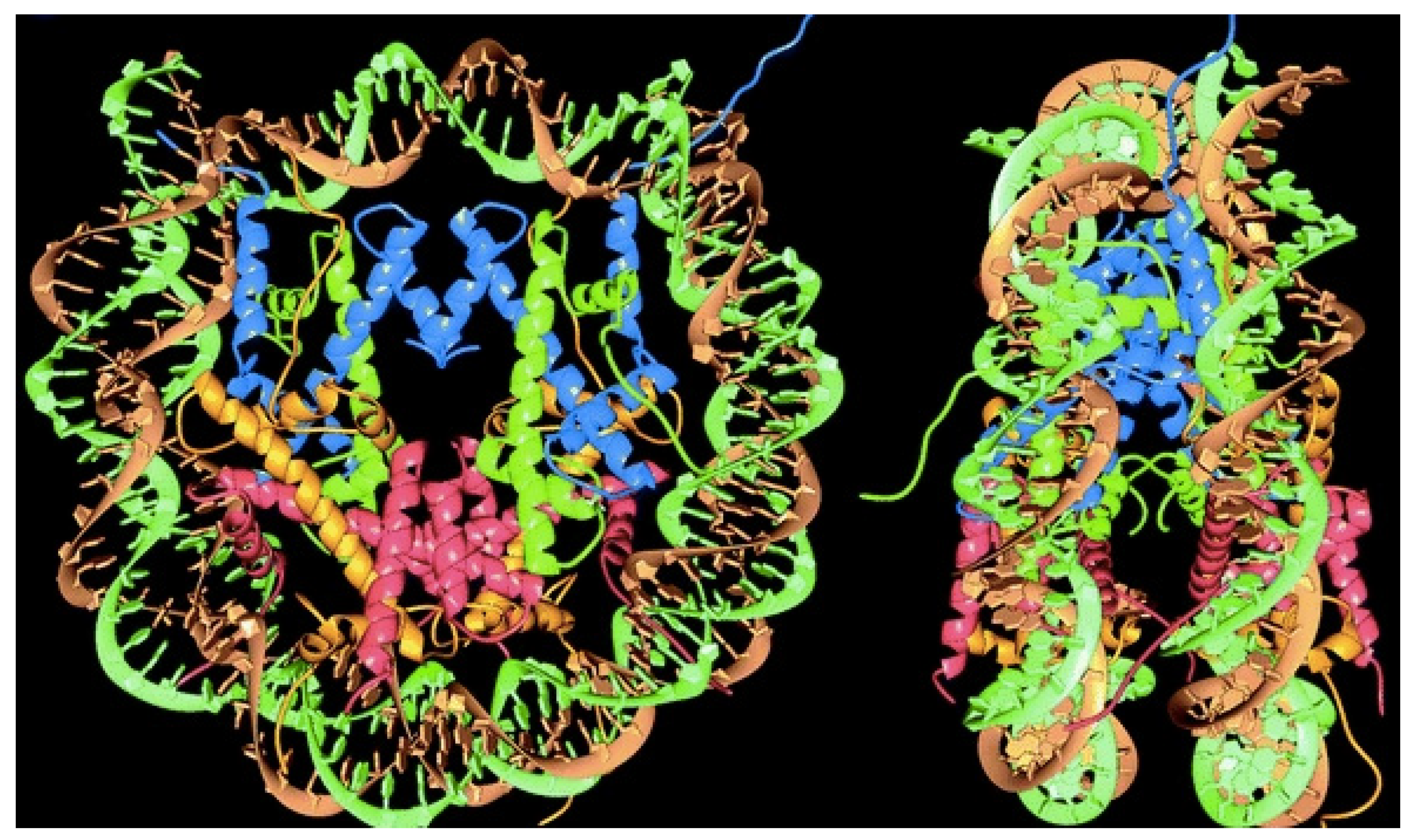
- Rohs, R.; West, S.M.; Sosinsky, A.; Liu, P.; Mann, R.S.; Honig, B. The Role of DNA Shape in Protein–DNA Recognition. Nature 2009, 461, 1248–1253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weaver, T.M.; Morrison, E.A.; Musselman, C.A. Reading More than Histones: The Prevalence of Nucleic Acid Binding among Reader Domains. Molecules 2018, 23, 2614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal Structure of the Nucleosome Core Particle at 2.8 Å Resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef]
- Widom, J. Toward a Unified Model of Chromatin Folding. Annu. Rev. Biophys. Biophys. Chem. 1989, 18, 365–395. [Google Scholar] [CrossRef]
- van Holde, K.; Zlatanova, J. Scanning Chromatin: A New Paradigm? J. Biol. Chem. 2006, 281, 12197–12200. [Google Scholar] [CrossRef] [Green Version]
- Blank, T.A.; Becker, P.B. The Effect of Nucleosome Phasing Sequences and DNA Topology on Nucleosome Spacing. J. Mol. Biol. 1996, 260, 1–8. [Google Scholar] [CrossRef]
- Blank, T.A.; Becker, P.B. Electrostatic Mechanism of Nucleosome Spacing. J. Mol. Biol. 1995, 252, 305–313. [Google Scholar] [CrossRef]
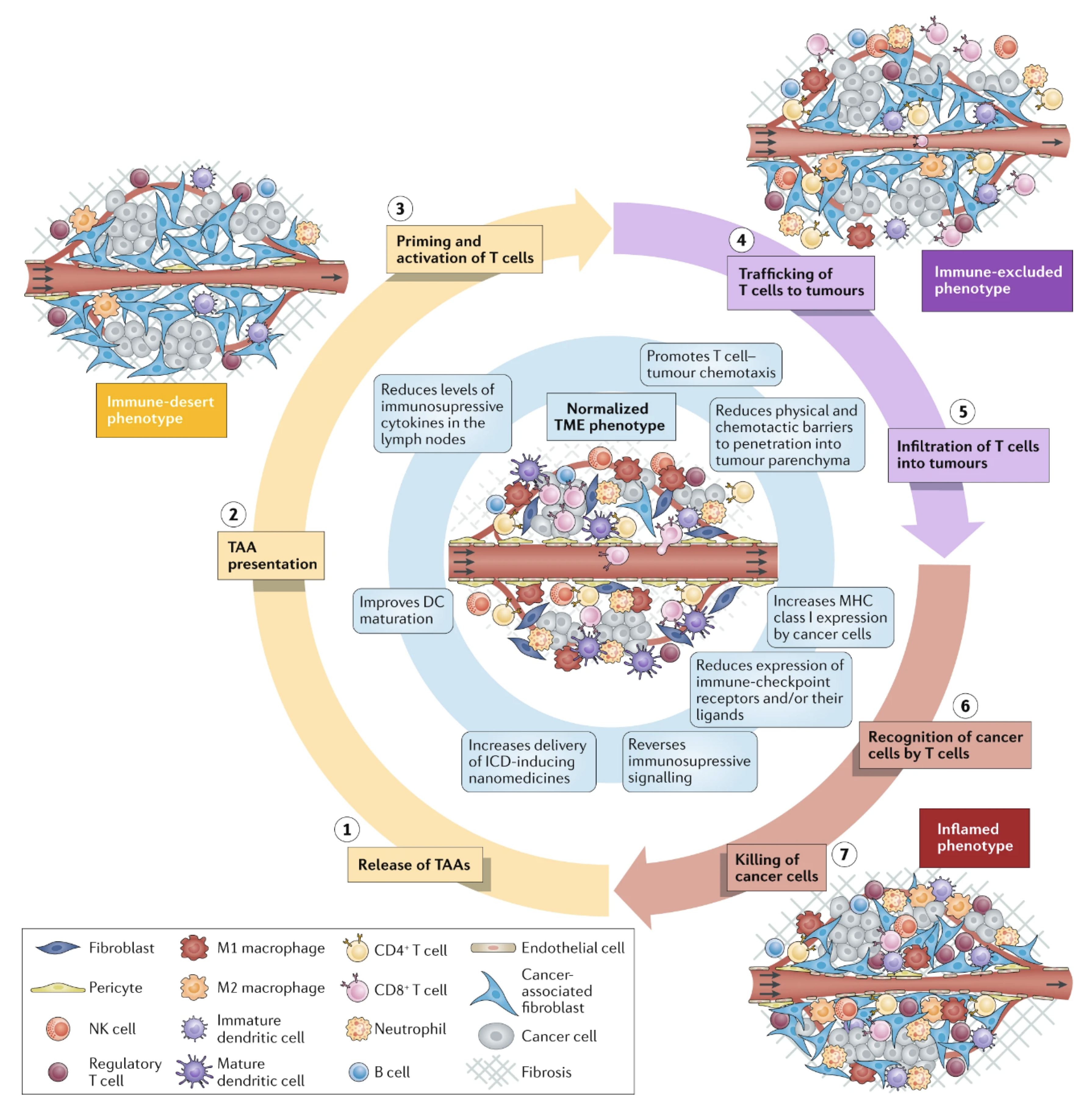
- Zuo, S.; Song, J.; Zhang, J.; He, Z.; Sun, B.; Sun, J. Nano-Immunotherapy for Each Stage of Cancer Cellular Immunity: Which, Why, and What? Theranostics 2021, 11, 7471–7487. [Google Scholar] [CrossRef]
- Vasan, N.; Baselga, J.; Hyman, D.M. A View on Drug Resistance in Cancer. Nature 2019, 575, 299–309. [Google Scholar] [CrossRef] [Green Version]
- Cao, J.; Huang, D.; Peppas, N.A. Advanced Engineered Nanoparticulate Platforms to Address Key Biological Barriers for Delivering Chemotherapeutic Agents to Target Sites. Adv. Drug Deliv. Rev. 2020, 167, 170–188. [Google Scholar] [CrossRef]
- Pérez-Betancourt, Y.; Távora, B.D.C.L.F.; Colombini, M.; Faquim-Mauro, E.L.; Carmona-Ribeiro, A.M. Simple Nanoparticles from the Assembly of Cationic Polymer and Antigen as Immunoadjuvants. Vaccines 2020, 8, 105. [Google Scholar] [CrossRef] [Green Version]
- Shalash, A.O.; Hussein, W.M.; Skwarczynski, M.; Toth, I. Key Considerations for the Development of Safe and Effective SARS-CoV-2 Subunit Vaccine: A Peptide-Based Vaccine Alternative. Adv. Sci. Weinh. Baden-Wurtt. Ger. 2021, 8, e2100985. [Google Scholar] [CrossRef]
- Martin, J.D.; Cabral, H.; Stylianopoulos, T.; Jain, R.K. Improving Cancer Immunotherapy Using Nanomedicines: Progress, Opportunities and Challenges. Nat. Rev. Clin. Oncol. 2020, 17, 251–266. [Google Scholar] [CrossRef] [PubMed]
- Kaps, L.; Schuppan, D. Targeting Cancer Associated Fibroblasts in Liver Fibrosis and Liver Cancer Using Nanocarriers. Cells 2020, 9, 2027. [Google Scholar] [CrossRef]
- Schiller, J.; Chackerian, B. Why HIV Virions Have Low Numbers of Envelope Spikes: Implications for Vaccine Development. PLOS Pathog. 2014, 10, e1004254. [Google Scholar] [CrossRef] [PubMed]
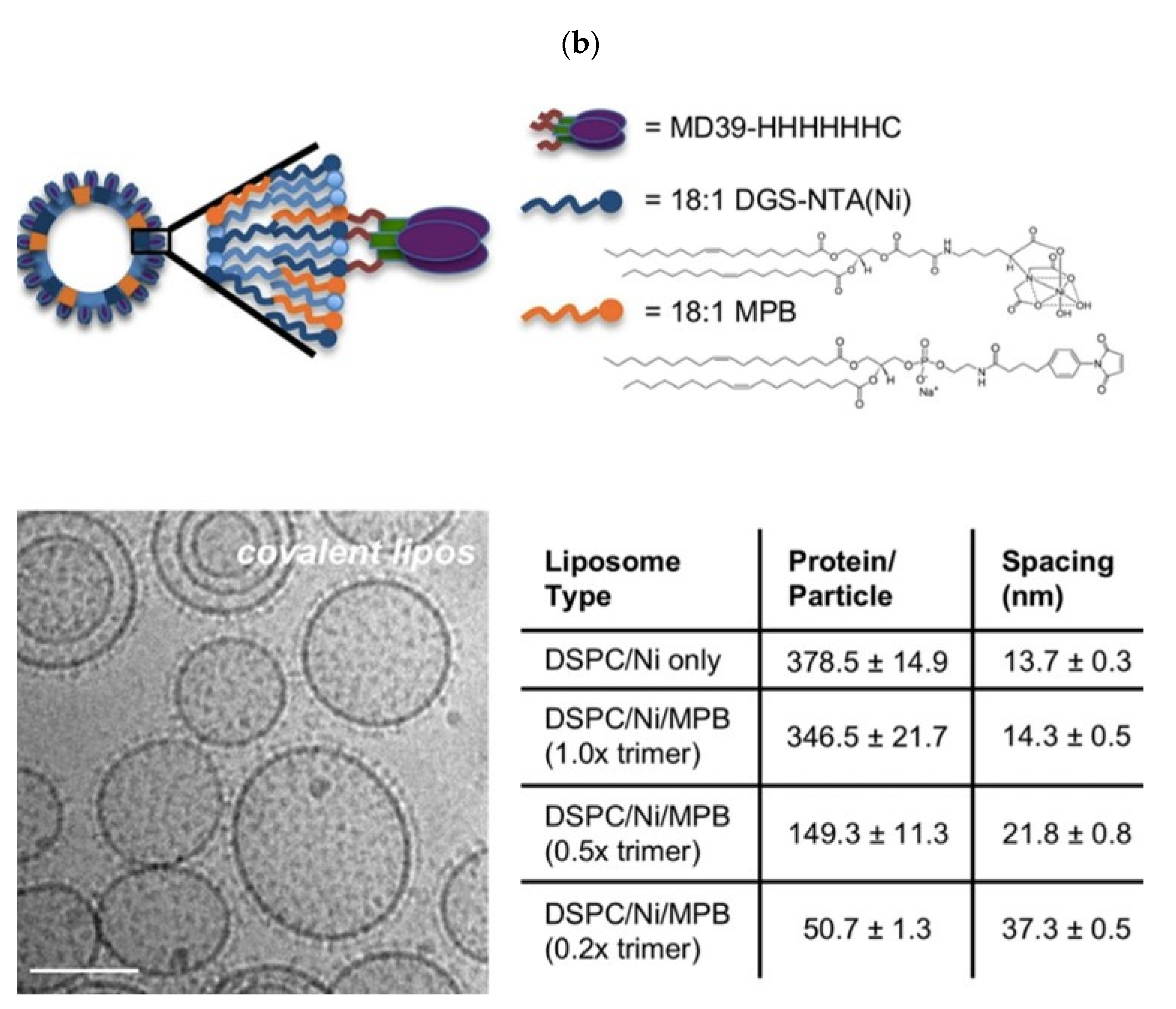
- Tokatlian, T.; Kulp, D.W.; Mutafyan, A.A.; Jones, C.A.; Menis, S.; Georgeson, E.; Kubitz, M.; Zhang, M.H.; Melo, M.B.; Silva, M.; et al. Enhancing Humoral Responses Against HIV Envelope Trimers via Nanoparticle Delivery with Stabilized Synthetic Liposomes. Sci. Rep. 2018, 8, 16527. [Google Scholar] [CrossRef]
- Starr, T.N.; Czudnochowski, N.; Liu, Z.; Zatta, F.; Park, Y.-J.; Addetia, A.; Pinto, D.; Beltramello, M.; Hernandez, P.; Greaney, A.J.; et al. SARS-CoV-2 RBD Antibodies That Maximize Breadth and Resistance to Escape. Nature 2021, 597, 97–102. [Google Scholar] [CrossRef]
- Cully, M. Broadly Neutralizing Anti-Coronavirus Antibodies. Nat. Rev. Drug Discov. 2021, 20, 665. [Google Scholar] [CrossRef] [PubMed]
- Tso, F.Y.; Lidenge, S.J.; Peña, P.B.; Clegg, A.A.; Ngowi, J.R.; Mwaiselage, J.; Ngalamika, O.; Julius, P.; West, J.T.; Wood, C. High Prevalence of Pre-Existing Serological Cross-Reactivity against Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) in Sub-Saharan Africa. Int. J. Infect. Dis. IJID Off. Publ. Int. Soc. Infect. Dis. 2021, 102, 577–583. [Google Scholar] [CrossRef]
- Pardi, N.; Tuyishime, S.; Muramatsu, H.; Kariko, K.; Mui, B.L.; Tam, Y.K.; Madden, T.D.; Hope, M.J.; Weissman, D. Expression Kinetics of Nucleoside-Modified MRNA Delivered in Lipid Nanoparticles to Mice by Various Routes. J. Control. Release Off. J. Control. Release Soc. 2015, 217, 345–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayaraman, M.; Ansell, S.M.; Mui, B.L.; Tam, Y.K.; Chen, J.; Du, X.; Butler, D.; Eltepu, L.; Matsuda, S.; Narayanannair, J.K.; et al. Maximizing the Potency of SiRNA Lipid Nanoparticles for Hepatic Gene Silencing in Vivo. Angew. Chem. 2012, 51, 8529–8533. [Google Scholar] [CrossRef]
- Maier, M.A.; Jayaraman, M.; Matsuda, S.; Liu, J.; Barros, S.; Querbes, W.; Tam, Y.K.; Ansell, S.M.; Kumar, V.; Qin, J.; et al. Biodegradable Lipids Enabling Rapidly Eliminated Lipid Nanoparticles for Systemic Delivery of RNAi Therapeutics. Mol. Ther. J. Am. Soc. Gene Ther. 2013, 21, 1570–1578. [Google Scholar] [CrossRef] [Green Version]
- Kon, E.; Elia, U.; Peer, D. Principles for Designing an Optimal MRNA Lipid Nanoparticle Vaccine. Curr. Opin. Biotechnol. 2021, 73, 329–336. [Google Scholar] [CrossRef]
- Tenchov, R.; Bird, R.; Curtze, A.E.; Zhou, Q. Lipid Nanoparticles─From Liposomes to MRNA Vaccine Delivery, a Landscape of Research Diversity and Advancement. ACS Nano 2021, 15, 16982–17015. [Google Scholar] [CrossRef] [PubMed]
- Schoenmaker, L.; Witzigmann, D.; Kulkarni, J.A.; Verbeke, R.; Kersten, G.; Jiskoot, W.; Crommelin, D.J.A. MRNA-Lipid Nanoparticle COVID-19 Vaccines: Structure and Stability. Int. J. Pharm. 2021, 601, 120586. [Google Scholar] [CrossRef]
- Hilgers, L.A.; Snippe, H. DDA as an Immunological Adjuvant. Res. Immunol. 1992, 143, 494–503; discussion 574-6. [Google Scholar] [CrossRef]
- De Gaspari, E.N. The Use of Monoclonal Antibodies to Neisseria Lactamica in an Antigen Selection to Neisseria Meningitides B Vaccine. Hybridoma 2008, 27, 387–393. [Google Scholar] [CrossRef]
- Carmona-Ribeiro, A.M.; Castuma, C.E.; Sesso, A.; Schreier, S. Bilayer Structure and Stability in Dihexadecyl Phosphate Dispersions. J. Phys. Chem. 1991, 95, 5361–5366. [Google Scholar] [CrossRef]
- Zetterberg, M.M.; Reijmar, K.; Pränting, M.; Engström, Å.; Andersson, D.I.; Edwards, K. PEG-Stabilized Lipid Disks as Carriers for Amphiphilic Antimicrobial Peptides. J. Control. Release Off. J. Control. Release Soc. 2011, 156, 323–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartelds, R.; Nematollahi, M.H.; Pols, T.; Stuart, M.C.A.; Pardakhty, A.; Asadikaram, G.; Poolman, B. Niosomes, an Alternative for Liposomal Delivery. PLoS ONE 2018, 13, e0194179. [Google Scholar] [CrossRef] [Green Version]
- Katare, R.; Gupta, P.N.; Mahor, S.; Rawat, A.; Khatri, K.; Katare, Y.; Panda, A.K.; Vyas, S.P. Development of Polysaccharide-Capped Niosomes for Oral Immunization of Tetanus Toxoid. J. Drug Deliv. Sci. Technol. 2006, 16, 167–172. [Google Scholar] [CrossRef]
- Uchegbu, I.F.; Florence, A.T. Non-Ionic Surfactant Vesicles (Niosomes): Physical and Pharmaceutical Chemistry. Adv. Colloid Interface Sci. 1995, 58, 1–55. [Google Scholar] [CrossRef]
- Jain, S.; Singh, P.; Mishra, V.; Vyas, S.P. Mannosylated Niosomes as Adjuvant-Carrier System for Oral Genetic Immunization against Hepatitis B. Immunol. Lett. 2005, 101, 41–49. [Google Scholar] [CrossRef]
- Jain, S.; Vyas, S.P. Mannosylated Niosomes as Adjuvant-Carrier System for Oral Mucosal Immunization. J. Liposome Res. 2006, 16, 331–345. [Google Scholar] [CrossRef]
- Irvine, D.J.; Hanson, M.C.; Rakhra, K.; Tokatlian, T. Synthetic Nanoparticles for Vaccines and Immunotherapy. Chem. Rev. 2015, 115, 11109–11146. [Google Scholar] [CrossRef] [Green Version]
- Xia, Y.; Song, T.; Hu, Y.; Ma, G. Synthetic Particles for Cancer Vaccines: Connecting the Inherent Supply Chain. Acc. Chem. Res. 2020, 53, 2068–2080. [Google Scholar] [CrossRef]
- Hernandez-Davies, J.E.; Felgner, J.; Strohmeier, S.; Pone, E.J.; Jain, A.; Jan, S.; Nakajima, R.; Jasinskas, A.; Strahsburger, E.; Krammer, F.; et al. Administration of Multivalent Influenza Virus Recombinant Hemagglutinin Vaccine in Combination-Adjuvant Elicits Broad Reactivity Beyond the Vaccine Components. Front. Immunol. 2021, 12, 692151. [Google Scholar] [CrossRef]
- Dias, L.D.S.; Silva, L.B.R.; Nosanchuk, J.D.; Taborda, C.P. Neutrophil Cells Are Essential for The Efficacy of a Therapeutic Vaccine against Paracoccidioidomycosis. J. Fungi 2021, 7, 416. [Google Scholar] [CrossRef]
- Wilson, K.L.; Pouniotis, D.; Hanley, J.; Xiang, S.D.; Ma, C.; Coppel, R.L.; Plebanski, M. A Synthetic Nanoparticle Based Vaccine Approach Targeting MSP4/5 Is Immunogenic and Induces Moderate Protection Against Murine Blood-Stage Malaria. Front. Immunol. 2019, 10, 331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, K.L.; Flanagan, K.L.; Prakash, M.D.; Plebanski, M. Malaria Vaccines in the Eradication Era: Current Status and Future Perspectives. Expert Rev. Vaccines 2019, 18, 133–151. [Google Scholar] [CrossRef] [PubMed]
- Kaur, A.; Kannan, D.; Mehta, S.K.; Singh, S.; Salunke, D.B. Synthetic Toll-like Receptor Agonists for the Development of Powerful Malaria Vaccines: A Patent Review. Expert Opin. Ther. Pat. 2018, 28, 837–847. [Google Scholar] [CrossRef] [PubMed]
- Duffy, P.E.; Patrick Gorres, J. Malaria Vaccines since 2000: Progress, Priorities, Products. NPJ Vaccines 2020, 5, 48. [Google Scholar] [CrossRef]
- Ferreira, T.C.S.; Sauter, I.P.; Borda-Samper, L.; Bentivoglio, E.; DaMata, J.P.; Taniwaki, N.N.; Orrego, P.R.; Araya, J.E.; Lincopan, N.; Cortez, M. Effect of DODAB Nano-Sized Cationic Bilayer Fragments against Leishmania Amazonensis. Molecules 2020, 25, 5741. [Google Scholar] [CrossRef]
- Shin, M.D.; Shukla, S.; Chung, Y.H.; Beiss, V.; Chan, S.K.; Ortega-Rivera, O.A.; Wirth, D.M.; Chen, A.; Sack, M.; Pokorski, J.K.; et al. COVID-19 Vaccine Development and a Potential Nanomaterial Path Forward. Nat. Nanotechnol. 2020, 15, 646–655. [Google Scholar] [CrossRef]
- Luo, M.; Samandi, L.Z.; Wang, Z.; Chen, Z.J.; Gao, J. Synthetic Nanovaccines for Immunotherapy. J. Controlled Release 2017, 263, 200–210. [Google Scholar] [CrossRef]
- Agger, E.M.; Rosenkrands, I.; Hansen, J.; Brahimi, K.; Vandahl, B.S.; Aagaard, C.; Werninghaus, K.; Kirschning, C.; Lang, R.; Christensen, D.; et al. Cationic Liposomes Formulated with Synthetic Mycobacterial Cordfactor (CAF01): A Versatile Adjuvant for Vaccines with Different Immunological Requirements. PLoS ONE 2008, 3, e3116. [Google Scholar] [CrossRef] [Green Version]
- Abdelwahab, W.M.; Riffey, A.; Buhl, C.; Johnson, C.; Ryter, K.; Evans, J.T.; Burkhart, D.J. Co-Adsorption of Synthetic Mincle Agonists and Antigen to Silica Nanoparticles for Enhanced Vaccine Activity: A Formulation Approach to Co-Delivery. Int. J. Pharm. 2021, 593, 120119. [Google Scholar] [CrossRef]
- Davidsen, J.; Rosenkrands, I.; Christensen, D.; Vangala, A.; Kirby, D.; Perrie, Y.; Agger, E.M.; Andersen, P. Characterization of Cationic Liposomes Based on Dimethyldioctadecylammonium and Synthetic Cord Factor from M. Tuberculosis (Trehalose 6,6′-Dibehenate)-a Novel Adjuvant Inducing Both Strong CMI and Antibody Responses. Biochim. Biophys. Acta 2005, 1718, 22–31. [Google Scholar] [CrossRef] [Green Version]
- Reddy, S.T.; Swartz, M.A.; Hubbell, J.A. Targeting Dendritic Cells with Biomaterials: Developing the next Generation of Vaccines. Trends Immunol. 2006, 27, 573–579. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, S.; Chen, J.-Y.; Chen, H.-W.; Hu, C.-M.J. Nanoparticle Vaccines Adopting Virus-like Features for Enhanced Immune Potentiation. Nanotheranostics 2017, 1, 244–260. [Google Scholar] [CrossRef] [Green Version]
- Al-Halifa, S.; Gauthier, L.; Arpin, D.; Bourgault, S.; Archambault, D. Nanoparticle-Based Vaccines Against Respiratory Viruses. Front. Immunol. 2019, 10, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moyle, P.M.; Hartas, J.; Henningham, A.; Batzloff, M.R.; Good, M.F.; Toth, I. An Efficient, Chemically-Defined Semisynthetic Lipid-Adjuvanted Nanoparticulate Vaccine Development System. Nanomed. Nanotechnol. Biol. Med. 2013, 9, 935–944. [Google Scholar] [CrossRef] [Green Version]
- Carmona-Ribeiro, A.M.; De Melo Carrasco, L.D. Novel Formulations for Antimicrobial Peptides. Int. J. Mol. Sci. 2014, 15, 18040–18083. [Google Scholar] [CrossRef] [Green Version]
- Skwarczynski, M.; Toth, I. Recent Advances in Peptide-Based Subunit Nanovaccines. Nanomedicine 2014, 9, 2657–2669. [Google Scholar] [CrossRef]
- Robinson, J.A. Folded Synthetic Peptides and Other Molecules Targeting Outer Membrane Protein Complexes in Gram-Negative Bacteria. Front. Chem. 2019, 7, 45. [Google Scholar] [CrossRef] [Green Version]
- Zerbe, K.; Moehle, K.; Robinson, J.A. Protein Epitope Mimetics: From New Antibiotics to Supramolecular Synthetic Vaccines. Acc. Chem. Res. 2017, 50, 1323–1331. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Wise, M.C.; Chokkalingam, N.; Walker, S.; Tello-Ruiz, E.; Elliott, S.T.C.; Perales-Puchalt, A.; Xiao, P.; Zhu, X.; Pumroy, R.A.; et al. In Vivo Assembly of Nanoparticles Achieved through Synergy of Structure-Based Protein Engineering and Synthetic DNA Generates Enhanced Adaptive Immunity. Adv. Sci. 2020, 7, 1902802. [Google Scholar] [CrossRef]
- Xu, Z.; Chokkalingam, N.; Tello-Ruiz, E.; Wise, M.C.; Bah, M.A.; Walker, S.; Tursi, N.J.; Fisher, P.D.; Schultheis, K.; Broderick, K.E.; et al. A DNA-Launched Nanoparticle Vaccine Elicits CD8+ T-Cell Immunity to Promote in Vivo Tumor Control. Cancer Immunol. Res. 2020, 8, 1354–1364. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-Y.; Noh, Y.-W.; Kang, T.H.; Kim, J.-E.; Kim, S.; Um, S.H.; Oh, D.-B.; Park, Y.-M.; Lim, Y.T. Synthetic Vaccine Nanoparticles Target to Lymph Node Triggering Enhanced Innate and Adaptive Antitumor Immunity. Biomaterials 2017, 130, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Carmona-Ribeiro, A.M.; Herrington, T.M. Phospholipid Adsorption onto Polystyrene Microspheres. J. Colloid Interface Sci. 1993, 156, 19–23. [Google Scholar] [CrossRef]
- Carmona-Ribeiro, A.M. Bilayer Vesicles and Liposomes as Interface Agents. Chem. Soc. Rev. 2001, 30, 241–247. [Google Scholar] [CrossRef]
- Melo, L.D.; Mamizuka, E.M.; Carmona-Ribeiro, A.M. Antimicrobial Particles from Cationic Lipid and Polyelectrolytes. Langmuir 2010, 26, 12300–12306. [Google Scholar] [CrossRef]
- Vieira, D.B.; Carmona-Ribeiro, A.M. Synthetic Bilayer Fragments for Solubilization of Amphotericin B. J. Colloid Interface Sci. 2001, 244, 427–431. [Google Scholar] [CrossRef]
- Vieira, D.B.; Carmona-Ribeiro, A.M. Cationic Nanoparticles for Delivery of Amphotericin B: Preparation, Characterization and Activity in Vitro. J. Nanobiotechnol. 2008, 6, 6. [Google Scholar] [CrossRef] [Green Version]
- Carmona-Ribeiro, A.M. Interactions between Cationic Liposomes and Drugs or Biomolecules. An. Acad. Bras. Ciênc. 2000, 72, 39–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribeiro, R.T.; Galvão, C.N.; Betancourt, Y.P.; Mathiazzi, B.I.; Carmona-Ribeiro, A.M. Microbicidal Dispersions and Coatings from Hybrid Nanoparticles of Poly (Methyl Methacrylate), Poly (Diallyl Dimethyl Ammonium) Chloride, Lipids, and Surfactants. Int. J. Mol. Sci. 2019, 20, 6150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galvão, C.N.; Sanches, L.M.; Mathiazzi, B.I.; Ribeiro, R.T.; Petri, D.F.S.; Carmona-Ribeiro, A.M. Antimicrobial Coatings from Hybrid Nanoparticles of Biocompatible and Antimicrobial Polymers. Int. J. Mol. Sci. 2018, 19, 2965. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Peppas, N.A. Hydrogels and Scaffolds for Immunomodulation. Adv. Mater. 2014, 26, 6530–6541. [Google Scholar] [CrossRef]
- Roth, G.A.; Gale, E.C.; Alcántara-Hernández, M.; Luo, W.; Axpe, E.; Verma, R.; Yin, Q.; Yu, A.C.; Lopez Hernandez, H.; Maikawa, C.L.; et al. Injectable Hydrogels for Sustained Codelivery of Subunit Vaccines Enhance Humoral Immunity. ACS Cent. Sci. 2020, 6, 1800–1812. [Google Scholar] [CrossRef]
- Gupta, D.; Gangwar, A.; Jyoti, K.; Sainaga Jyothi, V.G.S.; Sodhi, R.K.; Mehra, N.K.; Singh, S.B.; Madan, J. Self Healing Hydrogels: A New Paradigm Immunoadjuvant for Delivering Peptide Vaccine. Colloids Surf. B Biointerfaces 2020, 194, 111171. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liang, C.; Shi, F.; He, T.; Gong, C.; Wang, L.; Yang, Z. Cancer Vaccines Using Supramolecular Hydrogels of NSAID-Modified Peptides as Adjuvants Abolish Tumorigenesis. Nanoscale 2017, 9, 14058–14064. [Google Scholar] [CrossRef]
- Lima, E.G.; Gomes, L.R.; Carmona-Ribeiro, A.M. Stable Indomethacin Dispersions in Water from Drug, Ethanol, Cationic Lipid and Carboxymethyl-Cellulose. Pharm. Nanotechnol. 2016, 4, 126–135. [Google Scholar] [CrossRef]
- Rafael, D.; Melendres, M.M.R.; Andrade, F.; Montero, S.; Martinez-Trucharte, F.; Vilar-Hernandez, M.; Durán-Lara, E.F.; Schwartz, S., Jr.; Abasolo, I. Thermo-Responsive Hydrogels for Cancer Local Therapy: Challenges and State-of-Art. Int. J. Pharm. 2021, 606, 120954. [Google Scholar] [CrossRef]
- Kordalivand, N.; Tondini, E.; Lau, C.Y.J.; Vermonden, T.; Mastrobattista, E.; Hennink, W.E.; Ossendorp, F.; van Nostrum, C.F. Cationic Synthetic Long Peptides-Loaded Nanogels: An Efficient Therapeutic Vaccine Formulation for Induction of T-Cell Responses. J. Control. Release 2019, 315, 114–125. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carmona-Ribeiro, A.M. Supramolecular Nanostructures for Vaccines. Biomimetics 2022, 7, 6. https://doi.org/10.3390/biomimetics7010006
Carmona-Ribeiro AM. Supramolecular Nanostructures for Vaccines. Biomimetics. 2022; 7(1):6. https://doi.org/10.3390/biomimetics7010006
Chicago/Turabian StyleCarmona-Ribeiro, Ana Maria. 2022. "Supramolecular Nanostructures for Vaccines" Biomimetics 7, no. 1: 6. https://doi.org/10.3390/biomimetics7010006





