MdSGR2 Negatively Regulates Chlorophyll Degradation in Apple
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Fruit Color Determination
2.3. Determination of the Chlorophyll Content in the Peel
2.4. Transcriptome Sequencing Analysis
2.5. Real-Time Fluorescence Quantitative qRT-PCR
2.6. Subcellular Localization of MdSGR2
2.7. Transient Transformation of MdSGR2 in Tobacco
2.8. Silencing of MdSGR2 in Apple Peel
3. Results
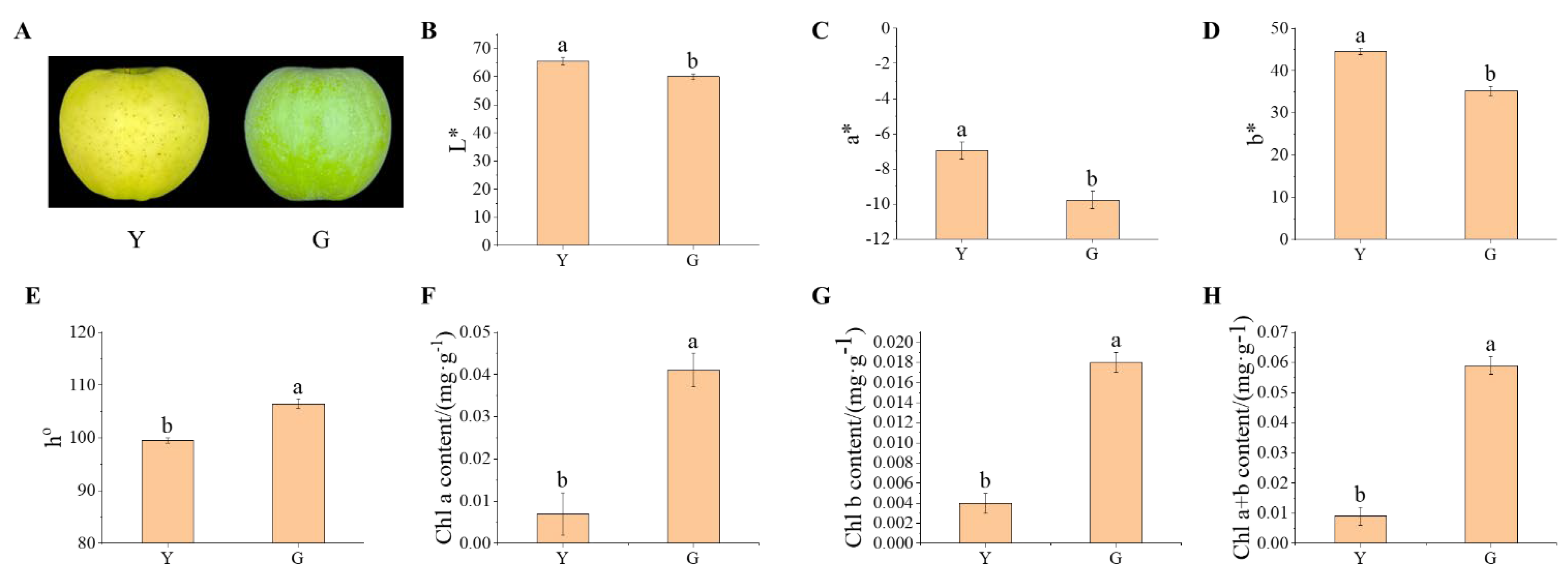
3.1. Difference Analysis of the Fruit Color Parameters and Chlorophyll in the Peel
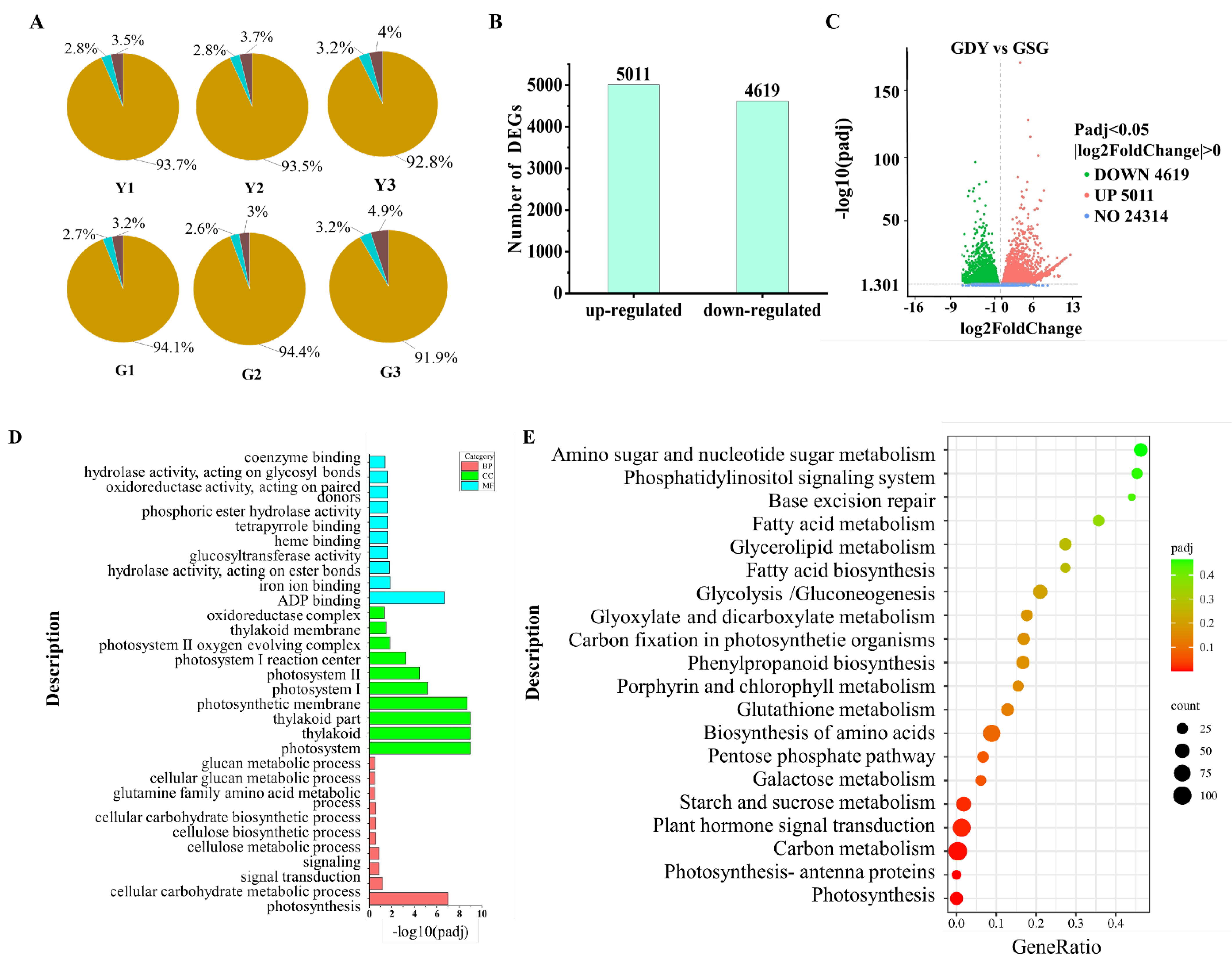
3.2. Transcriptome Sequencing and Quality Assessment Analysis
3.3. DGEs Analysis and Identification of Transcription Factors
3.4. GO Annotation and KEGG Pathway Enrichment Analysis
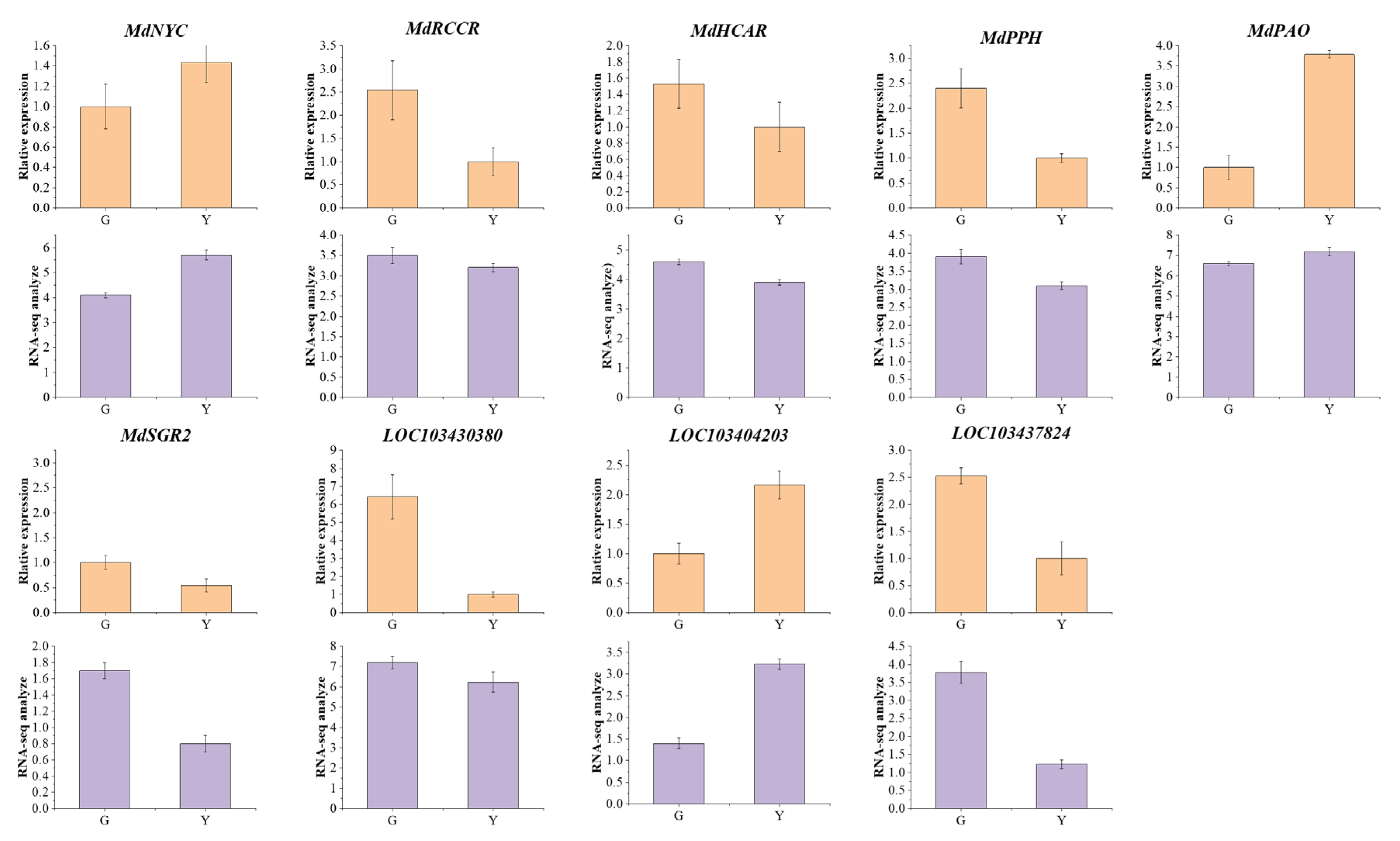
3.5. Verification Analysis of Representative Genes
3.6. Mining and Analysis of the DEGs in Chlorophyll Metabolic Pathways
3.7. Subcellular Location of MdSGR2
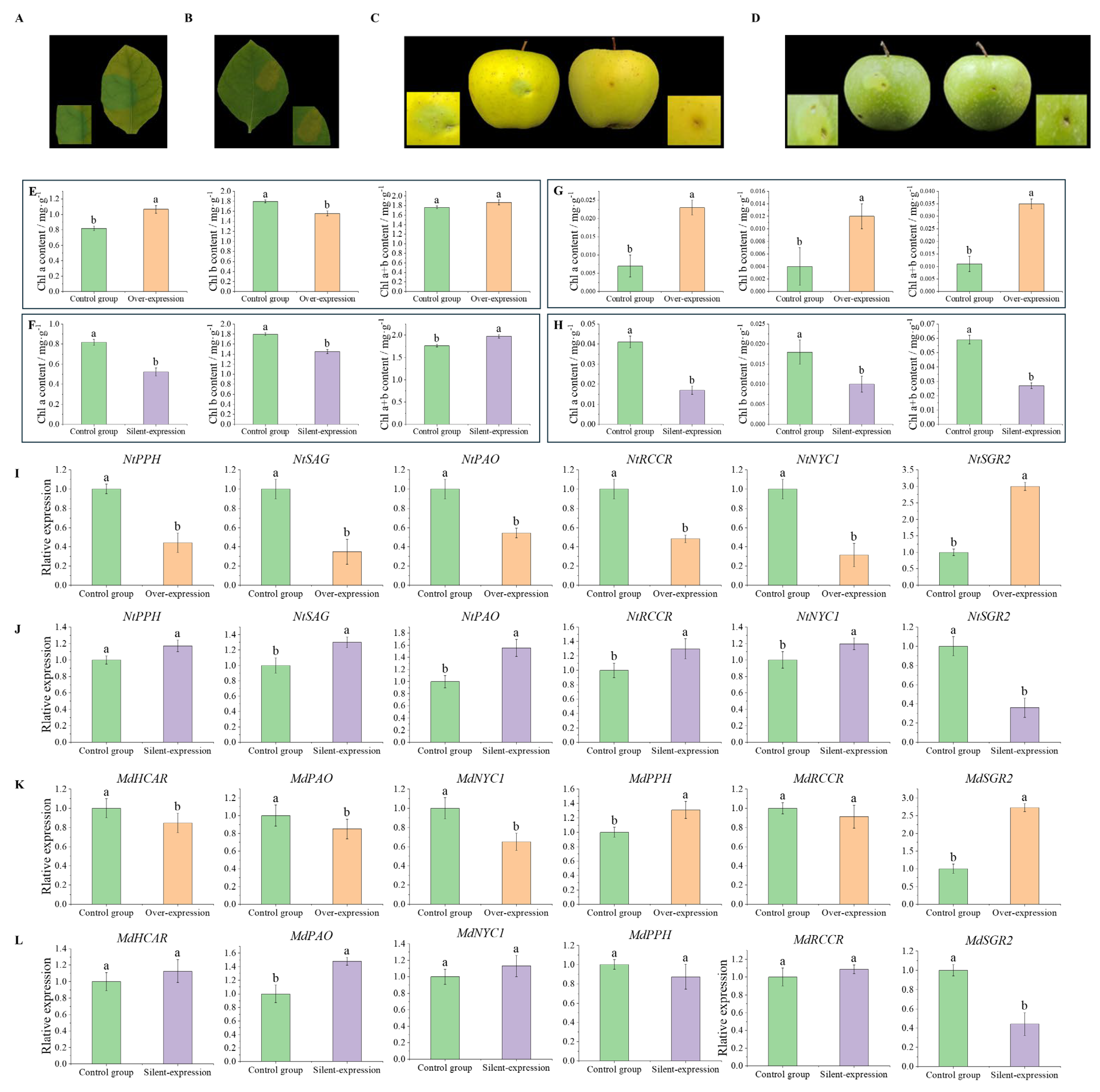
3.8. Transient Overexpression of MdSGR2 in Tobacco Leaf and Apple Peel Analysis
3.9. Instantaneous Silencing of MdSGR2 in the Tobacco Leaves and Apple Peel
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- An, J.-P.; Zhang, X.; Bi, S.Q.; You, C.-X.; Wang, X.-F.; Hao, Y. The ERF transcription factor MdERF38 promotes drought stress-induced anthocyanin biosynthesis in apple. Plant J. 2019, 101, 573–589. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Ji, H.; Huang, W.; Zhang, Z.; Zhu, K.; Zhu, S.; Chai, L.; Ye, J.; Deng, X. Transcription factor CrWRKY42 coregulates chlorophyll degradation and carotenoid biosynthesis in citrus. Plant Physiol. 2024, kiae048. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.-A.; Duan, S.; Jeong, H.-Y.; Lee, C.; Kang, I.-K.; Eom, S.H. Pigmentation and flavonoid metabolite diversity in immature ‘Fuji’ apple fruits in response to lights and methyl jasmonate. Int. J. Mol. Sci. 2022, 23, 1722. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Celton, J.-M.; Buck-Sorlin, G.; Balzergue, S.; Bucher, E.; Laurens, F. Skin color in apple fruit (Malus × domestica): Genetic and epigenetic insights. Epigenomes 2020, 4, 13. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.; Zhuo, M.; Abbas, F.; Hu, G.; Wang, H.; Huang, X. Transcription factor LcNAC002 coregulates chlorophyll degradation and anthocyanin biosynthesis in litchi. Plant Physiol. 2023, 192, 1913–1927. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xu, R.; Liu, Y.; You, C.-X.; An, J.-P. MdVQ10 promotes wound-triggered leaf senescence in association with MdWRKY75 and undergoes antagonistic modulation of MdCML15 and MdJAZs in apple. Plant J. 2023, 115, 1599–1618. [Google Scholar] [CrossRef]
- Pruzinská, A.; Tanner, G.; Anders, I.; Roca, M.; Hörtensteiner, S. Chlorophyll breakdown: Pheophorbide a oxygenase is a Rieske-type iron-sulfur protein, encoded by the accelerated cell death 1 gene. Proc. Natl. Acad. Sci. USA 2003, 100, 15259–15264. [Google Scholar] [CrossRef]
- Sun, L.; Xu, H.; Song, J.; Yang, X.; Wang, X.; Liu, H.; Pang, M.; Hu, Y.; Yang, Q.; Ning, X.; et al. OsNAC103, a NAC transcription factor, positively regulates leaf senescence and plant architecture in Rice. Rice 2024, 17, 15. [Google Scholar] [CrossRef]
- Taylor, L.; Nunes-Nesi, A.; Parsley, K.; Leiss, A.E.; Leach, G.E.; Coates, S.; Wingler, A.; Fernie, A.R.; Hibberd, J.M. Cytosolic pyruvate, orthophosphate dikinase functions in nitrogen remobilization during leaf senescence and limits individual seed growth and nitrogen content. Plant J. 2010, 62, 641–652. [Google Scholar] [CrossRef]
- Vom Dorp, K.; Hölzl, G.; Plohmann, C.; Eisenhut, M.; Abraham, M.; Weber, A.P.M.; Hanson, A.D.; Dörmann, P. Remobilization of phytol from chlorophyll degradation is essential for tocopherol synthesis and growth of Arabidopsis. Plant Cell 2015, 27, 2846–2859. [Google Scholar] [CrossRef]
- Zhao, Y.; Xu, W.; Wang, L.; Han, S.; Zhang, Y.; Liu, Q.; Liu, B.; Zhao, X. A maize necrotic leaf mutant caused by defect of coproporphyrinogen III oxidase in the porphyrin pathway. Genes 2022, 13, 272. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Gu, T.; Khan, I.; Zada, A.; Jia, T. Research progress in the interconversion, turnover and degradation of chlorophyll. Cells 2021, 10, 3134. [Google Scholar] [CrossRef] [PubMed]
- Christ, B.; Hörtensteiner, S. Mechanism and significance of chlorophyll breakdown. J. Plant Growth Regul. 2013, 33, 4–20. [Google Scholar] [CrossRef]
- Tanaka, R.; Tanaka, A. Chlorophyll cycle regulates the construction and destruction of the light-harvesting complexes. Biochim. Biophys. Acta 2011, 1807, 968–976. [Google Scholar] [CrossRef] [PubMed]
- Kusaba, M.; Ito, H.; Morita, R.; Iida, S.; Sato, Y.; Fujimoto, M.; Kawasaki, S.; Tanaka, R.; Hirochika, H.; Nishimura, M.; et al. Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 2007, 19, 1362–1375. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Morita, R.; Katsuma, S.; Nishimura, M.; Tanaka, A.; Kusaba, M. Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J. 2009, 57, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Dong, Y.; Yan, H.; Ge, W.; Shen, C.; Guan, J.; Liu, L.; Zhang, Y. Effects of 1-MCP on chlorophyll degradation pathway-associated genes expression and chloroplast ultrastructure during the peel yellowing of Chinese pear fruits in storage. Food Chem. 2012, 135, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Jiao, B.; Meng, Q.; Lv, W. Roles of stay-green (SGR) homologs during chlorophyll degradation in green plants. Bot. Stud. 2020, 25, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ou, L.; Liu, Z.; Lv, J.; Wang, J.; Song, J.; Yang, B.; Chen, W.; Yang, S.C.; Liu, W.; et al. A novel single-base mutation in CaSGR1 confers the stay-green phenotype in pepper (Capsicum annuum L.). Hortic. Plant J. 2022, 9, 293–305. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Park, S.-Y.; Paek, N.-C. The divergent roles of STAYGREEN (SGR) homologs in chlorophyll degradation. Mol. Cells 2015, 38, 390–395. [Google Scholar] [CrossRef]
- Uluisik, S.; Kiyak, A.; Kurt, F.; Filiz, E. STAY-GREEN (SGR) genes in tomato (Solanum lycopersicum): Genome-wide identification, and expression analyses reveal their involvements in ripening and salinity stress responses. Hortic. Environ. Biotechnol. 2022, 63, 557–569. [Google Scholar] [CrossRef]
- Yang, M.; Zhu, S.; Jiao, B.; Duan, M.; Meng, Q.; Ma, N.; Lv, W. SlSGRL, a tomato SGR-like protein, promotes chlorophyll degradation downstream of the ABA signaling pathway. Plant Cell 2020, 157, 316–327. [Google Scholar] [CrossRef]
- Armstead, I.; Donnison, I.S.; Aubry, S.; Harper, J.A.; Hörtensteiner, S.; James, C.L.; Mani, J.; Moffet, M.D.; Ougham, H.; Roberts, L.A.; et al. Cross-species identification of mendel’s/locus. Science 2007, 315, 73. [Google Scholar] [CrossRef] [PubMed]
- Barry, C.S.; McQuinn, R.P.; Chung, M.-Y.; Besuden, A.; Giovannoni, J.J. Amino acid substitutions in homologs of the STAY-GREEN protein are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper. Plant Physiol. 2008, 147, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Yu, J.W.; Park, J.-S.; Li, J.; Yoo, S.-C.; Lee, N.-Y.; Lee, S.-K.; Jeong, S.-W.; Seo, H.S.; Koh, H.-J.; et al. The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 2007, 19, 1649–1664. [Google Scholar] [CrossRef]
- Ren, G.; An, K.; Liao, Y.; Zhou, X.; Cao, Y.; Zhao, H.; Ge, X.; Kuai, B. Identification of a novel chloroplast protein AtNYE1 regulating chlorophyll degradation during leaf senescence in Arabidopsis. Plant Physiol. 2007, 144, 1429–1441. [Google Scholar] [CrossRef]
- Shang, X. Construction of integration QTL map and identification of candidate genes for stay-green in maize. Acta Pratac. Sin. 2012, 21, 175–185. [Google Scholar]
- Rong, H.L.; Tang, Y.; Zhang, H.; Wu, P.; Chen, Y.; Li, M.; Wu, G.; Jiang, H. The Stay-Green Rice like (SGRL) gene regulates chlorophyll degradation in rice. J. Plant Physiol. 2013, 170, 1367–1373. [Google Scholar] [CrossRef]
- Shin, D.; Lee, S.; Kim, T.-H.; Lee, J.-H.; Park, J.; Lee, J.; Lee, J.Y.; Cho, L.-H.; Choi, J.Y.; Lee, W.; et al. Natural variations at the Stay-Green gene promoter control lifespan and yield in rice cultivars. Nat. Commun. 2020, 11, 2819–2830. [Google Scholar] [CrossRef]
- Zhu, K.; Zheng, X.; Ye, J.; Huang, Y.; Chen, H.; Mei, X.; Xie, Z.; Cao, L.; Zeng, Y.; Larkin, R.M.; et al. Regulation of carotenoid and chlorophyll pools in hesperidia, anatomically unique fruits found only in Citrus. Plant Physiol. 2021, 187, 829–845. [Google Scholar] [CrossRef]
- Chen, X.; Li, J.; Yu, Y.; Kou, X.; Periakaruppan, R.; Chen, X.; Li, X. STAY-GREEN and light-harvesting complex II chlorophyll a/b binding protein are involved in albinism of a novel albino tea germplasm ‘Huabai 1’. Sci. Hortic. 2022, 293, 653–665. [Google Scholar] [CrossRef]
- Luo, Z.; Zhang, J.; Li, J.; Yang, C.; Wang, T.; Ouyang, B.; Li, H.; Giovannoni, J.J.; Ye, Z. A STAY-GREEN protein SlSGR1 regulates lycopene and β-carotene accumulation by interacting directly with SlPSY1 during ripening processes in tomato. New Phytol. 2013, 198, 442–452. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Deng, L.; Yan, B.W.; Pan, Y.; Luo, M.; Chen, X.; Hu, T.; Chen, G. Silencing of the LeSGR1 gene in tomato inhibits chlorophyll degradation and exhibits a stay-green phenotype. Biol. Plant. 2011, 55, 27–34. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Kim, D.; Kim, Y.-S.; Hörtensteiner, S.; Paek, N.-C. Arabidopsis STAYGREEN-LIKE (SGRL) promotes abiotic stress-induced leaf yellowing during vegetative growth. FEBS Lett. 2014, 588, 3830–3837. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, Y.; Park, S.-Y.; Kim, Y.-S.; Wang, S.-H.; Yoo, S.-C.; Hörtensteiner, S.; Paek, N.-C. Arabidopsis STAY-GREEN2 is a negative regulator of chlorophyll degradation during leaf senescence. Plant Mol. Biol. 2014, 7, 1288–1302. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, Y.; Schelbert, S.; Park, S.-Y.; Han, S.-H.; Lee, B.-D.; Andrès, C.B.; Kessler, F.; Hörtensteiner, S.; Paek, N.-C. STAY-GREEN and chlorophyll catabolic enzymes interact at light-harvesting complex II for chlorophyll detoxification during leaf senescence in arabidopsis. Plant Cell 2012, 24, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Gao, J. Experimental Supervision of Plant Physiology, 1st ed.; Higher Education Press: Beijing, China, 2006; pp. 74–76. [Google Scholar]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B.J. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Chen, Z.; Yu, L.; Liu, W.; Zhang, J.; Wang, N.; Chen, X. Research progress of fruit color development in apple (Malus domestica Borkh.). Plant Physiol. Biochem. 2021, 162, 267–279. [Google Scholar] [CrossRef]
- Wang, S.; Shi, X.; Liu, F.; Laborda, P. Effects of exogenous methyl jasmonate on quality and preservation of postharvest fruits. Food Chem. 2021, 353, 129482. [Google Scholar] [CrossRef]
- Zhang, A.; Zheng, J.; Chen, X.; Shi, X.; Wang, H.; Fu, Q. Comprehensive analysis of transcriptome and metabolome reveals the flavonoid metabolic pathway is associated with fruit peel coloration of melon. Molecules 2021, 26, 2830. [Google Scholar] [CrossRef]
- Zhu, K.; Chen, H.; Mei, X.; Lu, S.; Xie, H.; Liu, J.; Chai, L.; Xu, Q.; Wurtzel, E.T.; Ye, J.; et al. Transcription factor CsMADS3 coordinately regulates chlorophyll and carotenoid pools in Citrus hesperidium. Plant Physiol. 2023, 193, 519–536. [Google Scholar] [CrossRef] [PubMed]
- Lai, B.; Hu, B.; Qin, Y.; Zhao, J.; Wang, H.; Hu, G. Transcriptomic analysis of Litchi chinensis pericarp during maturation with a focus on chlorophyll degradation and flavonoid biosynthesis. BMC Genom. 2015, 16, 225. [Google Scholar] [CrossRef] [PubMed]
- Cackett, L.; Luginbuehl, L.H.; Schreier, T.B.; Lopez-Juez, E.; Hibberd, J.M. Chloroplast development in green plant tissues: The interplay between light, hormone, and transcriptional regulation. New Phytol. 2022, 233, 2000–2016. [Google Scholar] [CrossRef]
- Miller, T.E.; Beneyton, T.; Schwander, T.; Diehl, C.; Girault, M.; McLean, R.; Chotel, T.; Claus, P.; Cortina, N.S.; Baret, J.C.; et al. Light-powered CO2 fixation in a chloroplast mimic with natural and synthetic parts. Science 2020, 368, 649–654. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Dong, X.; Yu, L.; Zhang, Y. Response and function of Solanum lycopersicum L. SlSGR2 gene under cadmium stress. Horticulturae 2022, 8, 1002–1015. [Google Scholar] [CrossRef]
- Teng, K.; Chang, Z.; Xiao, G.; Guo, W.E.; Xu, L.X.; Chao, Y.; Han, L.B. Molecular cloning and characterization of a chlorophyll degradation regulatory gene (ZjSGR) from Zoysia japonica. Genet. Mol. Res. 2016, 15, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Han, L.; Pislariu, C.I.; Nakashima, J.; Fu, C.; Jiang, Q.; Quan, L.; Blancaflor, E.B.; Tang, Y.; Bouton, J.H.; et al. From model to crop: Functional analysis of a STAY-GREEN gene in the model legume medicago truncatula and effective use of the gene for alfalfa improvement. Plant Physiol. 2011, 157, 1483–1496. [Google Scholar] [CrossRef]
- Zhang, B.; Qu, D.; Yang, H.; Yang, Y.; Wang, F.; Zhu, Z.; Zhao, Z. Effects of MdMYB10 gene on phenylalanine metabolism in apple peel. Acta Hortic. Sin. 2018, 45, 1429–1440. [Google Scholar]
- Liu, K.; Zhao, Z.; Luo, R.; Chen, Y. Cloning and expression analysis of pheophorbide a oxygenase gene (MiPAO) and construction of silencing expression vector from mango. Acta Agric. Boreali-Sin. 2018, 33, 1–9. [Google Scholar]
- Liu, L.; Tan, P.; Jiang, H.; Chang, Z.; Teng, K. Overexpression of CbSGR, a stay-green gene from carex breviculmis accelerated chlorophyll degradation and senescence in arabidopsis thaliana. Grassl. China 2020, 42, 15–24. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, S.; Yao, P.; Kang, J.; Zheng, X.; Liu, C.; Gao, Y.; Zhang, D.; Zhang, X.; Hao, Y. MdSGR2 Negatively Regulates Chlorophyll Degradation in Apple. Horticulturae 2024, 10, 439. https://doi.org/10.3390/horticulturae10050439
Xue S, Yao P, Kang J, Zheng X, Liu C, Gao Y, Zhang D, Zhang X, Hao Y. MdSGR2 Negatively Regulates Chlorophyll Degradation in Apple. Horticulturae. 2024; 10(5):439. https://doi.org/10.3390/horticulturae10050439
Chicago/Turabian StyleXue, Shiyi, Pei Yao, Jiwei Kang, Xiong Zheng, Chang Liu, Yan Gao, Dehui Zhang, Xiaojun Zhang, and Yanyan Hao. 2024. "MdSGR2 Negatively Regulates Chlorophyll Degradation in Apple" Horticulturae 10, no. 5: 439. https://doi.org/10.3390/horticulturae10050439



