The Ins and Outs of miRNA-Mediated Gene Silencing during Neuronal Synaptic Plasticity
Abstract
:1. Introduction
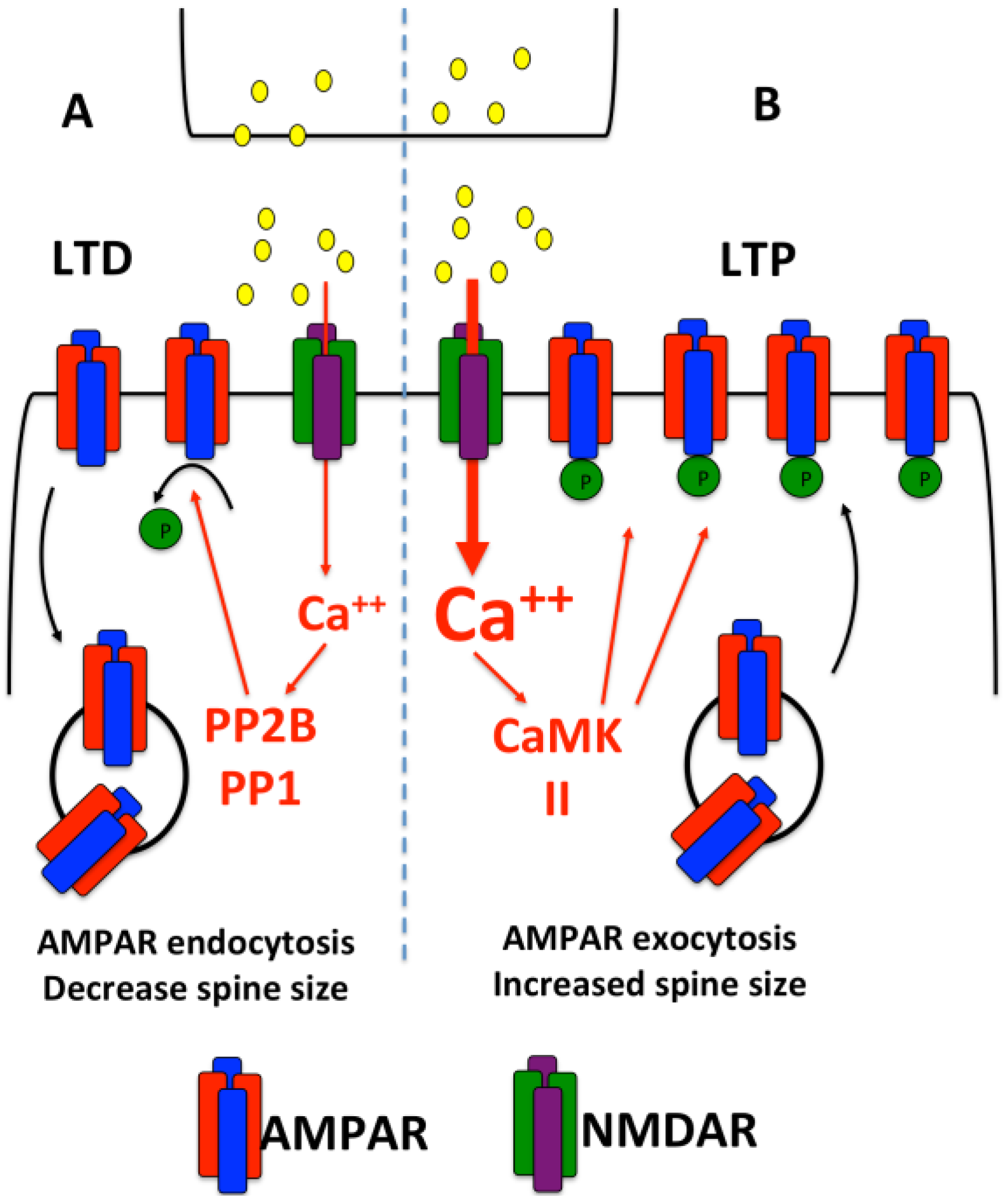
2. Dendritic Protein Synthesis
3. Dendiritc miRNAs
| miRNA | Target | Refs. |
|---|---|---|
| miR-134 | LIMK1, Pum2 | [40,54] |
| miR-132 | p250GAP | [55] |
| miR-125b | NR2A | [56] |
| miR-191 | Complexin-1, Complexin-2 | [37] |
| miR-135a | TPM2 | [37] |
| miR-501-3p | Gria1 | [57] |
| miR-138 | APT1 | [58] |
| miR-188 | NRP2 | [59] |
| miR-124 | Gria2 | [60] |

3.1. miR-134
3.2. miR-132
3.3. Other miRNAs
4. Other Non-Coding RNAs (ncRNAs) in Synaptic Plasticity
4.1. Long Non-Coding RNAs
4.2. Circular RNAs
5. RNA Binding Proteins and Synaptic Plasticity
6. Summary
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kim, J.J.; Diamond, D.M. The stressed hippocampus, synaptic plasticity and lost memories. Nat. Rev. Neurosci. 2002, 3, 453–462. [Google Scholar] [PubMed]
- Kauer, J.A.; Malenka, R.C. Synaptic plasticity and addiction. Nat. Rev. Neurosci. 2007, 8, 844–858. [Google Scholar] [CrossRef] [PubMed]
- Nabavi, S.; Fox, R.; Proulx, C.D.; Lin, J.Y.; Tsien, R.Y.; Malinow, R. Engineering a memory with ltd and ltp. Nature 2014, 511, 348–352. [Google Scholar] [CrossRef] [PubMed]
- Malenka, R.C.; Bear, M.F. Ltp and ltd: An embarrassment of riches. Neuron 2004, 44, 5–21. [Google Scholar] [CrossRef] [PubMed]
- Holtmaat, A.; Svoboda, K. Experience-dependent structural synaptic plasticity in the mammalian brain. Nat. Rev. Neurosci. 2009, 10, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Blitz, D.M.; Foster, K.A.; Regehr, W.G. Short-term synaptic plasticity: A comparison of two synapses. Nat. Rev. Neurosci. 2004, 5, 630–640. [Google Scholar] [CrossRef] [PubMed]
- Bliss, T.V.; Lomo, T. Long-lasting potentiation of synaptic transmission in the dentate area of the anaesthetized rabbit following stimulation of the perforant path. J. Physiol. 1973, 232, 331–356. [Google Scholar] [CrossRef] [PubMed]
- Luscher, C.; Malenka, R.C. Nmda receptor-dependent long-term potentiation and long-term depression (ltp/ltd). Cold Spring Harb. Perspect. Biol. 2012, 4, a005710. [Google Scholar] [CrossRef] [PubMed]
- Bannerman, D.M.; Sprengel, R.; Sanderson, D.J.; McHugh, S.B.; Rawlins, J.N.; Monyer, H.; Seeburg, P.H. Hippocampal synaptic plasticity, spatial memory and anxiety. Nat. Rev. Neurosci. 2014, 15, 181–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynch, M.A. Long-term potentiation and memory. Physiol. Rev. 2004, 84, 87–136. [Google Scholar] [CrossRef] [PubMed]
- Huganir, R.L.; Nicoll, R.A. Ampars and synaptic plasticity: The last 25 years. Neuron 2013, 80, 704–717. [Google Scholar] [CrossRef] [PubMed]
- Derkach, V.; Barria, A.; Soderling, T.R. Ca2+/calmodulin-kinase ii enhances channel conductance of alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionate type glutamate receptors. Proc. Natl. Acad. Sci. USA 1999, 96, 3269–3274. [Google Scholar] [CrossRef] [PubMed]
- Benke, T.A.; Luthi, A.; Isaac, J.T.; Collingridge, G.L. Modulation of ampa receptor unitary conductance by synaptic activity. Nature 1998, 393, 793–797. [Google Scholar] [PubMed]
- Ehlers, M.D. Reinsertion or degradation of ampa receptors determined by activity-dependent endocytic sorting. Neuron 2000, 28, 511–525. [Google Scholar] [CrossRef]
- Beattie, E.C.; Carroll, R.C.; Yu, X.; Morishita, W.; Yasuda, H.; von Zastrow, M.; Malenka, R.C. Regulation of ampa receptor endocytosis by a signaling mechanism shared with ltd. Nat. Neurosci. 2000, 3, 1291–1300. [Google Scholar] [PubMed]
- Mulkey, R.M.; Endo, S.; Shenolikar, S.; Malenka, R.C. Involvement of a calcineurin/inhibitor-1 phosphatase cascade in hippocampal long-term depression. Nature 1994, 369, 486–488. [Google Scholar] [CrossRef] [PubMed]
- Wiegert, J.S.; Oertner, T.G. Long-term depression triggers the selective elimination of weakly integrated synapses. Proc. Natl. Acad. Sci. USA 2013, 110, E4510–E4519. [Google Scholar] [CrossRef] [PubMed]
- Krichevsky, A.M.; King, K.S.; Donahue, C.P.; Khrapko, K.; Kosik, K.S. A microrna array reveals extensive regulation of micrornas during brain development. RNA 2003, 9, 1274–1281. [Google Scholar] [CrossRef] [PubMed]
- Miska, E.A.; Alvarez-Saavedra, E.; Townsend, M.; Yoshii, A.; Sestan, N.; Rakic, P.; Constantine-Paton, M.; Horvitz, H.R. Microarray analysis of microrna expression in the developing mammalian brain. Genome Biol. 2004, 5, R68. [Google Scholar] [CrossRef] [PubMed]
- Sempere, L.F.; Freemantle, S.; Pitha-Rowe, I.; Moss, E.; Dmitrovsky, E.; Ambros, V. Expression profiling of mammalian micrornas uncovers a subset of brain-expressed micrornas with possible roles in murine and human neuronal differentiation. Genome Biol. 2004, 5, R13. [Google Scholar] [CrossRef] [PubMed]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by micrornas: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Malmevik, J.; Petri, R.; Klussendorf, T.; Knauff, P.; Akerblom, M.; Johansson, J.; Soneji, S.; Jakobsson, J. Identification of the mirna targetome in hippocampal neurons using rip-seq. Sci. Rep. 2015, 5, 12609. [Google Scholar] [CrossRef] [PubMed]
- Holt, C.E.; Schuman, E.M. The central dogma decentralized: New perspectives on rna function and local translation in neurons. Neuron 2013, 80, 648–657. [Google Scholar] [CrossRef] [PubMed]
- Cajigas, I.J.; Tushev, G.; Will, T.J.; Tom Dieck, S.; Fuerst, N.; Schuman, E.M. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron 2012, 74, 453–466. [Google Scholar] [CrossRef] [PubMed]
- Steward, O.; Levy, W.B. Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J. Neurosci. 1982, 2, 284–291. [Google Scholar] [PubMed]
- Ostroff, L.E.; Fiala, J.C.; Allwardt, B.; Harris, K.M. Polyribosomes redistribute from dendritic shafts into spines with enlarged synapses during ltp in developing rat hippocampal slices. Neuron 2002, 35, 535–545. [Google Scholar] [CrossRef]
- Bourne, J.N.; Sorra, K.E.; Hurlburt, J.; Harris, K.M. Polyribosomes are increased in spines of ca1 dendrites 2 h after the induction of ltp in mature rat hippocampal slices. Hippocampus 2007, 17, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Moon, I.S.; Cho, S.J.; Seog, D.H.; Walikonis, R. Neuronal activation increases the density of eukaryotic translation initiation factor 4e mRNA clusters in dendrites of cultured hippocampal neurons. Exp. Mol. Med. 2009, 41, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Asaki, C.; Usuda, N.; Nakazawa, A.; Kametani, K.; Suzuki, T. Localization of translational components at the ultramicroscopic level at postsynaptic sites of the rat brain. Brain Res. 2003, 972, 168–176. [Google Scholar] [CrossRef]
- Marrs, G.S.; Green, S.H.; Dailey, M.E. Rapid formation and remodeling of postsynaptic densities in developing dendrites. Nat. Neurosci. 2001, 4, 1006–1013. [Google Scholar] [CrossRef] [PubMed]
- Batish, M.; van den Bogaard, P.; Kramer, F.R.; Tyagi, S. Neuronal mrnas travel singly into dendrites. Proc. Natl. Acad. Sci. USA 2012, 109, 4645–4650. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Tatavarty, V.; Korza, G.; Levin, M.K.; Carson, J.H. Multiplexed dendritic targeting of alpha calcium calmodulin-dependent protein kinase ii, neurogranin, and activity-regulated cytoskeleton-associated protein rnas by the a2 pathway. Mol. Biol. Cell 2008, 19, 2311–2327. [Google Scholar] [CrossRef] [PubMed]
- Kanai, Y.; Dohmae, N.; Hirokawa, N. Kinesin transports RNA: Isolation and characterization of an RNA-transporting granule. Neuron 2004, 43, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Kiebler, M.A.; Bassell, G.J. Neuronal RNA granules: Movers and makers. Neuron 2006, 51, 685–690. [Google Scholar] [CrossRef] [PubMed]
- Krichevsky, A.M.; Kosik, K.S. Neuronal RNA granules: A link between rna localization and stimulation-dependent translation. Neuron 2001, 32, 683–696. [Google Scholar] [CrossRef]
- Tubing, F.; Vendra, G.; Mikl, M.; Macchi, P.; Thomas, S.; Kiebler, M.A. Dendritically localized transcripts are sorted into distinct ribonucleoprotein particles that display fast directional motility along dendrites of hippocampal neurons. J. Neurosci. 2010, 30, 4160–4170. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Yu, D.; Gu, Q.H.; Yang, Y.; Tu, K.; Zhu, J.; Li, Z. Mir-191 and mir-135 are required for long-lasting spine remodelling associated with synaptic long-term depression. Nat. Commun. 2014, 5, 3263. [Google Scholar] [CrossRef] [PubMed]
- Aakalu, G.; Smith, W.B.; Nguyen, N.; Jiang, C.; Schuman, E.M. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron 2001, 30, 489–502. [Google Scholar] [CrossRef]
- Kacharmina, J.E.; Job, C.; Crino, P.; Eberwine, J. Stimulation of glutamate receptor protein synthesis and membrane insertion within isolated neuronal dendrites. Proc. Natl. Acad. Sci. USA 2000, 97, 11545–11550. [Google Scholar] [CrossRef] [PubMed]
- Schratt, G.M.; Tuebing, F.; Nigh, E.A.; Kane, C.G.; Sabatini, M.E.; Kiebler, M.; Greenberg, M.E. A brain-specific microRNA regulates dendritic spine development. Nature 2006, 439, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Weiler, I.J.; Irwin, S.A.; Klintsova, A.Y.; Spencer, C.M.; Brazelton, A.D.; Miyashiro, K.; Comery, T.A.; Patel, B.; Eberwine, J.; Greenough, W.T. Fragile x mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc. Natl. Acad. Sci. USA 1997, 94, 5395–5400. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, R.; Nagerl, U.V.; Bonhoeffer, T. Neuronal activity determines the protein synthesis dependence of long-term potentiation. Nat. Neurosci. 2006, 9, 478–480. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, R.; Nagerl, U.V.; Morris, R.G.; Bonhoeffer, T. Competing for memory: Hippocampal ltp under regimes of reduced protein synthesis. Neuron 2004, 44, 1011–1020. [Google Scholar] [CrossRef] [PubMed]
- Sutton, M.A.; Schuman, E.M. Dendritic protein synthesis, synaptic plasticity, and memory. Cell 2006, 127, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Scharf, M.T.; Woo, N.H.; Lattal, K.M.; Young, J.Z.; Nguyen, P.V.; Abel, T. Protein synthesis is required for the enhancement of long-term potentiation and long-term memory by spaced training. J. Neurophysiol. 2002, 87, 2770–2777. [Google Scholar] [PubMed]
- Hu, H.; Qin, Y.; Bochorishvili, G.; Zhu, Y.; van Aelst, L.; Zhu, J.J. Ras signaling mechanisms underlying impaired glur1-dependent plasticity associated with fragile x syndrome. J. Neurosci. 2008, 28, 7847–7862. [Google Scholar] [CrossRef] [PubMed]
- Jin, P.; Warren, S.T. Understanding the molecular basis of fragile x syndrome. Hum. Mol. Genet. 2000, 9, 901–908. [Google Scholar] [CrossRef] [PubMed]
- Brown, V.; Jin, P.; Ceman, S.; Darnell, J.C.; O'Donnell, W.T.; Tenenbaum, S.A.; Jin, X.; Feng, Y.; Wilkinson, K.D.; Keene, J.D.; et al. Microarray identification of fmrp-associated brain mrnas and altered mRNA translational profiles in fragile x syndrome. Cell 2001, 107, 477–487. [Google Scholar] [CrossRef]
- Sidorov, M.S.; Auerbach, B.D.; Bear, M.F. Fragile x mental retardation protein and synaptic plasticity. Mol. Brain 2013, 6, 15. [Google Scholar] [CrossRef] [PubMed]
- Osterweil, E.K.; Krueger, D.D.; Reinhold, K.; Bear, M.F. Hypersensitivity to mglur5 and erk1/2 leads to excessive protein synthesis in the hippocampus of a mouse model of fragile x syndrome. J. Neurosci. 2010, 30, 15616–15627. [Google Scholar] [CrossRef] [PubMed]
- Qin, M.; Kang, J.; Burlin, T.V.; Jiang, C.; Smith, C.B. Postadolescent changes in regional cerebral protein synthesis: An in vivo study in the fmr1 null mouse. J. Neurosci. 2005, 25, 5087–5095. [Google Scholar] [CrossRef] [PubMed]
- Lipton, J.; Sahin, M. Fragile x syndrome therapeutics: Translation, meet translational medicine. Neuron 2013, 77, 212–213. [Google Scholar] [CrossRef] [PubMed]
- Osterweil, E.K.; Chuang, S.C.; Chubykin, A.A.; Sidorov, M.; Bianchi, R.; Wong, R.K.; Bear, M.F. Lovastatin corrects excess protein synthesis and prevents epileptogenesis in a mouse model of fragile x syndrome. Neuron 2013, 77, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Fiore, R.; Khudayberdiev, S.; Christensen, M.; Siegel, G.; Flavell, S.W.; Kim, T.K.; Greenberg, M.E.; Schratt, G. Mef2-mediated transcription of the mir379–410 cluster regulates activity-dependent dendritogenesis by fine-tuning pumilio2 protein levels. EMBO J. 2009, 28, 697–710. [Google Scholar] [CrossRef] [PubMed]
- Vo, N.; Klein, M.E.; Varlamova, O.; Keller, D.M.; Yamamoto, T.; Goodman, R.H.; Impey, S. A camp-response element binding protein-induced microrna regulates neuronal morphogenesis. Proc. Natl. Acad. Sci. USA 2005, 102, 16426–16431. [Google Scholar] [CrossRef] [PubMed]
- Edbauer, D.; Neilson, J.R.; Foster, K.A.; Wang, C.F.; Seeburg, D.P.; Batterton, M.N.; Tada, T.; Dolan, B.M.; Sharp, P.A.; Sheng, M. Regulation of synaptic structure and function by fmrp-associated micrornas mir-125b and mir-132. Neuron 2010, 65, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Zhao, J.; Hu, T.; Luo, Y.; Zhu, J.; Li, Z. Mir-501–3p mediates the activity-dependent regulation of the expression of ampa receptor subunit glua1. J. Cell Biol. 2015, 208, 949–959. [Google Scholar] [CrossRef] [PubMed]
- Siegel, G.; Obernosterer, G.; Fiore, R.; Oehmen, M.; Bicker, S.; Christensen, M.; Khudayberdiev, S.; Leuschner, P.F.; Busch, C.J.; Kane, C.; et al. A functional screen implicates microrna-138-dependent regulation of the depalmitoylation enzyme apt1 in dendritic spine morphogenesis. Nat. Cell Biol. 2009, 11, 705–716. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Kim, J.H.; Kwon, O.B.; An, K.; Ryu, J.; Cho, K.; Suh, Y.H.; Kim, H.S. An activity-regulated microRNA, mir-188, controls dendritic plasticity and synaptic transmission by downregulating neuropilin-2. J. Neurosci. 2012, 32, 5678–5687. [Google Scholar] [CrossRef] [PubMed]
- Ho, V.M.; Dallalzadeh, L.O.; Karathanasis, N.; Keles, M.F.; Vangala, S.; Grogan, T.; Poirazi, P.; Martin, K.C. Glua2 mRNA distribution and regulation by mir-124 in hippocampal neurons. Mol. Cell Neurosci. 2014, 61, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bosch, M.; Hayashi, Y. Structural plasticity of dendritic spines. Curr. Opin. Neurobiol. 2012, 22, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Fortin, D.A.; Srivastava, T.; Soderling, T.R. Structural modulation of dendritic spines during synaptic plasticity. Neuroscientist 2012, 18, 326–341. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Homma, K.J.; Poo, M.M. Shrinkage of dendritic spines associated with long-term depression of hippocampal synapses. Neuron 2004, 44, 749–757. [Google Scholar] [CrossRef] [PubMed]
- Rocca, D.L.; Martin, S.; Jenkins, E.L.; Hanley, J.G. Inhibition of arp2/3-mediated actin polymerization by pick1 regulates neuronal morphology and ampa receptor endocytosis. Nat. Cell Biol. 2008, 10, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Higuchi, O.; Ohashi, K.; Nagata, K.; Wada, A.; Kangawa, K.; Nishida, E.; Mizuno, K. Cofilin phosphorylation by lim-kinase 1 and its role in rac-mediated actin reorganization. Nature 1998, 393, 809–812. [Google Scholar] [PubMed]
- Antoniou, A.; Baptista, M.; Carney, N.; Hanley, J.G. Pick1 links argonaute 2 to endosomes in neuronal dendrites and regulates miRNA activity. EMBO Rep. 2014, 15, 548–556. [Google Scholar] [CrossRef] [PubMed]
- Dobbin, M.M.; Madabhushi, R.; Pan, L.; Chen, Y.; Kim, D.; Gao, J.; Ahanonu, B.; Pao, P.C.; Qiu, Y.; Zhao, Y.; et al. Sirt1 collaborates with atm and hdac1 to maintain genomic stability in neurons. Nat. Neurosci. 2013, 16, 1008–1015. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, W.Y.; Mao, Y.W.; Graff, J.; Guan, J.S.; Pan, L.; Mak, G.; Kim, D.; Su, S.C.; Tsai, L.H. A novel pathway regulates memory and plasticity via sirt1 and mir-134. Nature 2010, 466, 1105–1109. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Mateos, E.M.; Engel, T.; Merino-Serrais, P.; McKiernan, R.C.; Tanaka, K.; Mouri, G.; Sano, T.; O'Tuathaigh, C.; Waddington, J.L.; Prenter, S.; et al. Silencing microrna-134 produces neuroprotective and prolonged seizure-suppressive effects. Nat. Med. 2012, 18, 1087–1094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Impey, S.; McCorkle, S.R.; Cha-Molstad, H.; Dwyer, J.M.; Yochum, G.S.; Boss, J.M.; McWeeney, S.; Dunn, J.J.; Mandel, G.; Goodman, R.H. Defining the creb regulon: A genome-wide analysis of transcription factor regulatory regions. Cell 2004, 119, 1041–1054. [Google Scholar] [CrossRef] [PubMed]
- Finkbeiner, S.; Tavazoie, S.F.; Maloratsky, A.; Jacobs, K.M.; Harris, K.M.; Greenberg, M.E. Creb: A major mediator of neuronal neurotrophin responses. Neuron 1997, 19, 1031–1047. [Google Scholar] [CrossRef]
- Magill, S.T.; Cambronne, X.A.; Luikart, B.W.; Lioy, D.T.; Leighton, B.H.; Westbrook, G.L.; Mandel, G.; Goodman, R.H. MicroRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc. Natl. Acad. Sci. USA 2010, 107, 20382–20387. [Google Scholar] [CrossRef] [PubMed]
- Hansen, K.F.; Sakamoto, K.; Wayman, G.A.; Impey, S.; Obrietan, K. Transgenic mir132 alters neuronal spine density and impairs novel object recognition memory. PLoS ONE 2010, 5, e15497. [Google Scholar] [CrossRef] [PubMed]
- Mellios, N.; Sur, M. The emerging role of microRNAs in schizophrenia and autism spectrum disorders. Front. Psychiatry 2012, 3, 39. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.H.; Zeier, Z.; Xi, L.; Lanz, T.A.; Deng, S.; Strathmann, J.; Willoughby, D.; Kenny, P.J.; Elsworth, J.D.; Lawrence, M.S.; et al. MicroRNA-132 dysregulation in schizophrenia has implications for both neurodevelopment and adult brain function. Proc. Natl. Acad. Sci. USA 2012, 109, 3125–3130. [Google Scholar] [CrossRef] [PubMed]
- Bats, C.; Groc, L.; Choquet, D. The interaction between stargazin and psd-95 regulates ampa receptor surface trafficking. Neuron 2007, 53, 719–734. [Google Scholar] [CrossRef] [PubMed]
- Muddashetty, R.S.; Nalavadi, V.C.; Gross, C.; Yao, X.; Xing, L.; Laur, O.; Warren, S.T.; Bassell, G.J. Reversible inhibition of psd-95 mrna translation by mir-125a, fmrp phosphorylation, and mglur signaling. Mol. Cell 2011, 42, 673–688. [Google Scholar] [CrossRef] [PubMed]
- Fischer, R.S.; Fowler, V.M. Tropomodulins: Life at the slow end. Trends Cell Biol. 2003, 13, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Salgado, J.M.; Ostroff, L.; Helton, T.D.; Robinson, C.G.; Harris, K.M.; Ehlers, M.D. Plasticity-induced growth of dendritic spines by exocytic trafficking from recycling endosomes. Neuron 2006, 52, 817–830. [Google Scholar] [CrossRef] [PubMed]
- Makino, H.; Malinow, R. Ampa receptor incorporation into synapses during ltp: The role of lateral movement and exocytosis. Neuron 2009, 64, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Flintoft, L. Non-coding RNA: Structure and function for lncrnas. Nat. Rev. Genet. 2013, 14, 598. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, H.; Burnett, J.C.; Rossi, J.J. The role of antisense long noncoding RNA in small RNA-triggered gene activation. RNA 2014, 20, 1916–1928. [Google Scholar] [CrossRef] [PubMed]
- Villegas, V.E.; Zaphiropoulos, P.G. Neighboring gene regulation by antisense long non-coding RNAs. Int. J. Mol. Sci. 2015, 16, 3251–3266. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The gencode v7 catalog of human long noncoding rnas: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Pestova, T.V.; Hellen, C.U.; Tiedge, H. Translational control by a small RNA: Dendritic bc1 RNA targets the eukaryotic initiation factor 4a helicase mechanism. Mol. Cell Biol. 2008, 28, 3008–3019. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Iacoangeli, A.; Popp, S.; Muslimov, I.A.; Imataka, H.; Sonenberg, N.; Lomakin, I.B.; Tiedge, H. Dendritic bc1 RNA: Functional role in regulation of translation initiation. J. Neurosci. 2002, 22, 10232–10241. [Google Scholar] [PubMed]
- Zalfa, F.; Giorgi, M.; Primerano, B.; Moro, A.; Di Penta, A.; Reis, S.; Oostra, B.; Bagni, C. The fragile x syndrome protein fmrp associates with bc1 RNA and regulates the translation of specific mrnas at synapses. Cell 2003, 112, 317–327. [Google Scholar] [CrossRef]
- Muslimov, I.A.; Banker, G.; Brosius, J.; Tiedge, H. Activity-dependent regulation of dendritic bc1 RNA in hippocampal neurons in culture. J. Cell Biol. 1998, 141, 1601–1611. [Google Scholar] [CrossRef] [PubMed]
- Skryabin, B.V.; Sukonina, V.; Jordan, U.; Lewejohann, L.; Sachser, N.; Muslimov, I.; Tiedge, H.; Brosius, J. Neuronal untranslated bc1 RNA: Targeted gene elimination in mice. Mol. Cell Biol. 2003, 23, 6435–6441. [Google Scholar] [CrossRef] [PubMed]
- Lewejohann, L.; Skryabin, B.V.; Sachser, N.; Prehn, C.; Heiduschka, P.; Thanos, S.; Jordan, U.; Dell'Omo, G.; Vyssotski, A.L.; Pleskacheva, M.G.; et al. Role of a neuronal small non-messenger rna: Behavioural alterations in bc1 rna-deleted mice. Behav. Brain Res. 2004, 154, 273–289. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Chuang, S.C.; Bianchi, R.; Zhao, W.; Lee, H.; Fenton, A.A.; Wong, R.K.; Tiedge, H. Bc1 regulation of metabotropic glutamate receptor-mediated neuronal excitability. J. Neurosci. 2009, 29, 9977–9986. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microrna sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- You, X.; Vlatkovic, I.; Babic, A.; Will, T.; Epstein, I.; Tushev, G.; Akbalik, G.; Wang, M.; Glock, C.; Quedenau, C.; et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 2015, 18, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Tu, J.C.; Xiao, B.; Naisbitt, S.; Yuan, J.P.; Petralia, R.S.; Brakeman, P.; Doan, A.; Aakalu, V.K.; Lanahan, A.A.; Sheng, M.; et al. Coupling of mglur/homer and psd-95 complexes by the shank family of postsynaptic density proteins. Neuron 1999, 23, 583–592. [Google Scholar] [CrossRef]
- Xiao, M.Y.; Gustafsson, B.; Niu, Y.P. Metabotropic glutamate receptors in the trafficking of ionotropic glutamate and GABA(A) receptors at central synapses. Curr. Neuropharmacol. 2006, 4, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Sheng, M.; Kim, E. The postsynaptic organization of synapses. Cold Spring Harb. Perspect Biol. 2011. [Google Scholar] [CrossRef] [PubMed]
- Cougot, N.; Bhattacharyya, S.N.; Tapia-Arancibia, L.; Bordonne, R.; Filipowicz, W.; Bertrand, E.; Rage, F. Dendrites of mammalian neurons contain specialized p-body-like structures that respond to neuronal activation. J. Neurosci. 2008, 28, 13793–13804. [Google Scholar] [CrossRef] [PubMed]
- Di Penta, A.; Mercaldo, V.; Florenzano, F.; Munck, S.; Ciotti, M.T.; Zalfa, F.; Mercanti, D.; Molinari, M.; Bagni, C.; Achsel, T. Dendritic lsm1/cbp80-mrnps mark the early steps of transport commitment and translational control. J. Cell Biol. 2009, 184, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Zeitelhofer, M.; Macchi, P.; Dahm, R. Perplexing bodies: The putative roles of p-bodies in neurons. RNA Biol. 2008, 5, 244–248. [Google Scholar] [CrossRef] [PubMed]
- Luchelli, L.; Thomas, M.G.; Boccaccio, G.L. Synaptic control of mRNA translation by reversible assembly of xrn1 bodies. J. Cell Sci. 2015, 128, 1542–1554. [Google Scholar] [CrossRef] [PubMed]
- Baez, M.V.; Luchelli, L.; Maschi, D.; Habif, M.; Pascual, M.; Thomas, M.G.; Boccaccio, G.L. Smaug1 mRNA-silencing foci respond to nmda and modulate synapse formation. J. Cell Biol. 2011, 195, 1141–1157. [Google Scholar] [CrossRef] [PubMed]
- Buxbaum, A.R.; Wu, B.; Singer, R.H. Single beta-actin mRNA detection in neurons reveals a mechanism for regulating its translatability. Science 2014, 343, 419–422. [Google Scholar] [CrossRef] [PubMed]
- Rajgor, D.; Shanahan, C.M. RNA granules and cytoskeletal links. Biochem. Soc. Trans. 2014, 42, 1206–1210. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rajgor, D.; Hanley, J.G. The Ins and Outs of miRNA-Mediated Gene Silencing during Neuronal Synaptic Plasticity. Non-Coding RNA 2016, 2, 1. https://doi.org/10.3390/ncrna2010001
Rajgor D, Hanley JG. The Ins and Outs of miRNA-Mediated Gene Silencing during Neuronal Synaptic Plasticity. Non-Coding RNA. 2016; 2(1):1. https://doi.org/10.3390/ncrna2010001
Chicago/Turabian StyleRajgor, Dipen, and Jonathan G. Hanley. 2016. "The Ins and Outs of miRNA-Mediated Gene Silencing during Neuronal Synaptic Plasticity" Non-Coding RNA 2, no. 1: 1. https://doi.org/10.3390/ncrna2010001





