The Role of MicroRNAs in Kidney Disease
Abstract
:1. MicroRNAs: Discovery, Classification and Physiological Function
| Diseases | MicroRNA | Gene Target | Pathway Affected | References |
|---|---|---|---|---|
| Cancer | ||||
| Chronic lymphocytic leukemia | miR-15a and miR-16 | BAZ2A, RNF41, RASSF5, MKK3 and LRIG1 | [18] | |
| Human hepatocellular carcinoma | miR-221 | CDKN1B | Cell cycle | [19] |
| Breast cancer | miR-21 | PDCD4 | Apoptosis | [20] |
| Lung cancer | let-7 miRNA | CDK1 | Proliferation | [21] |
| Pancreatic cancer | miR-34a | p53 | Apoptosis | [22] |
| Neuroblastoma | miR-34a | BCL2, MYCN | Apoptosis, Proliferation | [23] |
| Human colon cancer | miR-145 | IRS-1 | Growth and Proliferation | [24] |
| Esophageal cancer | miR-21 | Ran | Growth and Proliferation | [25] |
| Vascular Disease | ||||
| Myocardial infarction | miR-29 | collagens, fibrillins, and elastin | Fibrosis proteins | [26] |
| Peripheral arterial disease | miR-221 | Kip1 and Kip2 | High glucose-induced endothelial dysfunction | [27] |
| Cardiac failure | miR-1 | Bcl-2 | Ischematic heart tissue | [28] |
| Obesity | ||||
| miR-143 | ERK5 | Differentiation | [29] | |
| Amyotrophic Lateral Sclerosis (ALS) | ||||
| miR-23a | peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α) | Dysregulation in mitochondrial gene expression | [30] | |
| miR-29b | Muscle regeneration | [30] | ||
| mir-455 | Muscle wasting | [30] | ||
2. MicroRNA Biogenesis
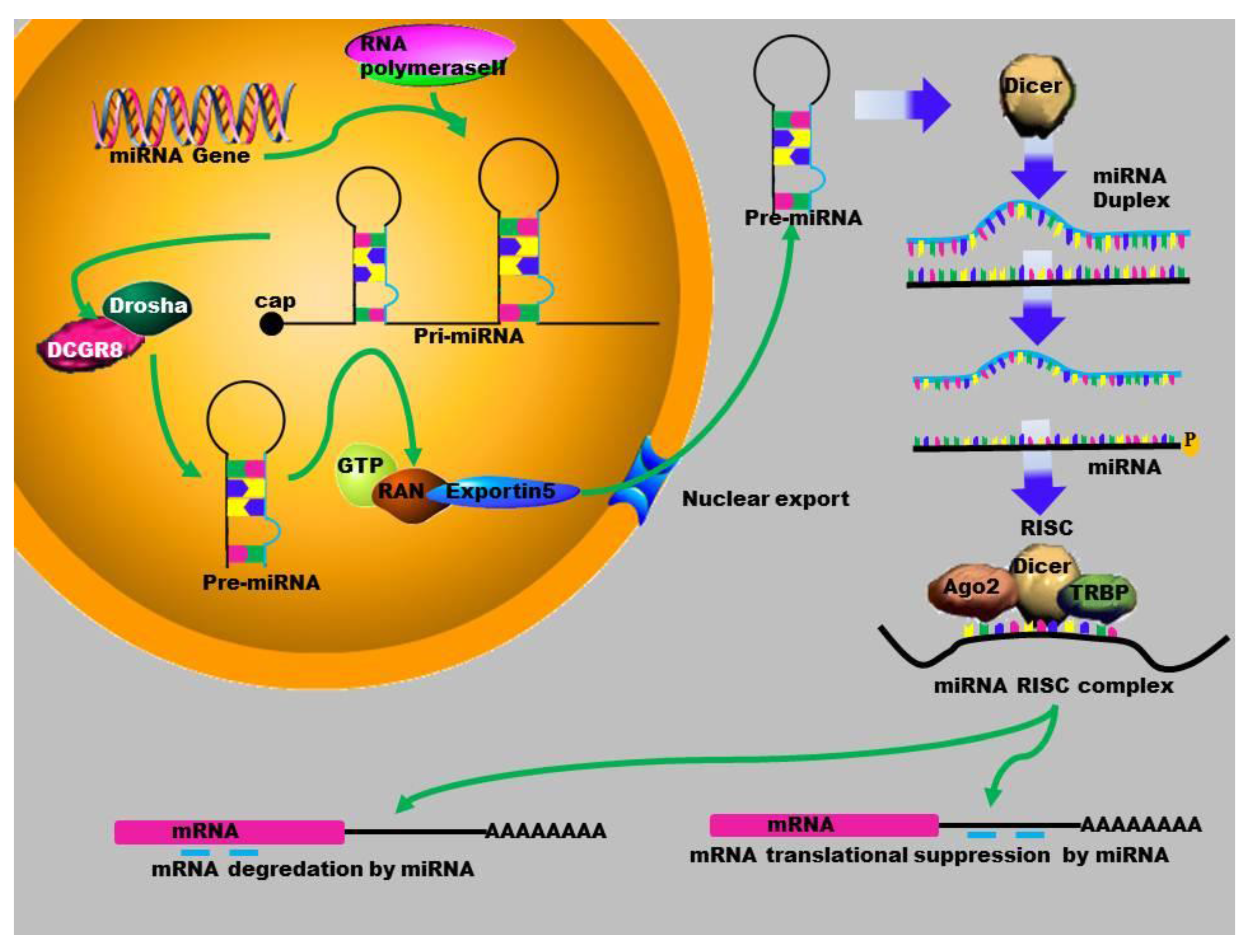
3. Gene Activation
4. MicroRNAs in Human Diseases
5. MicroRNA in Renal Diseases
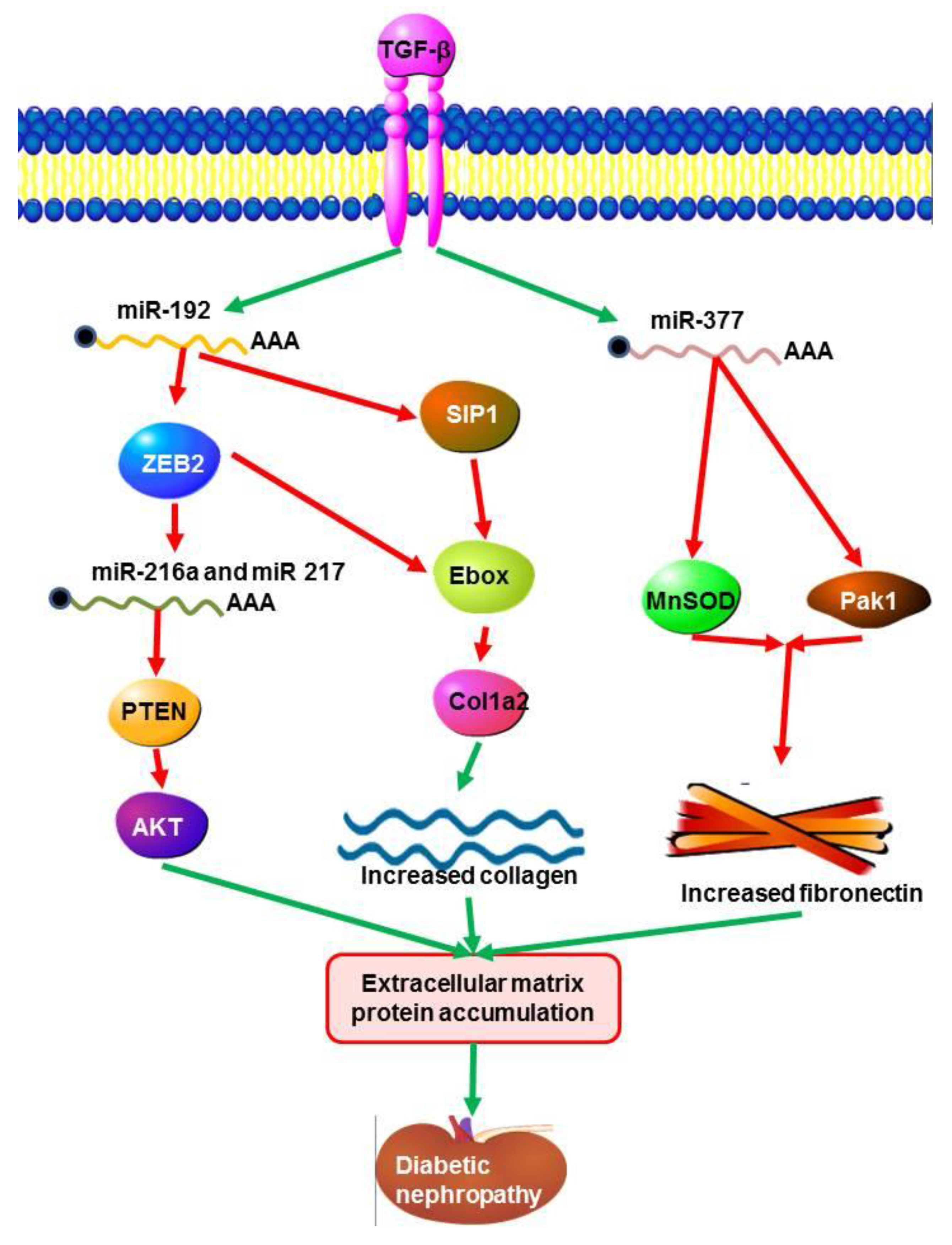
5.1. MicroRNAs in Diabetic Nephropathy (DN)
5.2. MicroRNAs in Renal Fibrosis
5.3. MicroRNAs in Lupus Nephritis (LN)
5.4. MicroRNA in Polycystic Kidney Diseases (PKD)
5.5. MicroRNA in Renal Cell Carcinoma (RCC)
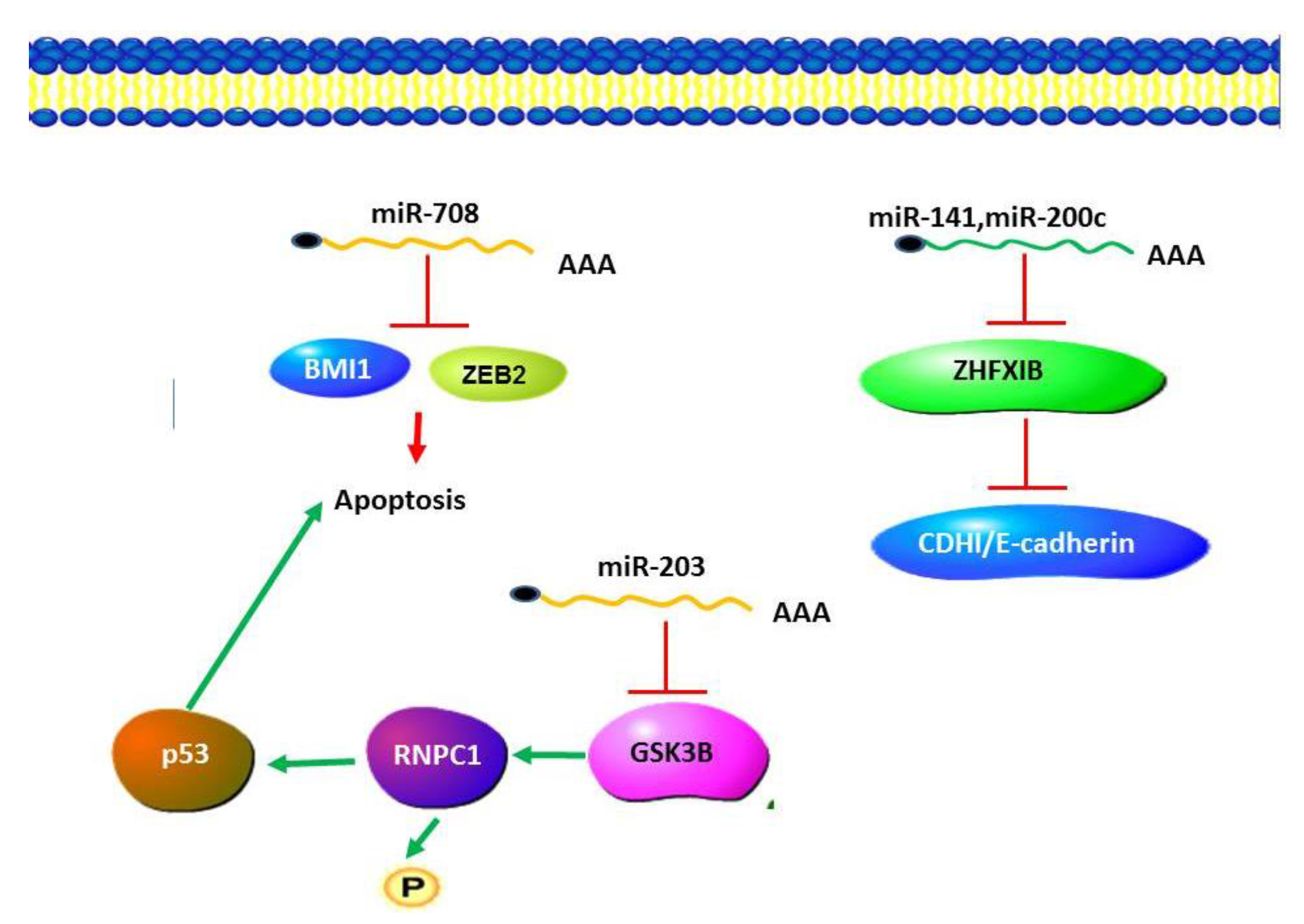
5.6. MicroRNAs in Wilm’s Tumor
5.7. MicroRNAs in IgA Nephropathy
5.8. MicroRNAs in Renal Ischemia/Reperfusion
5.9. MicroRNA in Allograft Acute Rejection
5.10. MicroRNA in Nephrotic Syndrome
5.11. MicroRNA in Human Immunodefiency Virus Associated Nephropathy (HIVAN)
5.12. MicroRNA in Hypertensive Nephrosclerosis
5.13. MicroRNA in Acute Pyelonephritis (APN)
5.14. Drug Associated Nephrotoxicity
6. Transforming Growth Factor (TGF-β)
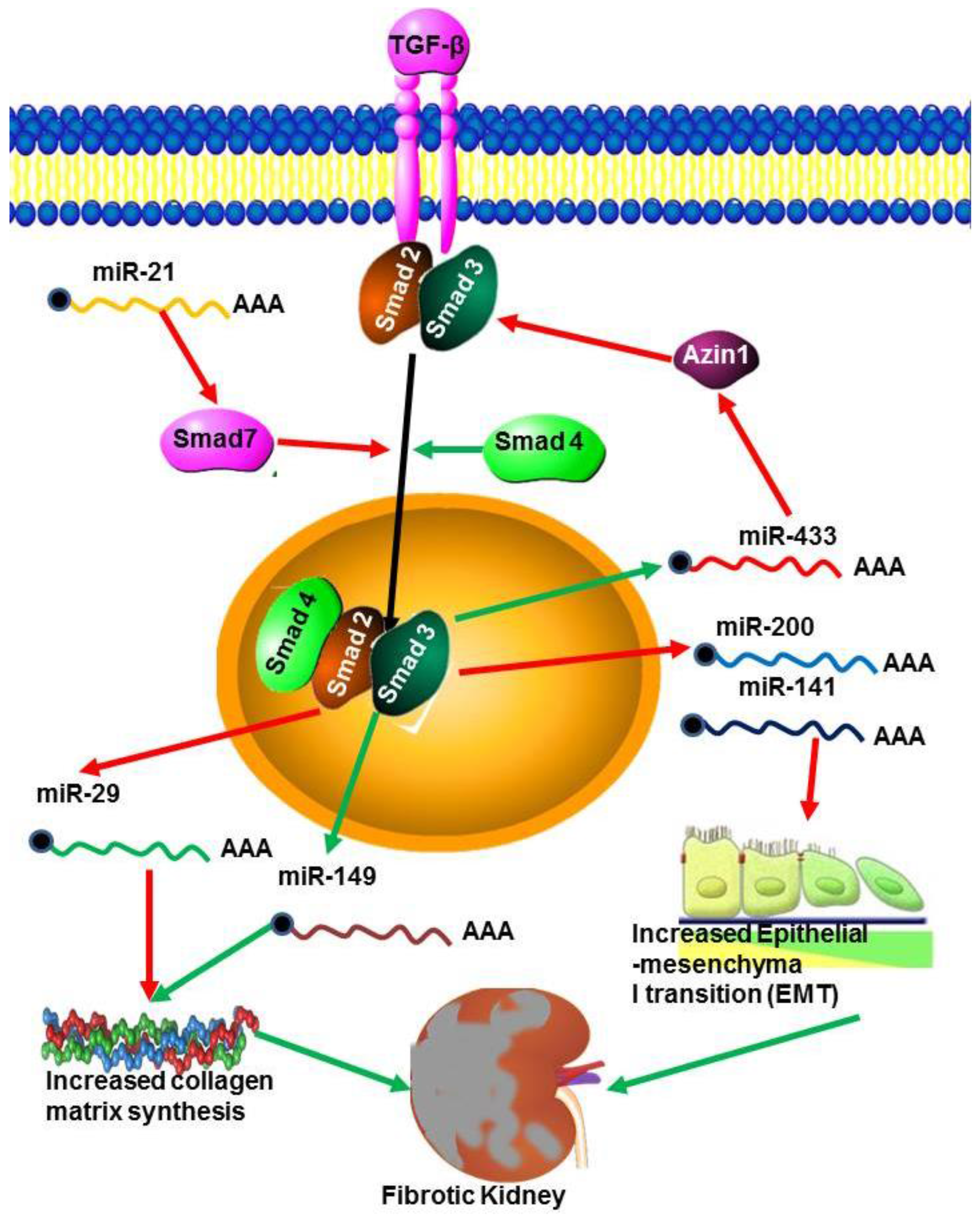
7. Important miRNAs
7.1. miR-29a
7.2. MiR-21
7.3. miR-200
8. MicroRNA in Renal Therapeutics
9. Biomarkers
10. Prospective
11. Conclusions
Acknowledgments
Conflicts of Interest
References
- Bottinger, E.P. TGF-beta in renal injury and disease 4. Semin. Nephrol. 2007, 27, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Du, T.; Zamore, P.D. MicroPrimer: The Biogenesis and Function of microRNA. Development 2005, 132, 4645–4652. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.K.; Zhu, J.Q.; Zhang, J.T.; Li, Q.; Li, Y.; He, J.; Qin, Y.W.; Jing, Q. Circulating microRNA: A novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur. Heart J. 2010, 31, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Small, E.M.; Olson, E.N. Pervasive roles of microRNAs in cardiovascular biology. Nature 2011, 469, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Reinhart, B.J.; Weinstein, E.G.; Rhoades, M.W.; Bartel, B.; Bartel, D.P. MicroRNAs in plants. Genes Dev. 2002, 16, 1616–1626. [Google Scholar] [CrossRef] [PubMed]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Bettinger, J.C.; Pasquinelli, A.E.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21 nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [PubMed]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, F.; Axtell, M.J. Analysis of Complementarity Requirements for Plant MicroRNA Targeting Using a Nicotiana benthamiana Quantitative Transient Assay. Plant Cell 2014, 26, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Phuah, N.H.; Nagoor, N.H. Regulation of microRNAs by natural agents: New strategies in cancer therapies. Biomed. Res. Int. 2014. [Google Scholar] [CrossRef] [PubMed]
- Costa, F.F. Non-coding RNAs: Lost in translation? Gene 2007, 386, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Nazarov, P.V.; Reinsbach, S.; Muller, A.; Nicot, A.; Philippidou, D.; Vallar, L.; Kreis, S. Interplay of microRNAs, transcription factors and target genes: Linking dynamic expression changes to function. Nucleic Acids Res. 2013, 41, 2817–2831. [Google Scholar] [CrossRef] [PubMed]
- Lai, A.; Kim, H.H.; Abdelmohsen, K.; Kuwano, Y.; Pullmann, R., Jr.; Srikantan, S.; Subrahmanyam, R.; Martindale, J.L.; Yang, X.; Ahmed, F. p16(INK4a) translation suppressed by miR-24. PLoS ONE 2008, 3, e1864. [Google Scholar]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Irio, M.; Croce, C. microRNA involvement in human cancer. Carcinogenesis 2012, 33, 1126–1133. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010, 466, 835–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jopling, C.L.; Yi, M.; Lancaster, A.M.; Lemon, S.M.; Sarnow, P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 2005, 309, 1577–1581. [Google Scholar] [CrossRef] [PubMed]
- Vo, N.K.; Cambronne, X.A.; Goodman, R.H. MicroRNA pathways in neural development and plasticity. Curr. Opin. Neurobiol. 2010, 20, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Hanlon, K.; Rudin, C.E.; Harries, L.W. Investigating the targets of MIR-15a and MIR-16-1 in patients with chronic lymphocytic leukemia (CLL). PLoS ONE 2009, 4, e7169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fornari, F.; Gramantieri, L.; Ferracin, M.; Veronese, A.; Sabbioni, S.; Calin, G.A.; Grazi, G.L.; Giovannini, C.; Croce, C.M.; Bolondi, L.; et al. miR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene 2008, 27, 5651–5661. [Google Scholar] [CrossRef] [PubMed]
- Frankel, L.B.; Christoffersen, N.R.; Jacobsen, A.; Lindow, M.; Krogh, A.; Lund, A.H. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J. Biol. Chem. 2008, 283, 1026–1033. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Lorenz, P.; Gross, G.; Ibrahim, S.; Kunz, M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008, 18, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.C.; Wentzel, E.A.; Kent, O.A.; Ramachandran, K.; Mullendore, M.; Lee, K.H.; Feldmann, G.; Yamakuchi, M.; Ferlito, M.; Lowenstein, C.J.; et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell 2007, 26, 745–752. [Google Scholar] [CrossRef] [PubMed]
- Cole, K.A.; Attiyeh, E.F.; Mosse, Y.P.; Laquaglia, M.J.; Diskin, S.J.; Brodeur, G.M.; Maris, J.M. A Functional Screen Identifies miR-34a as a Candidate Neuroblastoma Tumor Suppressor Gene. Mol. Cancer Res. 2008, 6, 735–742. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.M.; Yang, J.; Shen, X.Y.; Zhang, X.Y.; Meng, F.S.; Xu, J.T.; Zhang, B.F.; Gao, H.J. MicroRNA expression profile in non-cancerous colonic tissue associated with lymph node metastasis of colon cancer. J. Dig. Dis. 2009, 10, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Yang, Z.; Cao, M.; Xu, Y.; Li, J.; Chen, X.; Gao, Z.; Xin, J.; Zhou, S.; Zhou, Z.; et al. miR-203 supresses tumor growth and invasion and down-regulates miR-21 expresssion through Ran in esophangeal cancer. Cancer Lett. 2014, 341, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Van Rooij, E.; Sutherland, L.B.; Thatcher, J.E.; Dimaio, J.M.; Naseem, R.H.; Marshall, W.S.; Hill, J.A.; Olson, E.N. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc. Natl. Acad. Sci. USA 2008, 105, 13027–13032. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Yuan, P.; He, Y. Role of microRNAs in peripheral artery disease. Mol. Med. Rep. 2012, 6, 695–700. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zheng, J.; Sun, Y.; Wu, Z.; Liu, Z.; Huang, G. MicroRNA-1 regulates cardiomyocyte apoptosis by targeting Bcl-2. Int. Heart J. 2009, 50, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Hwang, S.J.; Bae, Y.C.; Jung, J.S. MiR-21 regulates adipogenic differentiation through the modulation of TGF-β signaling in mesenchymal stem cells derived from human adipose tissue. Stem Cells 2009, 27, 3093–3102. [Google Scholar]
- Russell, A.P.; Wada, S.; Vergani, L.; Hock, M.B.; Lamon, S.; Leger, B.; Ushida, T.; Cartoni, R.; Wadley, G.D.; Hespel, P.; et al. Disruption of skeletal muscle mitochondrial network genes and miRNAs in amyotrophic lateral sclerosis. Neurobiol. Dis. 2012, 49, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.; Sepp-Lorenzino, L.; Prisco, M.; Linsley, P.; deAngelis, T.; Baserga, R. MicroRNA 145 targets the insulin receptor substrate-1 and inhibits the growth of colon cancer cells. J. Biol. Chem. 2007, 282, 32582–32590. [Google Scholar] [CrossRef] [PubMed]
- Stefani, G.; Slack, F.J. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 2008, 9, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Ro, S.; Ma, H.Y.; Park, C.; Ortogero, N.; Song, R.; Hennig, G.W.; Zheng, H.; Lin, Y.M.; Moro, L.; Hsieh, J.T.; et al. The mitochondrial genome encodes abundant small noncoding RNAs. Cell Res. 2013, 23, 759–774. [Google Scholar] [CrossRef] [PubMed]
- Huang, V.; Li, L.C. miRNA goes nuclear. RNA Biol. 2012, 9, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Danger, R.; Braza, F.; Giral, M.; Soulilou, J.P.; Brouard, S. MicroRNAs, major players in B cells homeostasis and function. Front. Immunol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Geisler, S.; Coller, J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013, 14, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Place, R.F.; Li, L.C.; Pookot, D.; Noonan, E.J.; Dahiya, R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl. Acad. Sci. USA 2008, 105, 1608–1613. [Google Scholar] [CrossRef] [PubMed]
- Majid, S.; Dar, A.A.; Saini, S.; Yamamura, S.; Hirata, H.; Tanaka, Y.; Deng, G.; Dahiya, R. MicroRNA-205-directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer 2010, 116, 5637–5649. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.Y.; Manley, J.L. The TET family of proteins: Functions and roles in disease. J. Mol. Cell Biol. 2009, 1, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Friedlander, M.R.; Lizano, E.; Houben, A.J.; Bezdan, D.; Banez-Coronel, M.; Kudla, G.; Mateu-Huertas, E.; Kagerbauer, B.; González, J.; Chen, K.C.; et al. Evidence for the biogenesis of more than 1000 novel human microRNAs. Genome Biol. 2014, 15, R57. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.M.; Rao, D.S.; Chaudhuri, A.A.; Baltimore, D. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 2010, 10, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Dirks, J.; Remuzzi, G.; Horton, S.; Schieppati, A.; Adibul Hasan Rizvi, S. Chapter 36 Diseases of the Kidney and the Urinary System. In Disease Control Priorities in Developing Countries, 2nd ed.; Jamison, D.T., Breman, J.G., Measham, A.R., Eds.; World Bank: Washington, DC, USA, 2006. [Google Scholar]
- Sun, Y.; Koo, S.; White, N.; Peralta, E.; Esau, C.; Dean, N.M.; Perera, R.J. Development of a micro-array to detect human and mouse microRNAs and characterization of expression in human organs. Nucleic Acids Res. 2004, 32, e188. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Greene, A.S.; pietrusz, J.L.; Matus, I.R.; Liang, M. MicroRNA-target pairs in the rat kidney identified by microRNA microarray, proteomic, and bioinformatics analysis. Genome Res. 2008, 18, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Sevignani, C.; Dumitru, C.D.; Hyslop, T.; Noch, E.; Yendamuri, S.; Shimizu, M.; Rattan, S.; Bullrich, F.; Negrini, M.; et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 2004, 101, 2999–3004. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Croce, C.M. MicroRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 2012, 4, 143–159. [Google Scholar] [CrossRef] [PubMed]
- Pekarsky, Y.C.M.; Croce, C.M. Role of miR-15/16 in CLL. Cell Death Differ. 2014, 22, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Zhang, Y.; Wang, Z.; Wang, L.; Wei, X.; Zhang, B.; Wen, Z.; Fang, H.; Pang, Q.; Yi, F. Regulation of NADPH oxidase activity is associated with miRNA-25-mediated NOX 4 expression in experimental diabetic nephropathy. Am. J. Nephrol. 2010, 32, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Brattelid, T.; Aarnes, E.K.; Helgeland Guvaag, E.S.; Eichele, H.; Jonassen, A.K. Normalization strategy is critical for the outcome of miRNA expression analyses in the rat heart. Physiol. Genomics 2011, 43, 604–610. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, J.; Gupta, S.K.; Thum, T. Identification of cardiovascular microRNA targetomes. J. Mol. Cell. Cardiol. 2011, 51, 674–681. [Google Scholar] [CrossRef] [PubMed]
- Macconi, D.; Tomasoni, S.; Romagnani, P.; Trionfini, P.; Sangalli, F.; Mazzinghi, B.; Rizzo, P.; Lazzeri, E.; Abbate, M.; Remuzzi, G.; et al. MicroRNA-324-3p promotes renal fibrosis and is a ACE inhibition. J. Am. Soc. Nephrol. 2014, 23, 1496–1505. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Mi, Q.; Dong, Z. The regulation and function of microRNAs in kidney diseases. IUBMB Life 2013, 65, 602–614. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Pan, X.; Cobb, G.; Anderson, T. Plant microRNA: A small regulatory molecule with big impact. Dev. Biol. 2006, 289, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Bushati, N.; Cohen, S. MicroRNA functions. Annu. Rev. Cell Dev. Biol. 2007, 23, 175–205. [Google Scholar] [CrossRef] [PubMed]
- Van Rooij, E. The art of microRNA research. Circ. Res. 2011, 108, 219–234. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Guan, J.; Ping, Y.; Xu, C.; Huang, T.; Zhao, H.; Fan, H.; Li, Y.; Lv, Y.; Zhao, T.; et al. Prioritizing cancer-related key miRNA—Target interactions by integrative genomics. Nucleic Acids Res. 2012, 40, 7653–7665. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Li, C.X.; Lv, J.Y.; Li, Y.S.; Xiao, Y.; Shao, T.T.; Huo, X.; Li, X.; Zou, Y.; Han, Q.L.; et al. Prioritizing Candidate Disease miRNAs by Topological Features in the miRNA Target–Dysregulated Network: Case Study of Prostate Cancer. Mol. Cancer Ther. 2011, 10, 1857–1866. [Google Scholar] [CrossRef] [PubMed]
- Xin, F.; Li, M.; Balch, C.; Thomson, M.; Fan, M.; Liu, Y.; Hammond, S.M.; Kim, S.; Nephew, K.P. Computational analysis of microRNA profiles and their target genes suggests significant involvement in breast cancer antiestrogen resistance. Bioinformatics 2009, 25, 430–434. [Google Scholar] [CrossRef] [PubMed]
- O'Neill, L.A.; Sheedy, F.J.; McCoy, C.E. MicroRNAs: The fine-tuners of Toll-like receptor signalling. Nat. Rev. Immunol. 2011, 11, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Karin, M. Mammalian MAP kinase signalling cascades. Nature 2001, 410, 37–40. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, K.; Karolina, D.S.; Sepramaniam, S.; Armugam, A.; Wintour, E.M.; Bertram, J.F.; Jeyaseelan, K. Role of microRNAs in kidney homeostasis and disease. Kidney Int. 2012, 81, 617–627. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.; Kreidberg, J.A. MicroRNAs in renal development. Pediatr. Nephrol. 2013, 28, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Qu, L. The function of microRNAs in renal development and pathophysiology. J. Genet. Genomics 2013, 40, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, Y.; Minto, A.W.; Wang, J.; Shi, Q.; Li, X.; Quigg, R.J. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. FASEB J. 2008, 22, 4126–4135. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Zhang, J.; Wang, M.; Lanting, L.; Yuan, H.; Rossi, J.J.; Natarajan, R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc. Natl. Acad. Sci. USA 2007, 104, 3432–3437. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Komers, R.; Carew, R.; Winbanks, C.E.; Xu, B.; Herman-Edelstein, M.; Koh, P.; Thomas, M.; Jandeleit-Dahm, K.; Gregorevic, P.; et al. Suppression of microRNA-29 expression by TGF-β1 promotes collagen expression and renal fibrosis. J. Am. Soc. Nephrol. 2012, 23, 252–265. [Google Scholar] [CrossRef] [PubMed]
- Wynn, T.A.; Ramalingam, T.R. Mechanisms of fibrosis: Therapeutic translation for fibrotic disease. Nat. Med. 2012, 18, 1028–1040. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Chung, A.C.; Dong, Y.; Zhong, X.; Lan, H.Y. miR-433 promotes renal fibrosis by targeting the TGF-β/Smad3-Azin1 pathway. Kidney Int. 2013, 84, 1129–1144. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Herman-Edelstein, M.; Koh, P.; Burns, W.; Jandeleit-Dahm, K.; Watson, A.; Saleem, M.; Goodall, G.J.; Twigg, S.M.; Cooper, M.E.; et al. E-cadherin expression is regulated by miR-192/215 by a mechanism that is independent of the profibrotic effects of transforming growth factor-beta. Diabetes 2010, 59, 1794–1802. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Sui, W.; Lan, H.; Yan, Q.; Huang, H.; Huang, Y. Comprehensive analysis of microRNA expression patterns in renal biopsies of lupus nephritis patients. Rheumatol. Int. 2009, 29, 749–754. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Shen, N. MicroRNA Involvement in Lupus. Curr. Opin. Rheumatol. 2012, 24, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Tam, L.S.; Li, E.K.; Kwan, B.C.; Chow, K.M.; Luk, C.C.; Li, P.K.; Szeto, C.C. Serum and urinary free microRNA level in patients with systemic lupus erythematosus. Lupus 2011, 20, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Kwan, B.C.; Lai, F.M.; Tam, L.S.; Li, E.K.; Chow, K.M.; Wang, G.; Li, P.K.; Szeto, C.C. Glomerular and tubulointerstitial miR-638, miR-198 and miR-146a expression in lupus nephritis. Nephrology 2012, 17, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Te, J.L.; Dozmorov, I.M.; Guthridge, J.M.; Nguyen, K.L.; Cavett, J.W.; Kelly, J.A.; Bruner, G.R.; Harley, J.B.; Ojwang, J.O. Identification of unique microRNA signature associated with lupus nephritis. PLoS ONE 2010, 5, e10344. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Cleary, R.C.; Bogaert, Y.E. Combination of micro-RNA-192 and microRNA-27b from urinary exosomes differentiate between renal tubular damage and glomerular injury. J. Am. Soc. Nephrol. 2008, 19, 672A. [Google Scholar]
- Guan, J.; Wang, G.; Tam, L.S.; Kwan, B.C.H.; Li, E.K.M.; Chow, K.M.; Li, P.K.T.; Szeto, C.C. Urinary sediment ICAM-1 in lupus nephritis. Lupus 2012, 21, 1190–1195. [Google Scholar] [CrossRef] [PubMed]
- Ecder, T.; Schrier, R.W. Hypertension in autosomal-dominant polycystic kidney disease: Early occurrence and unique aspects. J. Am. Soc. Nephrol. 2001, 12, 194–200. [Google Scholar] [PubMed]
- Dweep, H.; Sticht, C.; Kharkar, A.; Pandey, P.; Gretz, N. Parallel analysis of mRNA and microRNA microarray profiles to explore functional regulatory patterns in polycystic kidney disease: Using PKD/Mhm rat model. PLoS ONE 2013, 8, e53780. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.; Bros, B.; Srivastava, P.K.; Bott, A.; Boehn, S.N.; Groene, H.J.; Gretz, N. Microarray-based approach identifies microRNAs and their target functional patterns in polycystic kidney disease. BMC Genomics 2008, 9, 624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, Y.C.; Blumenfeld, J.; Rennert, H. Autosomal dominant polycystic kidney disease: Genetics, mutations and microRNAs. Biochim. Biophys. Acta 2011, 1812, 1202–1212. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Li, Q.W.; Lv, X.Y.; Ai, J.Z.; Yang, Q.T.; Duan, J.J.; Bian, G.H.; Xiao, Y.; Wang, Y.D.; Zhang, Z.; et al. MicroRNA-17 post-transcriptionally regulates polycystic kidney disease-2 gene and promotes cell proliferation. Mol. Biol. Rep. 2010, 37, 2951–2958. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.O.; Masyuk, T.; Splinter, P.; Banales, J.M.; Masyuk, A.; Stroope, A.; Larusso, N. MicroRNA15a modulates expression of the cell-cycle regulator Cdc25A and affects hepatic cystogenesis in a rat model of polycystic kidney disease. J. Clin. Investig. 2008, 118, 3714–3724. [Google Scholar] [CrossRef] [PubMed]
- Lughezzani, G.; Jeldres, C.; Isbarn, H.; Perrotte, P.; Shariat, S.F.; Sun, M.; Widmer, H.; Arjane, P.; Peloquin, F.; Pharand, D.; et al. Tumor size is a determinant of the rate of stage T1 renal cell cancer synchronous metastasis. J. Urol. 2009, 182, 1287–1293. [Google Scholar] [CrossRef] [PubMed]
- Petillo, D.; Kort, E.J.; Anema, J.; Furge, K.A.; Yang, X.J.; Teh, B.T. MicroRNA profiling of human kidney cancer subtypes. Int. J. Oncol. 2009, 35, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Fridman, E.; Dotan, Z.; Barshack, I.; David, M.B.; Dov, A.; Tabak, S.; Zion, O.; Benjamin, S.; Benjamin, H.; Kuker, H.; et al. Accurate molecular classification of renal tumors using microRNA expression. J. Mol. Diagn. 2010, 12, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Youssef, Y.M.; White, N.M.; Grigull, J.; Krizova, A.; Samy, C.; Mejia-Guerrero, S.; Evans, A.; Yousef, G.M. Accurate molecular classification of kidney cancer subtypes using microRNA signature. Eur. Urol. 2011, 59, 721–730. [Google Scholar] [CrossRef] [PubMed]
- Saini, S.; Yamamura, S.; Majid, S.; Shahryari, v.; Hirata, H.; Tanaka, Y.; Dahiya, R. Regulatory role of miR-203in prostate cancer progression and metastasis. Clin. Cancer Res. 2011, 17, 5287–5298. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.; Campbell, C.E.; Bonetta, L.; McAndrews-Hill, M.S.; Chilton-MacNeill, S.; Coppes, M.J.; Law, D.J.; Feinberg, A.P.; Yeger, H.; Williams, B.R.G. Tissue, developmental, and tumor-specific expression of divergent transcripts in Wilms tumor. Science 1990, 250, 991–994. [Google Scholar] [CrossRef] [PubMed]
- Kort, E.J.; Farber, L.; Tretiakova, M.; Petillo, D.; Furge, K.A.; Yang, X.J.; Cornelius, A.; Teh, B.T. The E2F3-Oncomir-1 axis is activated in Wilms’ tumor. Cancer Res. 2008, 68, 4034–4038. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.A.; Bryan, K.; Williams, R.; Popov, S.; Vujanic, G.; Coulomb, A.; Boccon-Gibod, L.; Graf, N.; Pritchard-Jones, K.; O’Sullivan, M. Mirna profiles as a predictor of chemoresponsiveness in Wilms’ tumor blastema. PLoS ONE 2013, 8, e53417. [Google Scholar] [CrossRef] [PubMed]
- Wegert, J.; Ishaque, N.; Vardapour, R.; Geörg, C.; Gu, Z.; Bieg, M.; Ziegler, B.; Bausenwein, S.; Nourkami, N.; Ludwig, N.; et al. Mutations in the SIX1/2 pathway and the DROSHA/DGCR8 miRNA microprocessor complex underlie high-risk blastemal type Wilms tumors. Cancer Cell 2015, 27, 298–311. [Google Scholar] [CrossRef] [PubMed]
- Drake, K.M.; Ruteshouser, E.C.; Natrajan, R.; Harbor, P.; Wegert, J.; Gessler, M.; Pritchard-Jones, K.; Grundy, P.; Dome, J.; Huff, V.; et al. Loss of heterozygosity at 2q37 in sporadic Wilms’ tumor: Putative role for miR-562. Clin. Cancer Res. 2009, 15, 5985–5992. [Google Scholar] [CrossRef] [PubMed]
- Veronese, A.; Lupini, L.; Consiglio, J.; Visone, R.; Ferracin, M.; Fornari, F.; Zanesi, N.; Alder, H.; D’Elia, G.; Gramantieri, L.; et al. Oncogenic role of miR-483-5p at the IGF2/483 locus. Cancer 2010, 70, 3140–3149. [Google Scholar]
- Imam, J.S.; Buddavarapu, K.; Lee-Chang, J.S.; Ganapathy, S.; Camosy, C.; Chen, Y.; Rao, M.K. MicroRNA-185 suppresses tumor growth and progression by targeting the Six1 oncogene in human cancers. Oncogene 2010, 29, 4971–4979. [Google Scholar] [CrossRef] [PubMed]
- Takakura, S.; Mitsutake, N.; Nakashima, M.; Namba, H.; Saenko, V.A.; Rogounovitch, T.I.; Nakazawa, Y.; Hayashi, T.; Ohtsuru, A.; Yamashita, S. Oncogenic role of miR-17-92 cluster in anaplastic thyroid cancer cells. Cancer Sci. 2008, 99, 1147–1154. [Google Scholar] [CrossRef] [PubMed]
- Cloonan, N.; Brown, M.K.; Steptoe, A.L.; Wani, S.; Chan, W.L.; Forrest, AR.; Kolle, G.; Gabrielli, B.; Grimmond, S.M. The miR-17-5p microRNA is a key regulator of the G1/S phase cell cycle transition. Genome Biol. 2008, 9, R127. [Google Scholar] [CrossRef] [PubMed]
- Gebeshuber, C.A.; Kornauth, C.; Dong, L.; Sierig, R.; Seibler, J.; Reiss, M.; Tauber, S.; Bilban, M.; Wang, S.; Kain, R.; et al. Focal segmental glomerulosclerosis is induced by microRNA-193a and its downregulation of WT1. Nat. Med. 2013, 19, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Stanczyk, J.; Pedrioli, D.M.; Brentano, F.; Sanchez-Pernaute, O.; Kolling, C.; Gay, R.E.; Detmar, M.; Gay, S.; Kyburz, D. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheumatol. 2008, 58, 1001–1009. [Google Scholar] [CrossRef] [PubMed]
- Guerau-de-Arellano, M.; Smith, K.M.; Godlewski, J.; Liu, Y.; Winger, R.; Lawler, S.E.; Whitacre, C.C.; Racke, M.K.; Lovett-Racke, A.E. Micro-RNA dysregulation in multiple sclerosis favours pro-inflammatory T-cell-mediated autoimmunity. Brain 2011, 134, 3578–3589. [Google Scholar] [CrossRef] [PubMed]
- Ohl, K.; Tenbrock, K. Inflammatory cytokines in systemic lupus erythematosus. J. Biomed. Biotechnol. 2011. [Google Scholar] [CrossRef] [PubMed]
- Serino, G.; Sallustio, F.; Cox, S.N.; Pesce, F.; Schena, F.P. Abnormal miR-418b expression promotes aberrant glycosylation of IgA1 in IgA nephropathy. J. Am. Soc. Nephrol. 2012, 23, 814–824. [Google Scholar] [CrossRef] [PubMed]
- Allen, A.C.; Topham, P.S.; Harper, S.J.; Feehally, J. Leucocyte beta 1,3-galactosyltransferase activity in IgA nephropathy. Nephrol. Dial. Transplant. 1997, 12, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Zhou, Q.; Yang, L.C.; Li, Z.; Su, B.H.; Luo, H.; Fan, J.M. Peripheral B lymphocyte beta1,3-galactosyltransferase and chaperone expression in immunoglobulin A nephropathy. J. Intern. Med. 2005, 258, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Bellomo, R.; Kellum, J.A.; Ronco, C. Acute kidney injury. Lancet 2012, 380, 756–766. [Google Scholar] [CrossRef]
- Wei, Q.; Bhatt, K.; He, H.Z.; Mi, Q.S.; Hasse, V.H.; Dong, Z. Targeted deletion of Dicer from proximal tubules protects against renal ischemia-reperfusion injury. J. Am. Soc. Nephrol. 2010, 21, 756–761. [Google Scholar] [CrossRef] [PubMed]
- Godwin, J.G.; Ge, X.; Stephan, K.; Jurisch, A.; Tullius, S.G.; Iacomini, J. Identification of a microRNA signature of renal ischemia reperfusion injury. Proc. Natl. Acad. Sci. USA 2010, 107, 14339–14344. [Google Scholar] [CrossRef] [PubMed]
- Lorenzen, J.M.; Volkmann, I.; Fiedler, J.; Schmidt, M.; Scheffner, I.; Haller, H.; Gwinner, W.; Thum, T. Urinary miR-210 as a mediator of acute T-cell mediated rejection in renal allograft recipients. Am. J. Transplant. 2011, 11, 2221–2227. [Google Scholar] [CrossRef] [PubMed]
- Munshi, R.; Johnson, A.; Siew, E.D.; Ikizler, T.A.; Ware, L.B.; Wurfel, M.M.; Himmelfarb, J.; Zager, R.A. MCP-1 gene activation marks acute kidney injury. J. Am. Soc. Nephrol. 2011, 22, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Chau, B.; Xin, C.; Hartner, J.; Ren, S.; Castano, A.; Linn, G.; Li, J.; Tran, P.; Kaimal, V.; Huang, X.; et al. MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Sci. Transl. Med. 2012, 4. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, M.D.; Bagley, J.; Latz, J.; Godwin, J.G.; Ge, X.; Tullius, S.G.; Iacomini, J. MicroRNA expression data reveals a signature of kidney damage following ischemia reperfusion injury. PLoS ONE 2011, 6, e23011. [Google Scholar] [CrossRef] [PubMed]
- Anglicheaua, D.; Sharmaa, V.K.; Dinga, R.; Hummela, A.; Snopkowskia, C.; Dadhaniaa, D.; Seshane, S.V.; Suthanthirana, M. MicroRNA expression profiles predictive of human renal allograft status. Proc. Natl. Acad. Sci. USA 2009, 106, 13. [Google Scholar] [CrossRef] [PubMed]
- Sui, W.; Dai, Y.; Huang, Y.; Lan, H.; Yan, Q.; Huang, H. Microarray analysis of MicroRNA expression in acute rejection after renal transplantation. Transpl. Immunol. 2008, 19, 81. [Google Scholar] [CrossRef] [PubMed]
- Glowacki, F.; Savary, G.; Gnemmi, V.; Buob, D.; van der Hauwaert, C.; Lo-Guidice, J.M.; Bouyé, S.; Hazzan, M.; Pottier, N.; Perrais, M.; et al. Increased circulating miR-21 levels are associated with kidney fibrosis. PLoS ONE 2013, 8, e58014. [Google Scholar] [CrossRef] [PubMed]
- Zununi, S.; Ardalan, M. MicroRNA and Renal Allograft Monitoring. Nephrol. Urol. 2013, 5, 783–786. [Google Scholar] [CrossRef] [PubMed]
- Lorenzen, J.M.; Thum, T. Circulating and Urinary microRNAs in Kidney Disease. Clin. J. Am. Soc. Nephrol. 2012, 7, 1528–1533. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Wang, C.; Chen, X.; Zhong, T.; Cai, X.; Chen, S.; Shi, Y.; Hu, J.; Guan, X.; Xia, Z.; et al. Increased serum and urinary microRNAs in Children with idiopathic nephrotic syndrome. Clin. Chem. 2013. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Kwan, B.C.; Lai, F.M.; Chow, K.M.; Li, P.K.; Szeto, C.C. Urinary sediment miRNA levels in adult nephrotic syndrome. Clin. Chim. Acta 2013, 15, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Snyder, A.; Alsauskas, Z.C.; Leventhal, J.S.; Rosenstiel, P.E.; Gong, P.; Chan, J.J.; Barley, K.; He, J.C.; Klotman, M.E.; Ross, M.J.; et al. HIV-1 viral protein r induces ERK and caspase-8-dependent apoptosis in renal tubular epithelial cells. AIDS 2010, 24, 1107–1119. [Google Scholar] [CrossRef] [PubMed]
- Rao, T.K.; Filippone, E.J.; Nicastri, A.D.; Anthony, D.; Nicastri, M.D.; Landesman, S.H.; Frank, E.; Chen, C.K.; Friedman, E.A. Associated focal and segmental glomerulosclerosis in the acquired immunodeficiency syndrome. N. Eng. J. Med. 1984, 310, 669–673. [Google Scholar] [CrossRef] [PubMed]
- Lucas, G.M.; Lau, B.; Atta, M.G.; Fine, D.M.; Keruly, J.; Moore, R.D. Chronic kidney disease incidence and progression to end-stage renal disease, in HIV-infected individuals: A tale of two races. J. Infect. Dis. 2008, 197, 1548–1557. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, C.M.; Meliambro, K.; Klotman, P.E. Recent progress in HIV-Associated Nephropathy. Annu. Rev. Med. 2012, 633, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Li, H.; Wu, X.; Covarrubias, M.; Scherer, L.; Meinking, K.; Luk, B.; Chomcham, P.; Alluin, J.; Gombart, A.F.; et al. Interplay between HIV-1 infection and host MicroRNAs. Nucleic Acids Res. 2012, 40, 2181–2196. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.R.; Mukerjee, R.; Bagashev, A.; del Valle, L.; Chabrashvili, T.; Hawkins, B.J.; He, J.J.; Sawaya, B.E. HIV-1 Tat protein promotes neuronal dysfunction through disruption of microRNAs. J. Biol. Chem. 2011, 286, 41125–41134. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Nagilla, P.; Le, H.S.; Bunney, C.; Zych, C.; Thalamuthu, A.; Bar-Joseph, Z.; Mathavan, S.; Ayyavoo, V. Comparative expression profile of miRNA and mRNA in primary peripheral blood monoclear cells infected with human immunodeficiency virus (HIV-1). PLoS ONE 2011, 6, e22730. [Google Scholar] [CrossRef] [PubMed]
- Noorbakhsh, F.; Ramachandran, R.; Barsby, N.; Ellestad, K.K.; LeBlanc, A.; Dickie, P.; Baker, G.; Hollenberg, M.D.; Cohen, E.A.; Power, C. MicroRNA profiling reveals new aspects of HIV neurodegeneration: Caspase-6 regulates astrocyte survival. FASEB J. 2010, 4, 1799–1812. [Google Scholar] [CrossRef] [PubMed]
- Freedman, B.I.; Iskanda, R.S.S.; Appel, R.G. The link between hypertension and nephrosclerosis. Am. J. Kidney Dis. 1995, 25, 207–221. [Google Scholar] [CrossRef]
- Marques, F.Z.; Campain, A.E.; Tomaszewski, M.; Zukowska-Szczechowska, E.; Yang, Y.H.; Charchar, F.J.; Morris, B.J. Gene expression profiling reveals renin mRNA overexpression in human hypertensive kidneys and a role for microRNAs. Hypertension 2011, 58, 1093–1098. [Google Scholar] [CrossRef] [PubMed]
- Rollino, C.; Boero, R.; Ferro, M.; Anglesio, A.; Vaudano, G.P.; Cametti, A. Acute pyelonephritis: Analysis of 52 cases. Ren. Fail. 2002, 24, 601–608. [Google Scholar] [CrossRef] [PubMed]
- Oghumu, S.; Bracewell, A.; Nori, U.; Maclean, K.H.; Balada-Lasat, J.M.; Brodsky, S.; Pelletier, R.; Henry, M.; Satoskar, A.R.; Nadasdy, T.; et al. Acute Pyelonephritis in Renal Allografts—A New Role for MicroRNAs? Transplantation 2014, 97, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Pan, W.; Song, X.; Liu, Y.; Shao, X.; Tang, Y.; Liang, D.; He, D.; Wang, H.; Liu, W.; et al. The microRNA miR-23b suppresses IL-17 associated autoimmune inflammation by targeting TAB2, TAB3 and IKK-α. Nat. Med. 2013, 18, 1077–1086. [Google Scholar] [CrossRef] [PubMed]
- Saikumar, J.; Hoffmann, D.; Kim, T.M.; Gonzalez, V.R.; Zhang, Q.; Goering, P.L.; Brown, R.P.; Bijol, V.; Park, P.J.; Waikar, S.S.; et al. Expression, circulation and excretion profile microRNA-21-155 and 18a following acute kidney injury. Toxicol. Sci. 2012, 129, 256–267. [Google Scholar] [CrossRef] [PubMed]
- Pazhayattil, P.G.; Shirali, A.C. Drug-induced impairment of renal function. Int. J. Nephrol. Renovasc. Dis. 2014, 7, 457–468. [Google Scholar] [PubMed]
- Pellegrini, K.L.; Han, T.; Bijol, V.; Saikumar, J.; Cracium, F.L.; Chen, W.W.; Fuscoe, J.C.; Vaida, V.S. MicroRNA-155 deficient mice experience heightened kidney toxicity when dosed with cisplatin. Toxicol. Sci. 2014, 2, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.G.; Kim, J.G.; Kim, H.J.; Kwon, H.K.; Cho, I.J.; dal Choi, W.; Lee, H.W.; Kim, W.D.; Hwang, S.J.; Choi, S.; et al. Discovery of an integrative network of microRNAs and transcriptomics changes for acute kidney injury. Kidney Int. 2014, 86, 943–953. [Google Scholar] [CrossRef] [PubMed]
- Cybulsky, A.V. Growth factor pathways in proliferative glomerulonephritis. Curr. Opin. Nephrol. Hypertens. 2000, 9, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jing, Y.; Hao, J.; Frankfork, N.C.; Zhou, X.; Shen, B.; Liu, X.; Wang, L.; Li, R. MicroRNA-21 in the pathogenesis of acute kidney injury. Protein Cell 2013, 4, 813–819. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, K.; Nakatani, M.; hitachi, K.; Uezumi, A.; Sunada, Y.; Ageta, H.; Inokuchi, K. Activin sinailing as an emergimg target therapeutic intervention. Cell Commun. Signail. 2009. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Putta, S.; Wang, M.; Yuan, H.; Lanting, L.; Nair, I.; Gunn, A.; Nakagawa, Y.; Shimano, H.; Todorov, I.; et al. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat. Cell Biol. 2009, 11, 881–889. [Google Scholar] [CrossRef] [PubMed]
- Chung, A.C.K.; Huang, X.R.; Meng, X.; Lan, H.Y. miR-192medi-ates TGF-β/Smad3-driven renal fibrosis. J. Am. Soc. Nephrol. 2010, 21, 1317–1325. [Google Scholar] [CrossRef] [PubMed]
- López-Hernández, F.J.; López-Novoa, J.M. Role of TGF-β in chronic kidney disease: An integration of tubular, glomerular and vascular effects. Cell Tissue Res. 2012, 347, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.J.; Margue, C.; Behrmann, I.; Kreis, S. MiRNA-29: A microRNA Family with Tumor-Suppressing and Immune-Modulating Properties. Curr. Mol. Med. 2013, 13, 572–585. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.L.; Lee, P.H.; Hsu, Y.C.; Lei, C.C.; Ko, J.Y.; Chuang, P.C.; Huang, Y.T.; Wang, S.Y.; Wu, S.L.; Chen, Y.S.; et al. MicroRNA-29a promotion of nephrin acetylation ameliorates hyperglycemia-indiced podocytes dysfunction. J. Am. Soc. Nephrol. 2014, 25, 1698–1709. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Huang, X.R.; Wei, L.H.; Chung, A.C.; Yu, C.M.; Lan, H.Y. miR-29b as a therapeutic agent for angiotensinII-induced cardiac fibrosis by targeting TGF-beta/Smad3 signaling. Mol. Ther. 2014, 22, 974–985. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Chung, A.C.K.; Huang, X.R.; Meng, X.M.; Hui, D.S.C.; Yu, C.M.; Sung, J.J.; Lan, H.Y. TGF-β/Smad3 Signaling promotes renal fibrosis by inhibiting miR-29. J. Am. Soc. Nephrol. 2011, 22, 1462–1474. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Yong, T.Y.; Michael, M.Z.; Gleadle, J.M. Review: The role of microRNAs in kidney disease. Nephrology 2010, 15, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Peng, H.; Chen, J.; Chen, X.; Han, F.; Xu, X.; He, X.; Yan, N. MicroRNA-21 protects from mesangial cell proliferation induced by diabetic nephropathy in db/db mice. FEBS Lett. 2009, 583, 2009–2014. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Chung, A.C.; Chen, H.Y.; Dong, Y.; Meng, X.M.; Li, R.; Yang, W.; Hou, F.F.; Lan, H.Y. miR-21 is a key therapeutic target for renal injury in a mouse model of type2 diabetes. Diabetologia 2013, 56, 663–674. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Chung, A.C.K.; Chen, H.Y.; Meng, X.M.; Lan, H.Y. Smad3-Mediated upregulation of miR-21promotes renal fibrosis. J. Am. Soc. Nephrol. 2011, 22, 1668–1681. [Google Scholar] [CrossRef] [PubMed]
- Denby, L.; Ramdas, M.W.; McBride, J.; Wang, H.; Robinson, J.; McClure, J.; Crawford, W.; Lu, R.; Hillyard, D.Z.; Khanin, R.; Agami, R.; et al. miR-21 and miR-214 are consistency modulated during renal injury in rodent models. Am. J. Pathol. 2011, 179, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Huang, H.; Lv, X.; Wang, H.; Tang, Z.; Sun, H.; Li, Q.; Ai, J.; Tan, R.; Liu, Y.; et al. PKHD1 post-transcriptionally modulated by miR-365-1 inhibits cell-cell adhesion. Cell Biochem. Funct. 2012, 30, 382–389. [Google Scholar] [CrossRef] [PubMed]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The miR-200 family and miR-205 regulate epithelial to mesenchy-mal transition by targeting ZEB1andSIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Nakada, C.; Matsuura, K.; Tsukamoto, Y.; Tanigawa, M.; Yoshimoto, T.; Narimatsu, T.; Nguyen, L.T.; Hijiya, N.; Uchida, T.; Sato, F.; et al. Genome-wide microRNA expression profiling in renal cell carcinoma: Significant down-regulation of miR-141 and miR-200c. J. Pathol. 2008, 216, 418–427. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, R.L.; Hullinger, T.G.; Semus, H.M.; Dickinson, B.A.; Seto, A.G.; Lynch, J.M.; Stack, C.; Latimer, P.A.; Olson, E.N.; van Rooij, E. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation 2011, 124, 1537–1547. [Google Scholar] [CrossRef] [PubMed]
- Senanayake, U.; Das, S.; Vesely, P.; Alzoughbi, W.; Frohlich, L.F.; Chowdhury, P.; Leuschner, I.; Hoefler, G.; Guertl, B. Mir-192, mir-194, mir-215, mir-200c and mir-141 are downregulated and their common target ACVR2B is strongly expressed in renal child neoplasms. Carcinogenesis 2012, 33, 1014–1021. [Google Scholar] [CrossRef] [PubMed]
- Krützfeldt, J.; Rajewsky, N.; Braich, R.; Rajeev, K.G.; Tuschl, T.; Manoharan, M.; Stoffel, M. Silencing of microRNAs in vivo with antagomirs. Nature 2005, 438, 685–689. [Google Scholar] [CrossRef] [PubMed]
- Esau, C.; Davis, S.; Murray, S.F.; Yu, X.X.; Pandey, S.K.; Pear, M.; Watts, L.; Booten, S.L.; Graham, M.; McKay, R.; et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006, 3, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Elmén, J.; Lindow, M.; Schütz, S.; Lawrence, M.; Petri, A.; Obad, S.; Lind holm, M.; Hedtjärn, M.; Hansen, H.F.; Berger, U. LNA-mediated microRNA silencing in non-human primates. Nature 2008, 452, 896–899. [Google Scholar] [CrossRef] [PubMed]
- Spector, Y.; Fridman, E.; Rosenwald, S.; Zilber, S.; Huang, Y.; Barshack, I.; Zion, O.; Mitchell, H.; Sanden, M.; Meiri, E. Development and validation of microRNAs-based diagnostic assay for classification of renal cell carcinomas. Mol. Oncol. 2013, 7, 732–738. [Google Scholar] [CrossRef] [PubMed]
- Skommer, J.; Rana, I.; Marques, F.Z.; Zhu, W.; Du, Z.; Charchar, F.J. Mall molecules, big effects: The role of microRNAs in regulation of cardiomyocyte death. Cell Death Dis. 2014, 5, e1325. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Kriegel, A.J.; Liu, Y.; Usa, K.; Mladinov, D.; Liu, H.; Fang, Y.; Ding, X.; Liang, M. Delayed ischemic preconditioning contributes to renal protection by upregulation of miR-21. Kidney Int. 2012, 82, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Zarjou, A.; Yang, S.; Abraham, E.; Agarwal, A.; Liu, G. Identification of a microRNA signature in renal fibrosis: Role of miR-21. Am. J. Physiol. Ren. Physiol. 2013, 301, F793–F801. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Shen, J.; Medico, L.; Wang, D.; Ambrosone, C.B.; Liu, S. A pilot study of circulating miRNAs as potential biomarkers of early stage breast cancer. PLoS ONE 2010, 5, e1373. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O'Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Zubakov, D.; Boersma, A.W.; Choi, Y.; van Kuijk, P.F.; Wiemer, E.A.; Kayser, M. MicroRNA markers for forensic body fluid identification obtained from microarray screening and quantitative RT-PCR confirmation. Int. J. Leg. Med. 2010, 124, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Gottardo, F.; Liu, C.G.; Ferracin, M.; Calin, G.A.; Fassan, M.; Bassi, P.; Sevignani, C.; Byrne, D.; Negrini, M.; Pagano, F.; et al. Micro-RNA profiling in kidney and bladder cancers. Urol. Oncol. 2007, 25, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Cortez, M.A.; Calin, G.A. MicroRNA identification in plasma and serum: A new tool to diagnose and monitor diseases. Exp. Opin. Biol. Ther. 2009, 9, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebet, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Benway, C.J.; Bagley, J.; Iacomini, J. MicroRNA-494 Promotes Cyclosporine-Induced Nephrotoxicity and Epithelial to Mesenchymal Transition by Inhibiting PTEN. Am. J. Transplant. 2015, 15, 1682–1691. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mukhadi, S.; Hull, R.; Mbita, Z.; Dlamini, Z. The Role of MicroRNAs in Kidney Disease. Non-Coding RNA 2015, 1, 192-221. https://doi.org/10.3390/ncrna1030192
Mukhadi S, Hull R, Mbita Z, Dlamini Z. The Role of MicroRNAs in Kidney Disease. Non-Coding RNA. 2015; 1(3):192-221. https://doi.org/10.3390/ncrna1030192
Chicago/Turabian StyleMukhadi, Sydwell, Rodney Hull, Zukile Mbita, and Zodwa Dlamini. 2015. "The Role of MicroRNAs in Kidney Disease" Non-Coding RNA 1, no. 3: 192-221. https://doi.org/10.3390/ncrna1030192






