Use of Aleuria alantia Lectin Affinity Chromatography to Enrich Candidate Biomarkers from the Urine of Patients with Bladder Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Cell Lines
2.2. Patient Samples
2.3. Lectin ELISAs
2.4. Lectin Dot Blots
2.5. Lectin Affinity Chromatography
2.6. Filter-Aided Sample Preparation and Tryptic Digestion
2.7. Stable Isotope Labelling
2.8. Peptide Fractionation and LC-MS/MS
2.9. Peptide Identification and Analysis
3. Results
3.1. Lectin Binding to Urothelial Cell Line Lysates
3.2. Lectin Binding Properties of Urinary Proteins
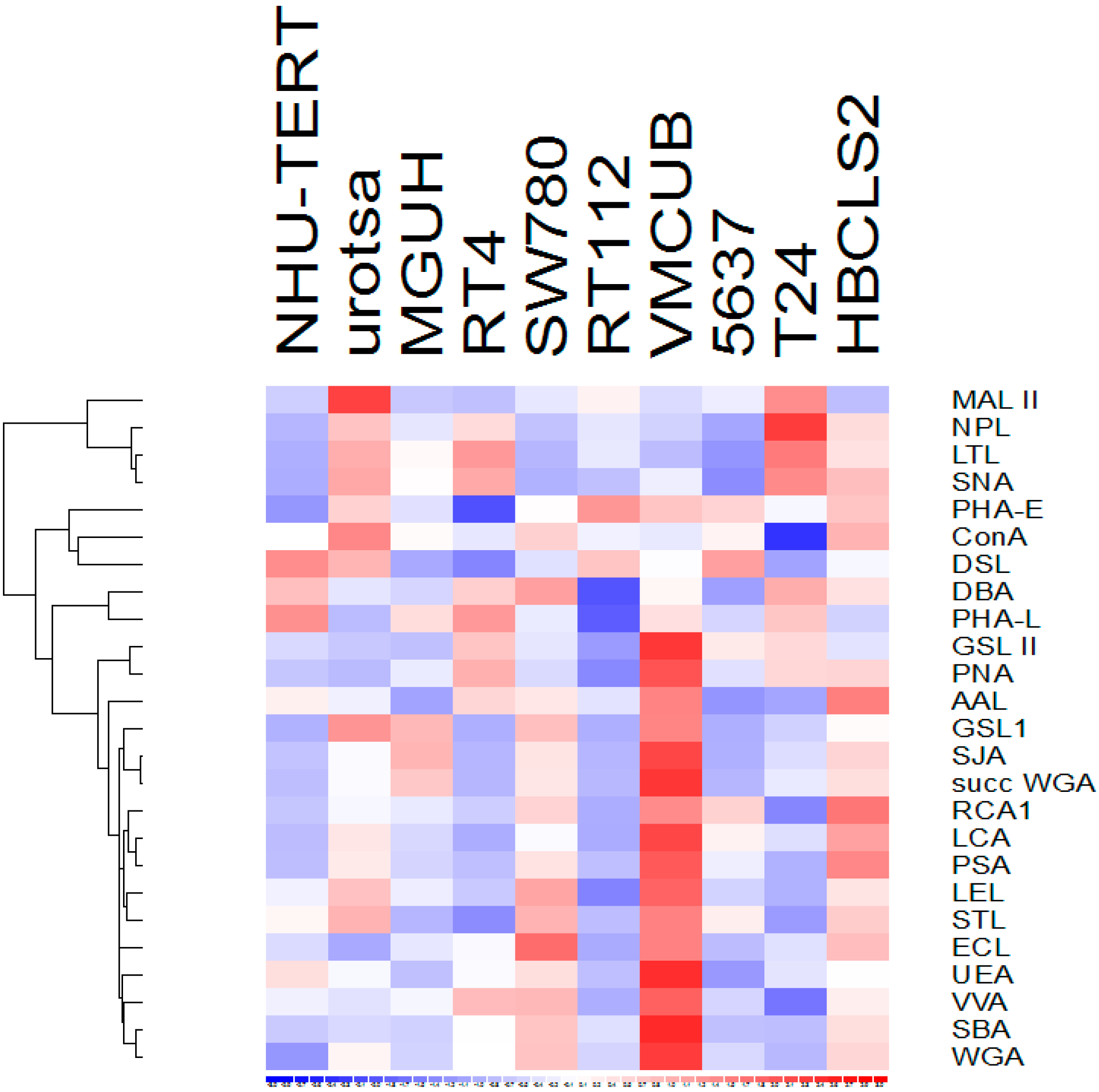
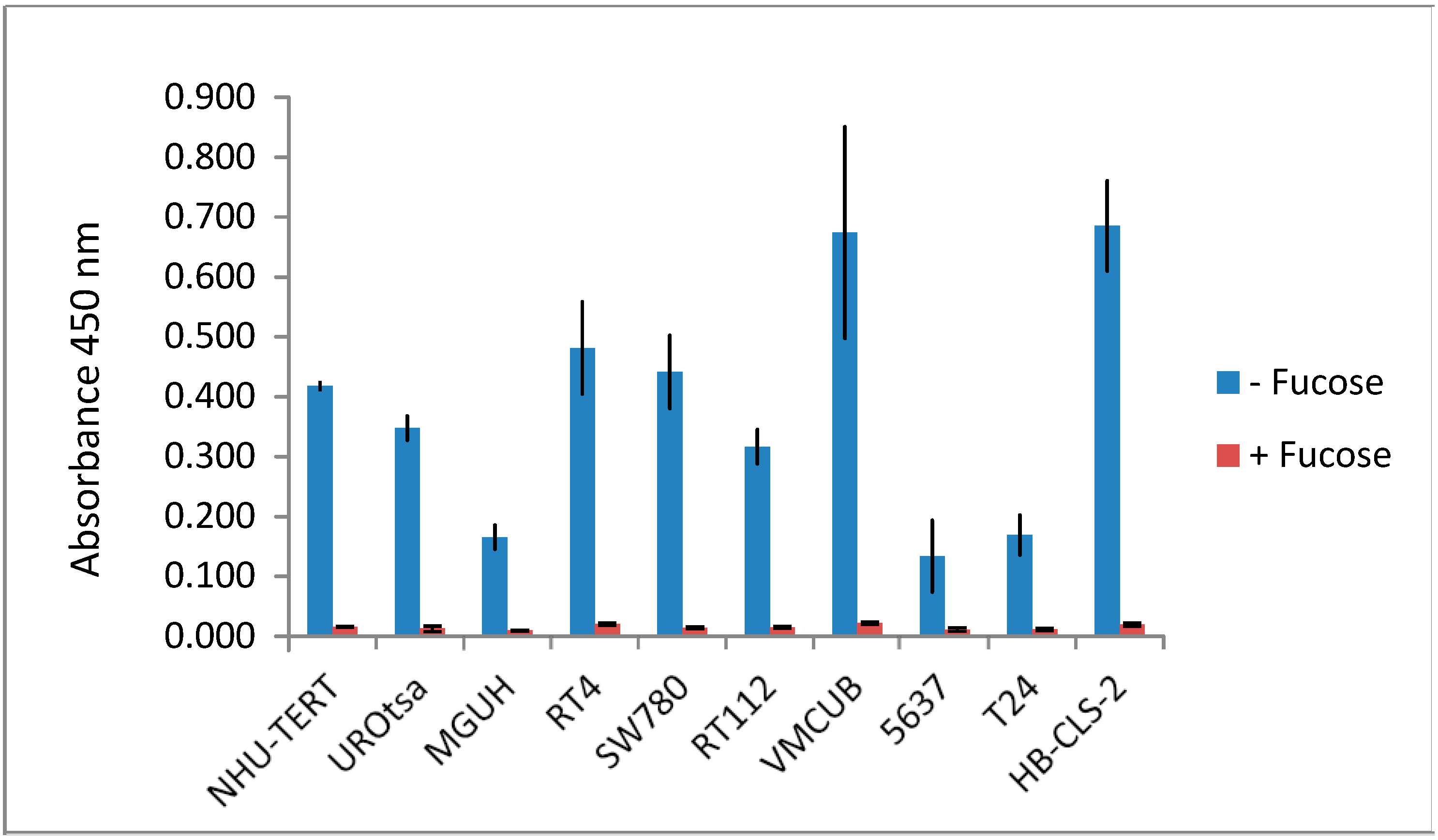
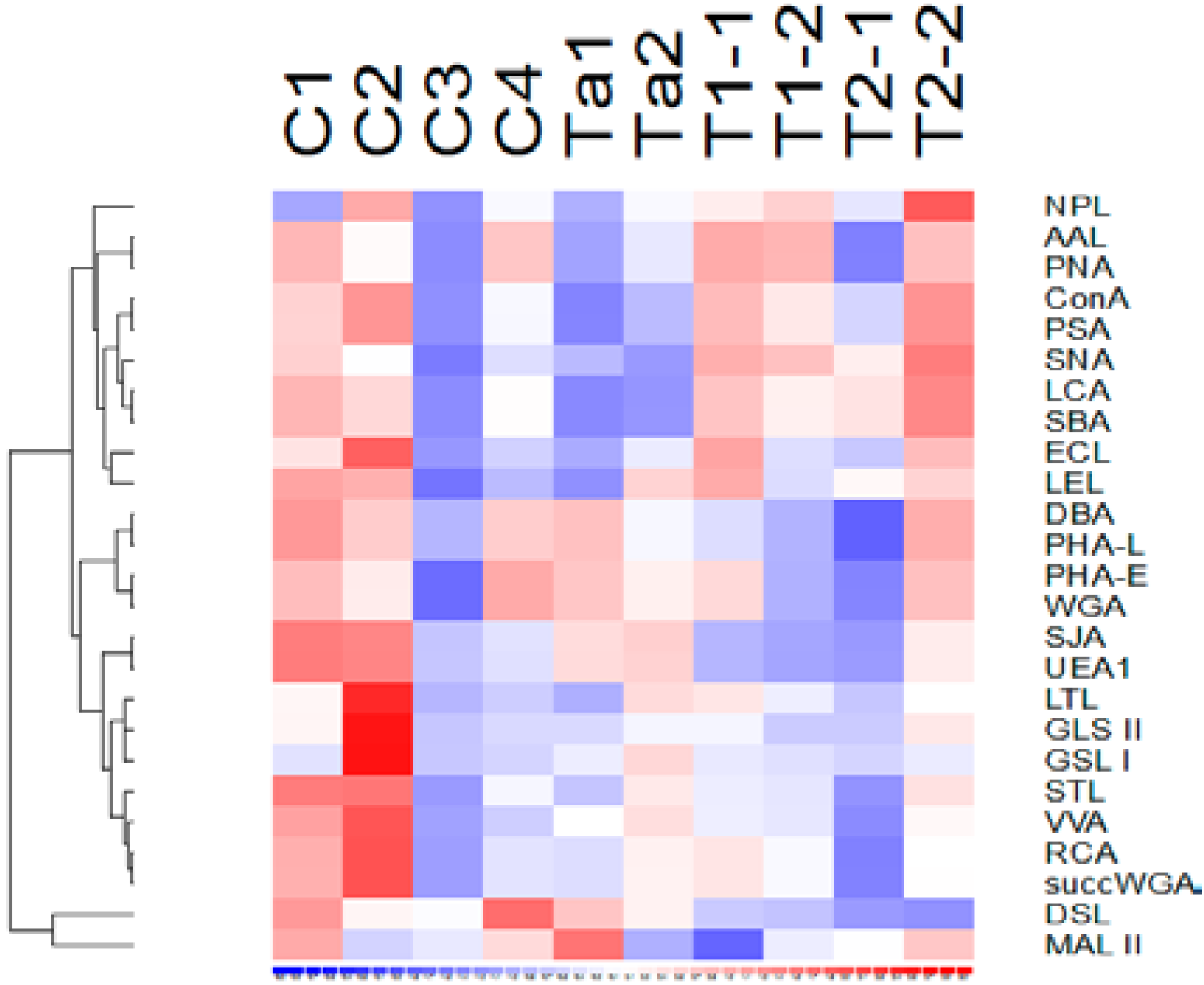
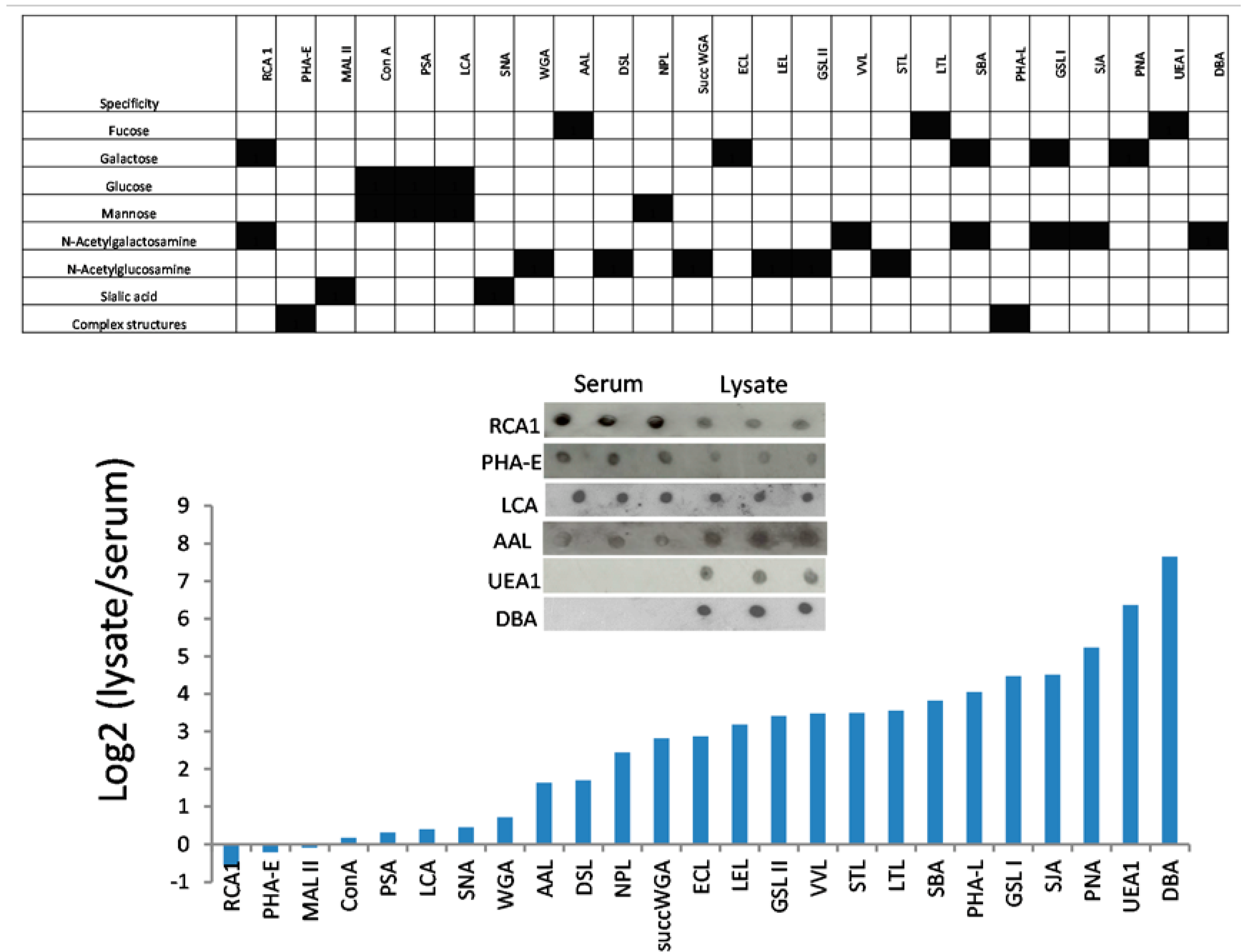
3.3. Lectins with Selectivity for Urothelial Glycans Relative to Plasma Glycans
3.4. Glycoproteome Analysis of Pooled pTa UBC Patient Urine
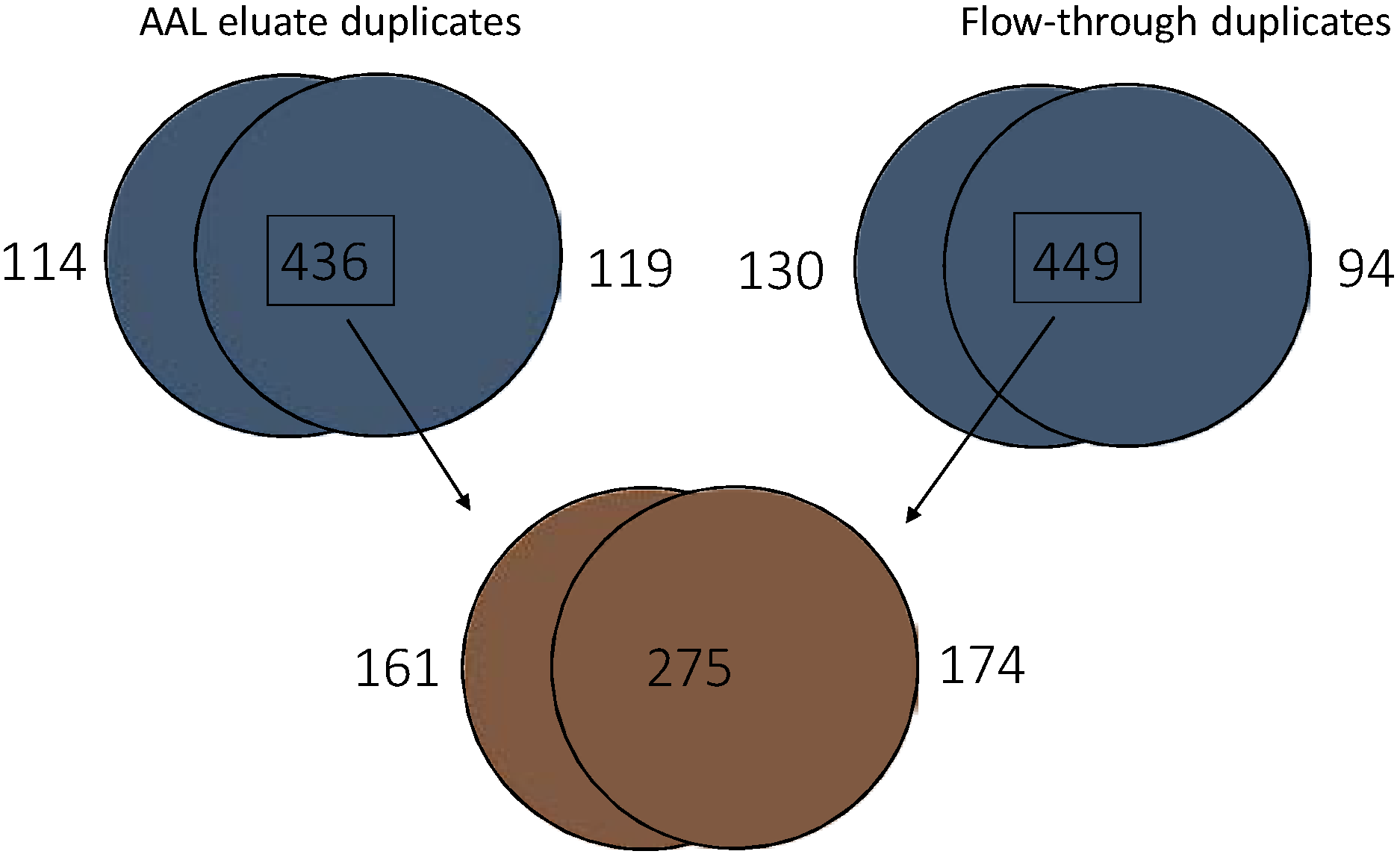
3.5. Quantitative Comparison of AAL Binding Proteins in the Urine of Control Subject and Patients with G1 pTa Bladder Cancer
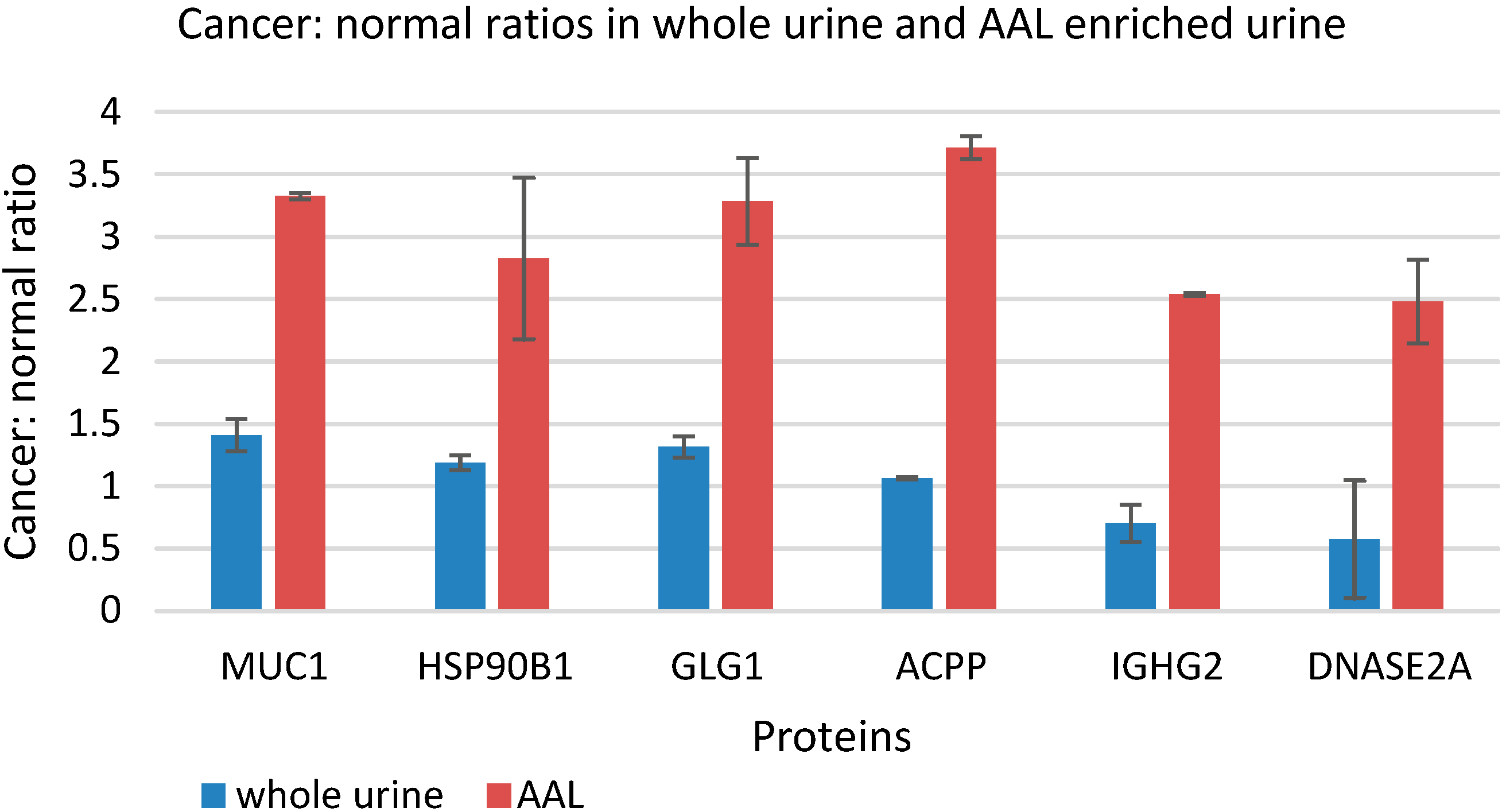
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Knowles, M.; Hurst, C. Molecular biology of bladder cancer: New insights into pathogenesis and clinical diversity. Nat. Rev. Cancer 2015, 15, 25–41. [Google Scholar] [CrossRef] [PubMed]
- Sylvester, R.; van der Meijden, A.; Oosterlinck, W.; Witjes, J.; Bouffioux, C.; Denis, L.; Newling, D.W.; Kurth, K. Predicting recurrence and progression in individual patients with stage Ta T1 bladder cancer using EORTC risk tables: A combined analysis of 2596 patients from seven EORTC trials. Eur. Urol. 2006, 49, 466–477. [Google Scholar] [CrossRef] [PubMed]
- Babjuk, M.; Burger, M.; Zigeuner, R.; Shariat, S.; van Rhijn, B.; Compérat, E.; Sylvester, R.J.; Kaasinen, E.; Böhle, A.; Palou Redorta, J.; et al. EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder: Update 2013. Eur. Urol. 2013, 64, 639–653. [Google Scholar] [CrossRef] [PubMed]
- Schmitz-Dräger, B.; Droller, M.; Lokeshwar, V.; Lotan, Y.; Hudson, M.; van Rhijn, B.; Marberger, M.J.; Fradet, Y.; Hemstreet, G.P.; Malmstrom, P.U.; et al. Molecular markers for bladder cancer screening, early diagnosis, and surveillance: The WHO/ICUD consensus. Urol. Int. 2015, 94, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Miah, S.; Dudziec, E.; Drayton, R.; Zlotta, A.; Morgan, S.; Rosario, D.; Hamdy, F.C.; Catto, J.W. An evaluation of urinary microRNA reveals a high sensitivity for bladder cancer. Br. J. Cancer 2012, 107, 123–128. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, P.; Sharples, K.; Dalphin, M.; Davidson, P.; Gilling, P.; Cambridge, L.; Harvey, J.; Toro, T.; Giles, N.; Luxmanan, C.; et al. A multigene urine test for the detection and stratification of bladder cancer in patients presenting with hematuria. J. Urol. 2012, 188, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Hurst, C.; Platt, F.; Knowles, M. Comprehensive mutation analysis of the TERT promoter in bladder cancer and detection of mutations in voided urine. Eur. Urol. 2014, 65, 367–369. [Google Scholar] [CrossRef] [PubMed]
- Pasikanti, K.; Esuvaranathan, K.; Ho, P.; Mahendran, R.; Kamaraj, R.; Wu, Q.; Chiong, E.; Chan, E.C. Noninvasive urinary metabonomic diagnosis of human bladder cancer. J. Proteome Res. 2010, 9, 2988–2995. [Google Scholar] [CrossRef] [PubMed]
- Wood, S.; Knowles, M.; Thompson, D.; Selby, P.; Banks, R. Proteomic studies of urinary biomarkers for prostate, bladder and kidney cancers. Nat. Rev. Urol. 2013, 10, 206–218. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, H.; Domanski, D.; Smith, D.; Liang, K.; Wu, C.; Chen, C.L.; Chung, T.; Chen, M.C.; Chang, Y.S.; et al. Multiplexed quantification of 63 proteins in human urine by multiple reaction monitoring-based mass spectrometry for discovery of potential bladder cancer biomarkers. J. Proteomics 2012, 75, 3529–3545. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Lin, T.; Tsai, C.; Wu, C.; Chung, T.; Chien, K.; Wu, M.; Chang, Y.S.; Yu, J.S.; Chen, Y.T. Identification of potential bladder cancer markers in urine by abundant-protein depletion coupled with quantitative proteomics. J. Proteomics 2013, 85, 28–43. [Google Scholar] [CrossRef] [PubMed]
- Bryan, R.; Wei, W.; Shimwell, N.; Collins, S.; Hussain, S.; Billingham, L.; Murray, P.G.; Deshmukh, N.; James, N.D.; Wallace, D.M.; et al. Assessment of high-throughput high-resolution MALDI-TOF-MS of urinary peptides for the detection of muscle-invasive bladder cancer. Proteomics Clin. Appl. 2011, 5, 493–503. [Google Scholar] [CrossRef] [PubMed]
- Schiffer, E.; Vlahou, A.; Petrolekas, A.; Stravodimos, K.; Tauber, R.; Geschwend, J.; Neuhaus, J.; Stolzenburg, J.U.; Conaway, M.R.; Mischak, H.; et al. Prediction of muscle-invasive bladder cancer using urinary proteomics. Clin. Cancer Res. 2009, 15, 4935–4943. [Google Scholar] [CrossRef] [PubMed]
- Vlahou, A.; Schellhammer, P.F.; Mendrinos, S.; Patel, K.; Kondylis, F.; Gong, L.; Nasim, S.; Wright, G.L., Jr. Development of a novel proteomic approach for the detection of transitional cell carcinoma of the bladder in urine. Am. J. Pathol. 2001, 158, 1491–1502. [Google Scholar] [CrossRef]
- Kreunin, P.; Zhao, J.; Rosser, C.; Urquidi, V.; Lubman, D.; Goodison, S. Bladder cancer associated glycoprotein signatures revealed by urinary proteomic profiling. J. Proteome Res. 2007, 6, 2631–2639. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Feng, S.; Shedden, K.; Xie, X.; Liu, Y.; Rosser, C.; Lubman, D.M.; Goodison, S. Urinary glycoprotein biomarker discovery for bladder cancer detection using LC/MS-MS and label-free quantification. Clin. Cancer Res. 2011, 17, 3349–3359. [Google Scholar] [CrossRef] [PubMed]
- Ohyama, C. Glycosylation in bladder cancer. Int. J. Clin. Oncol. 2008, 13, 308–313. [Google Scholar] [CrossRef] [PubMed]
- Meany, D.; Chan, D. Aberrant glycosylation associated with enzymes as cancer biomarkers. Clin. Proteomics 2011, 8, 7. [Google Scholar] [CrossRef] [PubMed]
- Dwek, M.; Jenks, A.; Leathem, A. A sensitive assay to measure biomarker glycosylation demonstrates increased fucosylation of prostate specific antigen (PSA) in patients with prostate cancer compared with benign prostatic hyperplasia. Clin. Chim. Acta 2010, 411, 1935–1939. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhu, J.; Yin, H.; Buckanovich, R.; Lubman, D. Analysis of glycan variation on glycoproteins from serum by the reverse lectin-based ELISA assay. J. Proteome Res. 2014, 13, 2197–2204. [Google Scholar] [CrossRef] [PubMed]
- Abbott, K.; Lim, J.; Wells, L.; Benigno, B.; McDonald, J.; Pierce, M. Identification of candidate biomarkers with cancer-specific glycosylation in the tissue and serum of endometrioid ovarian cancer patients by glycoproteomic analysis. Proteomics 2010, 10, 470–481. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Mallory, T.; Satomura, S. AFP-L3: A new generation of tumor marker for hepatocellular carcinoma. Clin. Chem. Acta 2001, 313, 15–19. [Google Scholar] [CrossRef]
- Bryan, R.; Regan, H.; Pirrie, S.; Devall, A.; Cheng, K.; Zeegers, M.; James, N.D.; Knowles, M.A.; Ward, D.G. Protein shedding in urothelial bladder cancer: Prognostic implications of soluble urinary EGFR and EpCAM. Br. J. Cancer 2015, 112, 1052–1058. [Google Scholar] [CrossRef] [PubMed]
- Zeegers, M.; Bryan, R.; Langford, C.; Billingham, L.; Murray, P.; Deshmukh, N.; Hussain, S.; James, N.; Wallace, D.M.; Cheng, K.K. The West Midlands Bladder Cancer Prognosis Programme: Rationale and design. BJU Int. 2010, 105, 784–788. [Google Scholar] [CrossRef] [PubMed]
- Smyth, G. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004, 3, 3. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Tan, Z.; Lu, W.; Guo, J.; Yu, H.; Yu, J.; Sun, C.; Qi, X.; Li, Z.; Guan, F. Quantitative glycome analysis of N-glycan patterns in bladder cancer vs normal bladder cells using an integrated strategy. J. Proteome Res. 2015, 14, 639–653. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, M.; Amano, M.; Kitamura, H.; Tsukamoto, T.; Masumori, N.; Hirose, K.; Ohashi, T.; Nishimura, S.I. N- and O-glycome analysis of serum and urine from bladder cancer patients using a high-throughput glycoblotting method. J. Glycomics Lipidomics 2013, 3. [Google Scholar] [CrossRef]
- Langkilde, N.; Wolf, H.; Orntoft, T. Binding of wheat and peanut lectins to human transitional cell carcinomas. Correlation with histopathologic grade, invasion, and DNA ploidy. Cancer 1989, 15, 849–853. [Google Scholar] [CrossRef]
- Neal, D.; Charlton, R.; Bennett, M. Histochemical study of lectin binding in neoplastic and non-neoplastic urothelium. Br. J. Urol. 1987, 60, 399–401. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Jiang, K.; Zhang, Q.; Guo, K.; Liu, Y. Serum fucosylated paraoxonase 1 as a potential glycobiomarker for clinical diagnosis of early hepatocellular carcinoma using ELISA Index. Glycoconj. J. 2015, 32, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Melnick, J.; Dul, J.; Argon, Y. Sequential interaction of the chaperones BiP and GRP94 with immunoglobulin chains in the endoplasmic reticulum. Nature 1994, 370, 373–375. [Google Scholar] [CrossRef] [PubMed]
- LeRoy, B.; Painter, A.; Sheppard, H.; Popiolek, L.; Samuel-Foo, M.; Andacht, T. Protein expression profiling of normal and neoplastic canine prostate and bladder tissue. Vet. Comp. Oncol. 2007, 5, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Muniyan, S.; Chaturvedi, N.; Dwyer, J.; Lagrange, C.; Chaney, W.; Lin, M. Human prostatic Acid phosphatase: Structure, function and regulation. Int. J. Mol. Sci. 2013, 14, 10438–10464. [Google Scholar] [CrossRef] [PubMed]
- White, K.; Rodemich, L.; Nyalwidhe, J.; Comunale, M.; Clements, M.; Lance, R.; Schellhammer, P.F.; Mehta, A.S.; Semmes, O.J.; Drake, R.R. Glycomic characterization of prostate-specific antigen and prostatic acid phosphatase in prostate cancer and benign disease seminal plasma fluids. J. Proteome Res. 2009, 8, 620–630. [Google Scholar] [CrossRef] [PubMed]
- Heyderman, E.; Brown, B.; Richardson, T. Epithelial markers in prostatic, bladder, and colorectal cancer: An immunoperoxidase study of epithelial membrane antigen, carcinoembryonic antigen, and prostatic acid phosphatase. J. Clin. Pathol. 1984, 37, 1363–1369. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Lam, T.; N’Dow, J. Significance of MUC1 in bladder cancer. BJU Int. 2015, 115, 161–162. [Google Scholar] [CrossRef] [PubMed]
- Simms, M.; Hughes, O.; Limb, M.; Price, M.; Bishop, M. MUC1 mucin as a tumour marker in bladder cancer. BJU Int. 1999, 84, 350–352. [Google Scholar] [CrossRef] [PubMed]
- Stojnev, S.; Ristic-Petrovic, A.; Velickovic, L.; Krstic, M.; Bogdanovic, D.; Khanh do, T.; Ristic, A.; Conic, I.; Stefanovic, V. Prognostic significance of mucin expression in urothelial bladder cancer. Int. J. Clin. Exp. Pathol. 2014, 7, 4945–4958. [Google Scholar] [PubMed]
- Xiang, S.; Zhou, S.; Guan, W.; Hu, Z.; Zhuang, Q.; Ye, Z. Value of quantitative examination of urine MUC1 in bladder transitional cell carcinoma. Di Yi Jun Yi Da Xue Xue Bao 2005, 25, 998–1000. [Google Scholar] [PubMed]
- Miyaoka, Y.; Kato, H.; Ebato, K.; Saito, S.; Miyata, N.; Imamura, T.; Miyajima, A. Retention in the Golgi apparatus and expression on the cell surface of Cfr/Esl-1/Glg-1/MG-160 are regulated by two distinct mechanisms. Biochem. J. 2011, 440, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Antoine, M.; Köhl, R.; Tag, C.; Gressner, A.; Hellerbrand, C.; Kiefer, P. Secreted cysteine-rich FGF receptor derives from posttranslational processing by furin-like prohormone convertases. Biochem. Biophys. Res. Commun. 2009, 382, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Oksvold, P.; Fagerberg, L.; Lundberg, E.; Jonasson, K.; Forsberg, M.; Zwahlen, M.; Kampf, C.; Wester, K.; Hober, S.; et al. Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 2010, 28, 1248–1250. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ambrose, S.R.; Gordon, N.S.; Goldsmith, J.C.; Wei, W.; Zeegers, M.P.; James, N.D.; Knowles, M.A.; Bryan, R.T.; Ward, D.G. Use of Aleuria alantia Lectin Affinity Chromatography to Enrich Candidate Biomarkers from the Urine of Patients with Bladder Cancer. Proteomes 2015, 3, 266-282. https://doi.org/10.3390/proteomes3030266
Ambrose SR, Gordon NS, Goldsmith JC, Wei W, Zeegers MP, James ND, Knowles MA, Bryan RT, Ward DG. Use of Aleuria alantia Lectin Affinity Chromatography to Enrich Candidate Biomarkers from the Urine of Patients with Bladder Cancer. Proteomes. 2015; 3(3):266-282. https://doi.org/10.3390/proteomes3030266
Chicago/Turabian StyleAmbrose, Sarah R., Naheema S. Gordon, James C. Goldsmith, Wenbin Wei, Maurice P. Zeegers, Nicholas D. James, Margaret A. Knowles, Richard T. Bryan, and Douglas G. Ward. 2015. "Use of Aleuria alantia Lectin Affinity Chromatography to Enrich Candidate Biomarkers from the Urine of Patients with Bladder Cancer" Proteomes 3, no. 3: 266-282. https://doi.org/10.3390/proteomes3030266





