Comparative Proteomics of Mouse Tears and Saliva: Evidence from Large Protein Families for Functional Adaptation
Abstract
:1. Introduction
2. Experimental Section
2.1. Tear Fluid Sources and Treatment
2.2. Proteomics Analysis
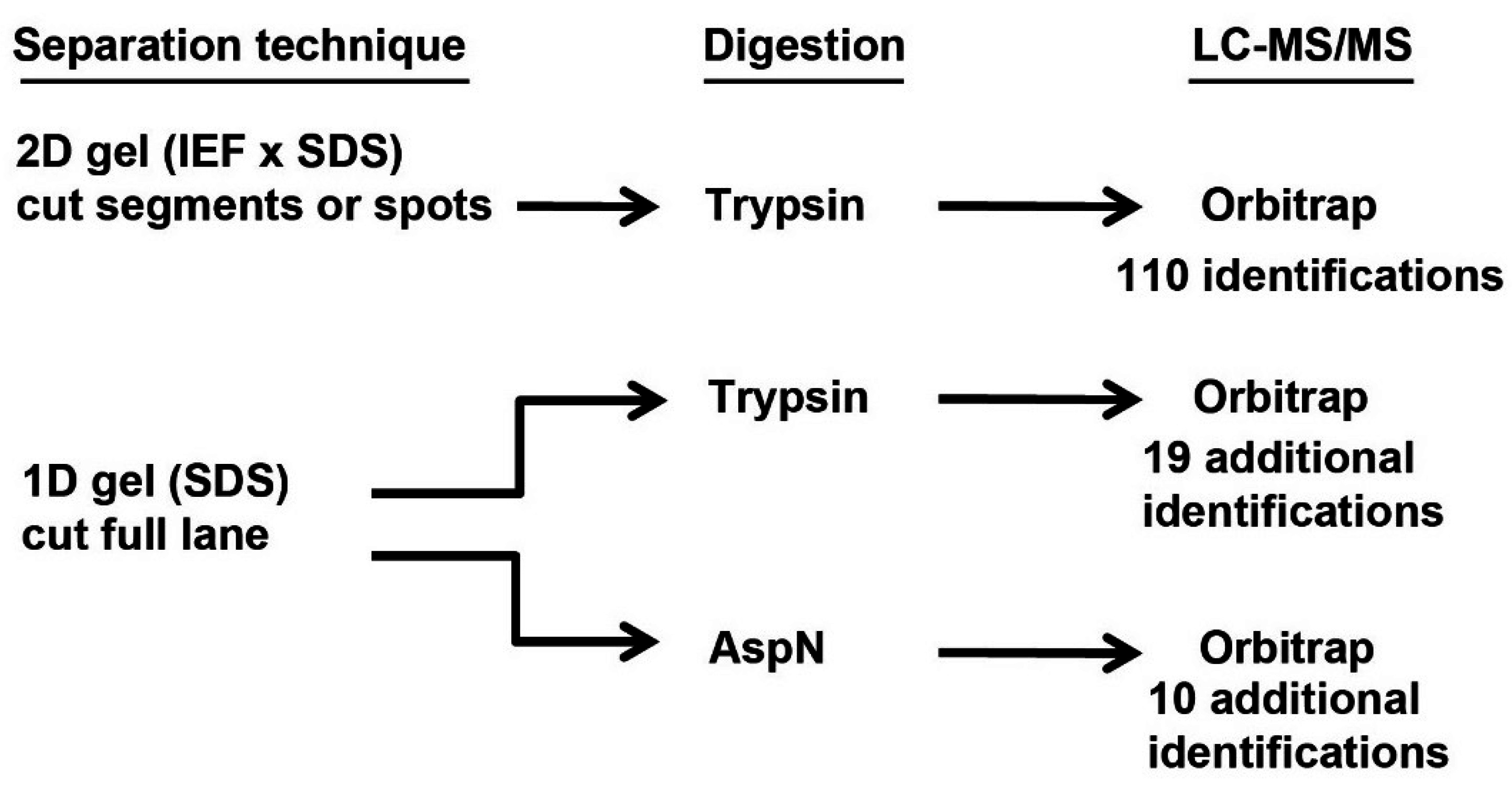
2.3. Protein Identification from Proteomic Data
3. Results and Discussion
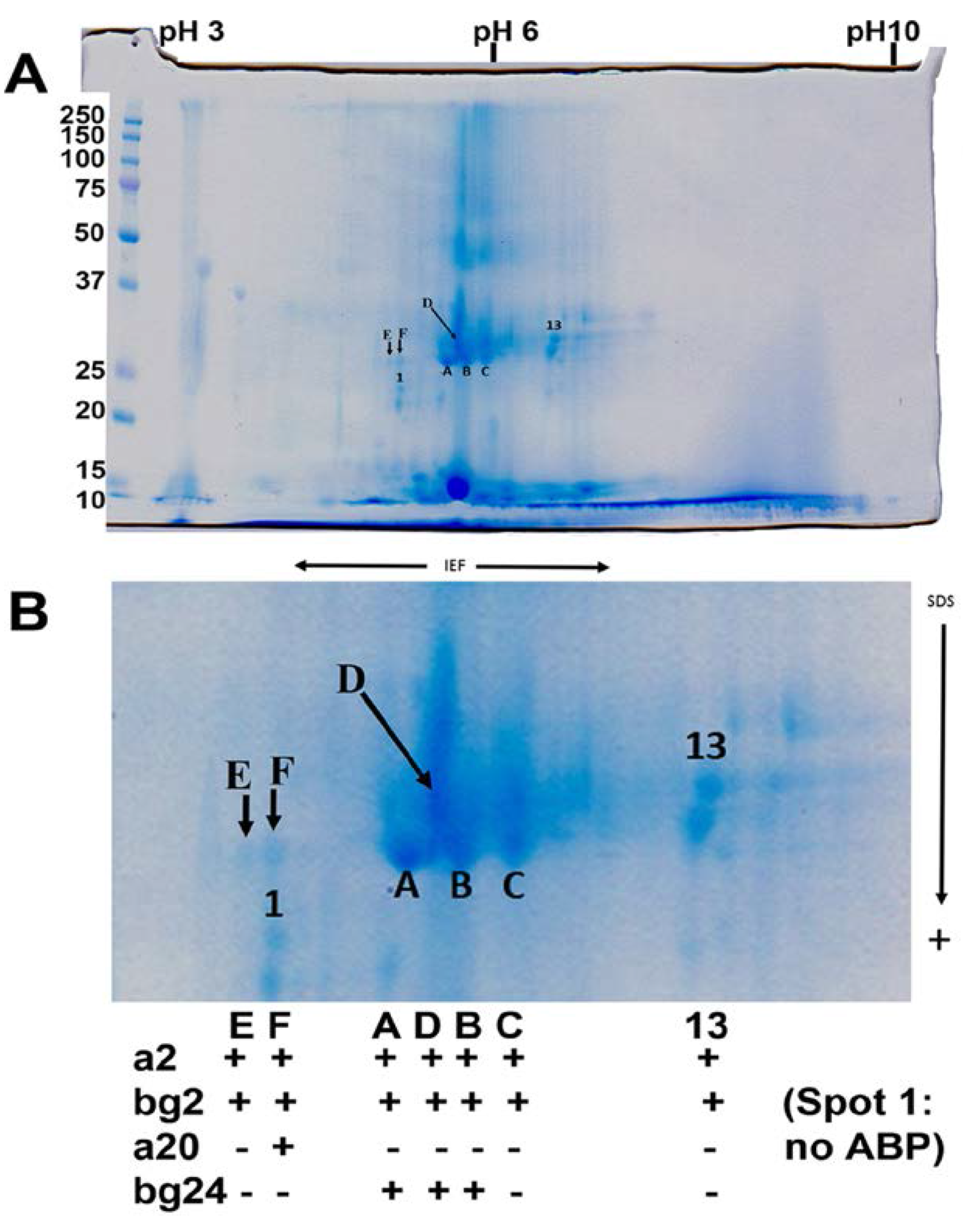
3.1. Protein Identifications in Tears Made under Three Different Conditions
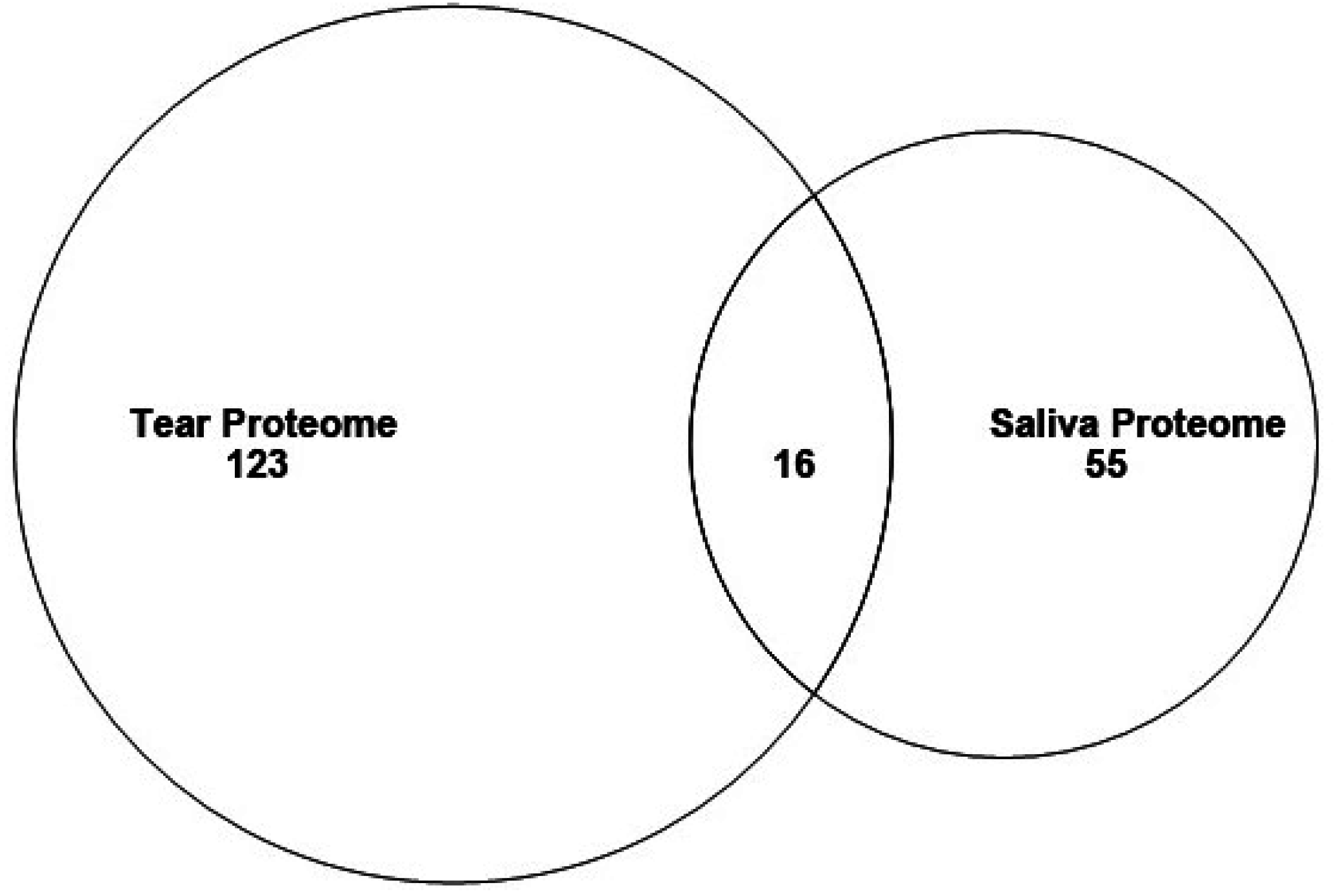
3.2. Proteins in Mouse Tear and Saliva Proteomes
| Protein Name | Uniprot Number |
|---|---|
| 16.5 kDa submandibular gland glycoprotein | Q9DA65 |
| Carbonic anhydrase 6 | P18761 |
| Deoxyribonuclease-1 | P49183 |
| Kallikrein-1 | P15947 |
| Keratin intermediate filament 16a | Q9EQD6 |
| Keratin, type II cytoskeletal 5 | Q922U2 |
| Lactoperoxidase | Q91WA0 |
| Major urinary protein 5 | P11591 |
| Nucleobindin-2 | P81117 |
| Polymeric immunoglobulin receptor | O70570 |
| Prolactin-inducible protein homolog | P02816 |
| Protein LEG1 homolog (Mus musculus) | Q8C6C9 |
| Serum albumin | P07724 |
| Titin isoform N2-A (fragment in saliva) | E9Q8N1 |
| Trypsinogen 7 | Q9CPN9 |
| WAP four-disulfide core domain protein 12 | Q9JHY3 |
3.3. Major Protein Groups in Mouse Tears and in Mouse Saliva
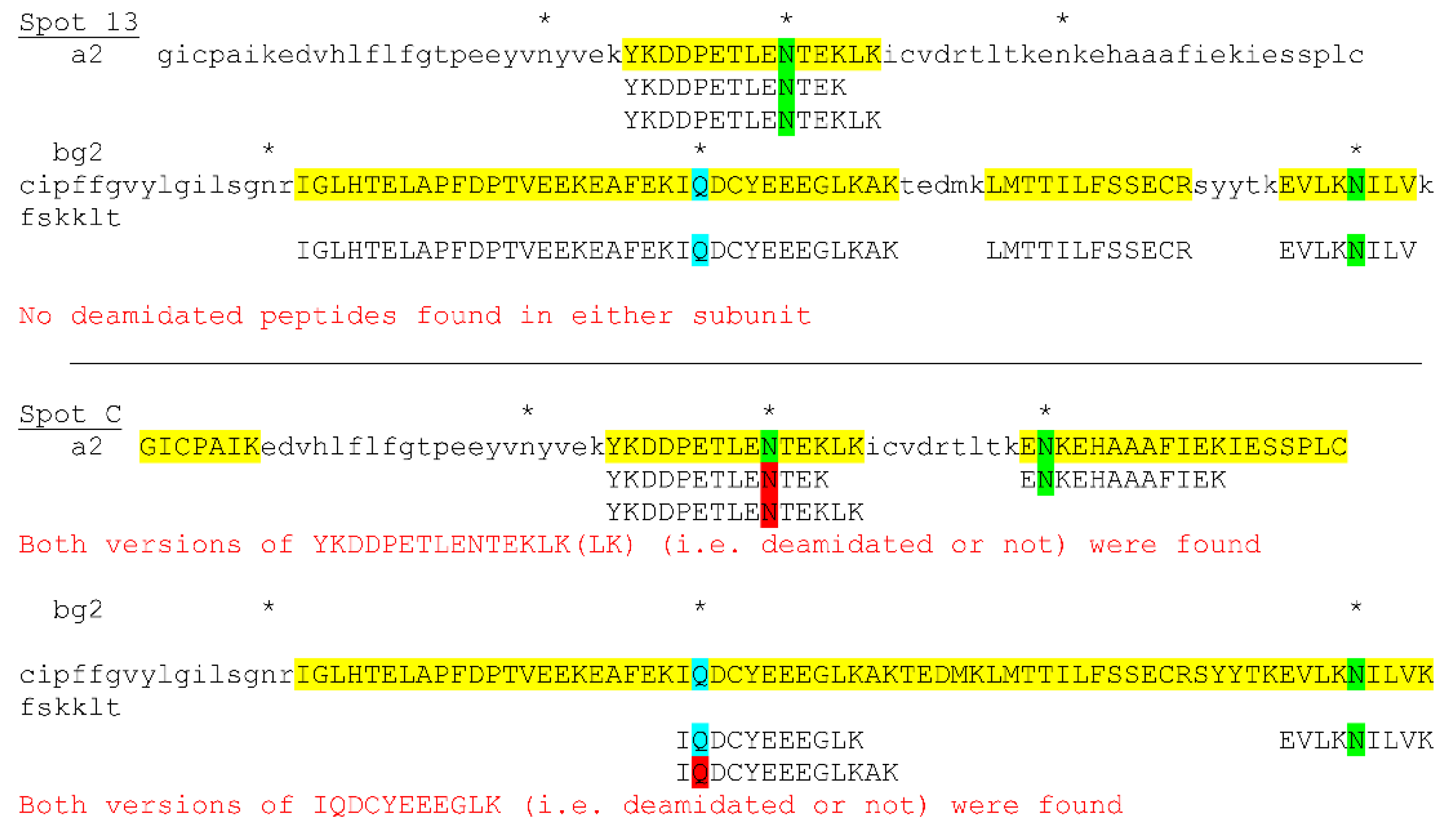
3.4. Posttranslational Modifications of Proteins in Tears
3.5. Comparative Proteomics and Adaptation of Two Exocrine Secretions, Tears and Saliva
4. Conclusions
Supplementary Materials
Supplementary File 1Supplementary File 2Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gonzalez, N.; Iloro, I.; Duran, J.A.; Elortza, F.; Suarez, T. Evaluation of inter-day and inter-individual variability of tear peptide/protein profiles by MALDI-TOF MS analyses. Mol. Vis. 2012, 18, 1572–1582. [Google Scholar] [PubMed]
- Farandos, N.M.; Yetisen, A.K.; Monteiro, M.J.; Lowe, C.R.; Yun, S.H. Contact lens sensors in ocular diagnostics. Adv. Healthc. Mater. 2015, 4, 792–810. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, Y.; Dogru, M.; Tsubota, K. Laboratory findings in tear fluid analysis. Clin. Chim. Acta. 2006, 369, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Beuerman, R.W. Tear analysis in ocular surface diseases. Prog. Retin. Eye Res. 2012, 31, 527–550. [Google Scholar] [CrossRef] [PubMed]
- Tiffany, J.M. The normal tear film. Dev. Ophthalmol. 2008, 41, 1–20. [Google Scholar] [PubMed]
- Moshirfar, M.; Pierson, K.; Hanamaikai, K.; Santiago-Caban, L.; Muthappan, V.; Passi, S.F. Artificial tears potpourri: A literature review. Clin. Ophthalmol. 2014, 8, 1419–1433. [Google Scholar] [PubMed]
- Karnati, R.; Laurie, D.E.; Laurie, G.W. Lacritin and the tear proteome as natural replacement therapy for dry eye. Exp. Eye Res. 2013, 117, 39–52. [Google Scholar] [CrossRef]
- Allen, R.C. Hereditary disorders affecting the lacrimal system. Curr. Opin. Ophthalmol. 2014, 25, 424–431. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Casals, M.; Brito-Zeron, P.; Siso-Almirall, A.; Bosch, X. Primary sjogren syndrome. BMJ 2012, 344, e3821. [Google Scholar] [CrossRef] [PubMed]
- Crow, J.F. C. C. Little, cancer and inbred mice. Genetics 2002, 161, 1357–1361. [Google Scholar] [PubMed]
- Strong, L.C. The establishment of the C3H inbred strain of mice for the study of spontaneous carcinoma of the mammary gland. Genetics 1935, 20, 586–591. [Google Scholar] [CrossRef] [PubMed]
- Lavoie, T.N.; Lee, B.H.; Nguyen, C.Q. Current concepts: Mouse models of sjogren's syndrome. J. Biomed. Biotechnol. 2011, 2011, 549107. [Google Scholar] [CrossRef] [PubMed]
- Karn, R.C.; Chung, A.G.; Laukaitis, C.M. Shared and unique proteins in human, mouse and rat saliva proteomes: Footprints of functional adaptation. Proteomes 2013, 1, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Karn, R.C.; Laukaitis, C.M. Positive selection shaped the convergent evolution of independently expanded kallikrein subfamilies expressed in mouse and rat saliva proteomes. PLoS ONE 2011, 6, e20979. [Google Scholar] [CrossRef] [PubMed]
- Laukaitis, C.M.; Heger, A.; Blakley, T.D.; Munclinger, P.; Ponting, C.P.; Karn, R.C. Rapid bursts of androgen-binding protein (ABP) gene duplication occurred independently in diverse mammals. BMC Evol. Biol. 2008, 8, 46. [Google Scholar] [CrossRef] [PubMed]
- Kimoto, H.; Sato, K.; Nodari, F.; Haga, S.; Holy, T.E.; Touhara, K. Sex- and strain-specific expression and vomeronasal activity of mouse esp family peptides. Curr. Biol. 2007, 17, 1879–1884. [Google Scholar] [CrossRef] [PubMed]
- Logan, D.W.; Marton, T.F.; Stowers, L. Species specificity in major urinary proteins by parallel evolution. PLoS One 2008, 3, e3280. [Google Scholar] [CrossRef] [PubMed]
- Mudge, J.M.; Armstrong, S.D.; McLaren, K.; Beynon, R.J.; Hurst, J.L.; Nicholson, C.; Robertson, D.H.; Wilming, L.G.; Harrow, J.L. Dynamic instability of the major urinary protein gene family revealed by genomic and phenotypic comparisons between c57 and 129 strain mice. Genome Biol. 2008, 9, R91. [Google Scholar] [CrossRef] [PubMed]
- Karn, R.C. Sex-limited effects of the expression of the db gene in mice during puberty. Biochem. Genet. 1981, 19, 355–371. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Nagasaki, T. Lacrimal gland as the major source of mouse tear factors that are cytotoxic to corneal keratocytes. Exp. Eye Res. 2003, 77, 297–304. [Google Scholar] [CrossRef]
- Karn, R.C.; Chung, A.G.; Laukaitis, C.M. Did androgen-binding protein paralogs undergo neo- and/or subfunctionalization as the ABP gene region expanded in the mouse genome? PLoS One 2014, 9, e115454. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Jensen, O.N.; Podtelejnikov, A.V.; Neubauer, G.; Shevchenko, A.; Mortensen, P.; Mann, M. A strategy for identifying gel-separated proteins in sequence databases by ms alone. Biochem. Soc. Trans. 1996, 24, 893–896. [Google Scholar] [CrossRef] [PubMed]
- Rabilloud, T.; Vaezzadeh, A.R.; Potier, N.; Lelong, C.; Leize-Wagner, E.; Chevallet, M. Power and limitations of electrophoretic separations in proteomics strategies. Mass Spectrom. Rev. 2009, 28, 816–843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andon, N.L.; Hollingworth, S.; Koller, A.; Greenland, A.J.; Yates, J.R., III; Haynes, P.A. Proteomic characterization of wheat amyloplasts using identification of proteins by tandem mass spectrometry. Proteomics 2002, 2, 1156–1168. [Google Scholar] [CrossRef]
- Qian, W.J.; Monroe, M.E.; Liu, T.; Jacobs, J.M.; Anderson, G.A.; Shen, Y.; Moore, R.J.; Anderson, D.J.; Zhang, R.; Calvano, S.E.; et al. Quantitative proteome analysis of human plasma following in vivo lipopolysaccharide administration using 16o/18o labeling and the accurate mass and time tag approach. Mol. Cell. Proteom. 2005, 4, 700–709. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.; Nesvizhskii, A.I.; Kolker, E.; Aebersold, R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002, 74, 5383–5392. [Google Scholar] [CrossRef] [PubMed]
- Nesvizhskii, A.I.; Keller, A.; Kolker, E.; Aebersold, R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003, 75, 4646–4658. [Google Scholar] [CrossRef] [PubMed]
- Karn, R.C. Genetic Control of Mammalian Salivary Proteins. In Frontiers of Oral Physiology; Ferguson, D.B., Ed.; Karger: Basel, Switzerland, 1991; Volume 8, pp. 117–140. [Google Scholar]
- Laukaitis, C.; Karn, R.C. Recognition of subspecies status mediated by androgen-binding protein (ABP) in the evolution of incipient reinforcement on the european house mouse hybrid zone. In Evolution of the house mouse; Macholan, M., Baird, S.J.E., Munclinger, P., Piálek, J., Eds.; Cambridge University Press: Cambridge, UK, 2012; Chapter 7. [Google Scholar]
- Kimoto, H.; Haga, S.; Sato, K.; Touhara, K. Sex-specific peptides from exocrine glands stimulate mouse vomeronasal sensory neurons. Nature 2005, 437, 898–901. [Google Scholar] [CrossRef] [PubMed]
- Kimoto, H.; Touhara, K. Induction of c-Fos expression in mouse vomeronasal neurons by sex-specific non-volatile pheromone(s). Chem. Senses. 2005, 30 (Suppl 1), i146–147. [Google Scholar] [CrossRef] [PubMed]
- Hurst, J.L. Female recognition and assessment of males through scent. Behav. Brain Res. 2009, 200, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Karn, R.C.; Laukaitis, C.M. The roles of gene duplication, gene conversion and positive selection in rodent esp and mup pheromone gene families with comparison to the abp family. PLoS One 2012, 7, e47697. [Google Scholar] [CrossRef] [PubMed]
- Karn, R.C. Evolution of rodent pheromones: A review of the abps with comparison to the esps and the mups. Int. J. Biochem. Res. Rev. 2013, 3, 328–363. [Google Scholar] [CrossRef] [PubMed]
- Hurst, L.D.; Smith, N.G. Do essential genes evolve slowly? Curr. Biol. 1999, 9, 747–750. [Google Scholar] [CrossRef]
- Hughes, A.L.; Nei, M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 1988, 335, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Jensen, J.D.; Wong, A.; Aquadro, C.F. Approaches for identifying targets of positive selection. Trends Genet. 2007, 23, 568–577. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Gojobori, T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986, 3, 418–426. [Google Scholar] [PubMed]
- Nielsen, R.; Bustamante, C.; Clark, A.G.; Glanowski, S.; Sackton, T.B.; Hubisz, M.J.; Fledel-Alon, A.; Tanenbaum, D.M.; Civello, D.; White, T.J.; et al. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol 2005, 3, e170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.H.; Bielawski, J.P. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 2000, 15, 496–503. [Google Scholar] [CrossRef]
- Emes, R.D.; Goodstadt, L.; Winter, E.E.; Ponting, C.P. Comparison of the genomes of human and mouse lays the foundation of genome zoology. Hum. Mol. Genet. 2003, 12, 701–709. [Google Scholar] [CrossRef] [PubMed]
- Karn, R.C.; Laukaitis, C.M. The mechanism of expansion and the volatility it created in three pheromone gene clusters in the mouse (Mus musculus) genome. Genome Biol. Evol. 2009, 1, 494–503. [Google Scholar] [CrossRef] [PubMed]
- Laukaitis, C.M.; Critser, E.S.; Karn, R.C. Salivary androgen-binding protein (ABP) mediates sexual isolation in mus musculus. Evolution 1997, 51, 2000–2005. [Google Scholar] [CrossRef]
- Talley, H.M.; Laukaitis, C.M.; Karn, R.C. Female preference for male saliva: Implications for sexual isolation of mus musculus subspecies. Evolution 2001, 55, 631–634. [Google Scholar] [CrossRef]
- Bímová, B.V.; Macholán, M.; Baird, S.E.B.; Munclinger, P.; Laukaitis, C.M.; Karn, R.C.; Luzynski, K.; Tucker, P.; Piálek, J. Reinforcement selection acting on the european house mouse hybrid zone. Mol. Ecol. 2011, 20, 2403–2424. [Google Scholar] [CrossRef] [PubMed]
- Abe, T.; Touhara, K. Structure and function of a peptide pheromone family that stimulate the vomeronasal sensory system in mice. Biochem. Soc. Trans. 2014, 42, 873–877. [Google Scholar] [CrossRef] [PubMed]
- Haga, S.; Hattori, T.; Sato, T.; Sato, K.; Matsuda, S.; Kobayakawa, R.; Sakano, H.; Yoshihara, Y.; Kikusui, T.; Touhara, K. The male mouse pheromone esp1 enhances female sexual receptive behaviour through a specific vomeronasal receptor. Nature 2010, 466, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Chamero, P.; Marton, T.F.; Logan, D.W.; Flanagan, K.; Cruz, J.R.; Saghatelian, A.; Cravatt, B.F.; Stowers, L. Identification of protein pheromones that promote aggressive behaviour. Nature 2007, 450, 899–902. [Google Scholar] [CrossRef] [PubMed]
- Stowers, L.; Holy, T.E.; Meister, M.; Dulac, C.; Koentges, G. Loss of sex discrimination and male-male aggression in mice deficient for TRP2. Science 2002, 295, 1493–1500. [Google Scholar] [CrossRef] [PubMed]
- Clissold, P.M.; Hainey, S.; Bishop, J.O. Messenger rnas coding for mouse major urinary proteins are differentially induced by testosterone. Biochem. Genet. 1984, 22, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.J.; Ghazal, P.; Bingham, R.W.; Barrett, D.; Bishop, J.O. Sequence structures of a mouse major urinary protein gene and pseudogene compared. EMBO J. 1985, 4, 3159–3165. [Google Scholar] [PubMed]
- Mucignat-Caretta, C.; Caretta, A.; Cavaggioni, A. Acceleration of puberty onset in female mice by male urinary proteins. J. Physiol. 1995, 486, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Bímová, B.; Karn, R.C.; Pialek, J. The role of salivary androgen-binding protein in reproductive isolation between two subspecies of house mouse: Mus musculus musculus and mus musculus domesticus. Biol. J. Linn. Soc. 2005, 84, 349–361. [Google Scholar] [CrossRef]
- Karn, R.C.; Laukaitis, C.M. Selection shaped the evolution of mouse androgen-binding protein (ABP) function and promoted the duplication of Abp genes. Biochem. Soc. Trans. 2014, 42, 851–860. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karn, R.C.; Laukaitis, C.M. Comparative Proteomics of Mouse Tears and Saliva: Evidence from Large Protein Families for Functional Adaptation. Proteomes 2015, 3, 283-297. https://doi.org/10.3390/proteomes3030283
Karn RC, Laukaitis CM. Comparative Proteomics of Mouse Tears and Saliva: Evidence from Large Protein Families for Functional Adaptation. Proteomes. 2015; 3(3):283-297. https://doi.org/10.3390/proteomes3030283
Chicago/Turabian StyleKarn, Robert C., and Christina M. Laukaitis. 2015. "Comparative Proteomics of Mouse Tears and Saliva: Evidence from Large Protein Families for Functional Adaptation" Proteomes 3, no. 3: 283-297. https://doi.org/10.3390/proteomes3030283





