2-DE Mapping of the Blue Mussel Gill Proteome: The Usual Suspects Revisited
Abstract
:1. Introduction
2. Experimental Section
2.1. Chemicals
2.2. Animals and Sample Preparation
2.3. Gel Analysis
2.4. Mass Spectrometry and Protein Identification
3. Results and Discussion
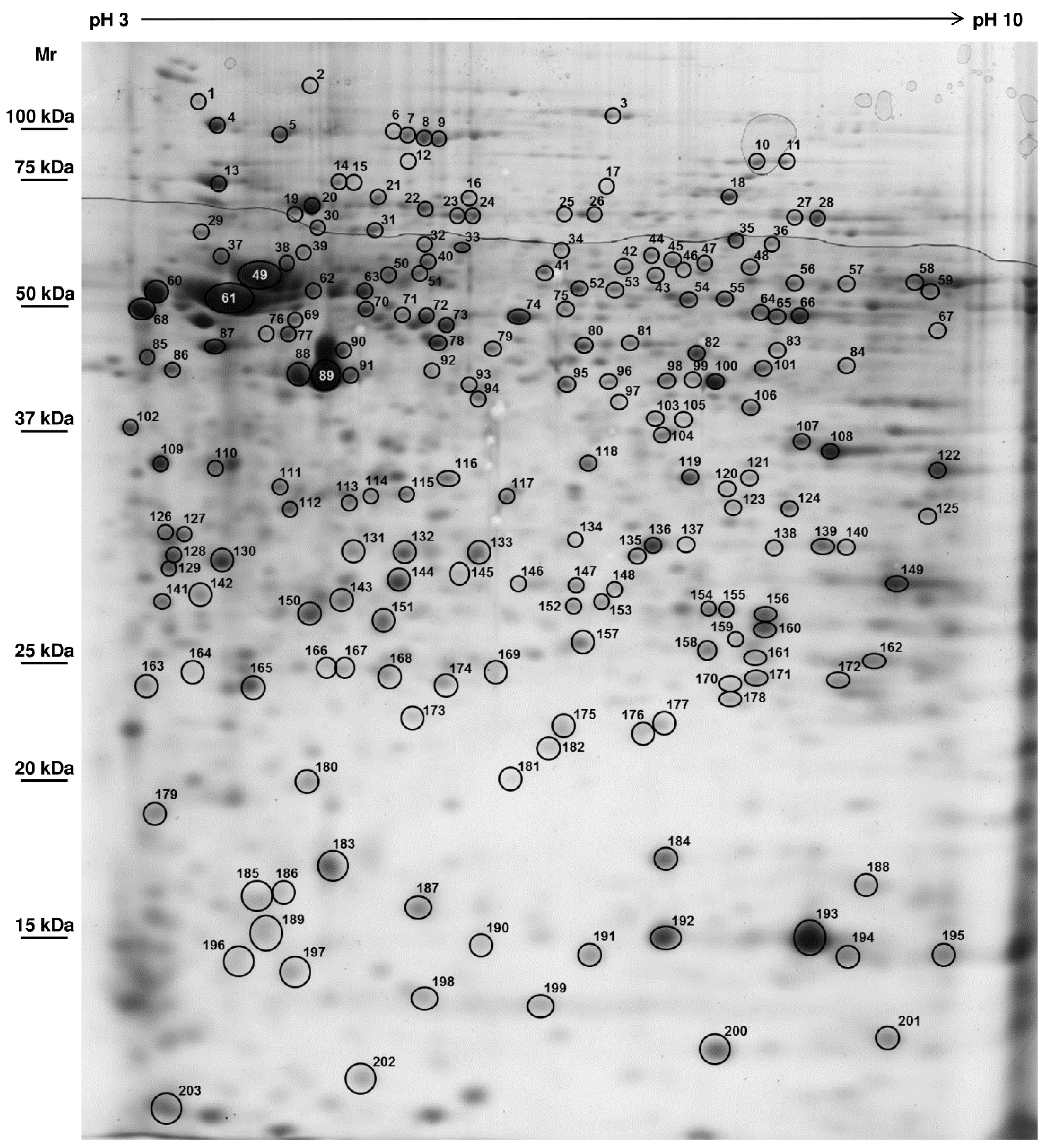
| N° | Name | Mr obs. | pI obs. | species | access number | Mr calc. | pI calc. | score | seq. | cov. | rel. Ab. | SD |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Metabolism; Carbohydrate metabolism; Amino sugar and nucleotide sugar metabolism | ||||||||||||
| 79 | fumarylacetoacetate hydrolase, similar | 46744 | 5.76 | Trichoplax adhaerens | gi|195998011 | 46138 | 5.89 | 85 | 2 | 5 | 0.86 | 0.08 |
| 125 | GDP-L-fucose synthetase | 32148 | 8.21 | Crassostrea gigas | gi|405958300 | 35147 | 6.41 | 76 | 3 | 8 | 1.77 | 0.21 |
| 172 | glucosamine phosphate isomerase | 24602 | 7.51 | Idiogaryops pumilis | gi|262304349 | 19748 | 5.39 | 66 | 2 | 13 | 1.91 | 0.18 |
| 103 | UDP-glucose 4-epimerase | 38721 | 6.31 | Crassostrea gigas | gi|405968861 | 37674 | 6.72 | 113 | 4 | 14 | 0.92 | 0.13 |
| 34 | UDP-N-acetylglucosamine pyrophosphorylase, provisional | 60863 | 5.97 | Capitella teleta | gi|443696999 | 57560 | 6.14 | 99 | 2 | 5 | 0.47 | 0.05 |
| 42 | UDP-N-acetylglucosamine pyrophosphorylase, provisional | 57755 | 6.18 | Capitella teleta | gi|443696999 | 57560 | 6.14 | 98 | 2 | 5 | 0.67 | 0.12 |
| Metabolism; Carbohydrate metabolism; Glycolysis / Gluconeogenesis | ||||||||||||
| 74 | enolase | 49971 | 5.84 | Tomocerus sp. jcrjws1 | gi|8101744 | 41585 | 5.37 | 190 | 4 | 16 | 4.02 | 0.29 |
| 93 | fructose-bisphosphate aldolase | 41755 | 5.69 | Crassostrea gigas | gi|405964948 | 43741 | 5.88 | 131 | 2 | 8 | 0.87 | 0.06 |
| 104 | fructose-bisphosphate aldolase | 37405 | 6.34 | Mytilus edulis | gi|46909221 | 21776 | 5.86 | 186 | 4 | 23 | 0.97 | 0.07 |
| 122 | glyceraldehyde-3-phosphate dehydrogenase | 35098 | 8.37 | Crassostrea gigas | gi|405957058 | 36402 | 6.95 | 195 | 4 | 9 | 4.78 | 0.37 |
| 156 | glyceraldehyde-3-phosphate dehydrogenase A (EC 1.2.1.12) | 27219 | 6.91 | Urticina eques | gi|124264159 | 32082 | 6.51 | 70 | 2 | 8 | 3.96 | 0.18 |
| 37 | NADPH-dependent aldehyde reductase, putative | 60059 | 4.94 | Mytilus galloprovincialis | FL493052 | 29121 | 5.54 | 135 | 3 | 16 | 2.71 | 0.19 |
| 27 | phosphoenolpyruvate carboxykinase | 67990 | 7.12 | Loa loa | gi|312080904 | 72497 | 6.52 | 76 | 4 | 4 | 1.03 | 0.14 |
| 28 | phosphoenolpyruvate carboxykinase | 67990 | 7.22 | Loa loa | gi|312080904 | 72497 | 6.52 | 76 | 4 | 4 | 1.21 | 0.14 |
| 101 | phosphoglycerate kinase | 43890 | 6.85 | Caenorhabditis brenneri | gi|341896690 | 44295 | 6.28 | 291 | 6 | 18 | 2.30 | 0.31 |
| 84 | phosphoglycerate kinase | 43890 | 7.41 | Crassostrea gigas | gi|405963233 | 44217 | 7.59 | 85 | 4 | 13 | 1.18 | 0.11 |
| 163 | phosphoglycerate mutase 1 | 25755 | 4.55 | Pelodictyon phaeoclathratiforme | Q3VP85_9CHLB | 28466 | 5.20 | 72 | 2 | 8 | 2.90 | 0.38 |
| 159 | triosephosphate isomerase | 25844 | 6.75 | Mytilus edulis | gi|46909461 | 16417 | 4.93 | 233 | 5 | 33 | 1.88 | 0.14 |
| 157 | triosephosphate isomerase, partial | 25666 | 6.04 | Mytilus edulis | gi|46909461 | 16417 | 4.93 | 330 | 6 | 31 | 1.74 | 0.07 |
| Metabolism; Carbohydrate metabolism; Citrate cycle (TCA cycle) | ||||||||||||
| 10 | aconitase 2, mitochondrial isoform 2, similar | 83797 | 6.8 | Strongylocentrotus purpuratus | gi|115735566 | 65256 | 4.96 | 105 | 3 | 6 | 0.70 | 0.07 |
| 11 | aconitase 2, mitochondrial isoform 2, similar | 83797 | 6.97 | Strongylocentrotus purpuratus | gi|115936456 | 84808 | 5.49 | 219 | 4 | 7 | 0.64 | 0.09 |
| 82 | citrate synthase, mitochondrial, predicted | 45275 | 6.49 | Strongylocentrotus purpuratus | gi|390339579 | 51662 | 6.09 | 101 | 3 | 7 | 1.67 | 0.17 |
| 54 | dihydrolipoamide dehydrogenase | 52991 | 6.55 | Trichoplax adhaerens | gi|196005079 | 48079 | 6.74 | 105 | 2 | 5 | 1.35 | 0.14 |
| 98 | isocitrate dehydrogenase | 42584 | 6.36 | Crassostrea gigas | gi|48476117 | 51365 | 8.52 | 245 | 5 | 14 | 1.17 | 0.11 |
| 99 | isocitrate dehydrogenase | 42584 | 6.47 | Crassostrea gigas | gi|48476117 | 51365 | 8.52 | 216 | 7 | 17 | 0.52 | 0.04 |
| 100 | isocitrate dehydrogenase | 42584 | 6.60 | Crassostrea gigas | gi|48476117 | 51365 | 8.52 | 445 | 9 | 20 | 2.94 | 0.21 |
| 161 | isocitrate dehydrogenase | 24686 | 6.88 | Mytilus trossulus | gi|385268539 | 50918 | 6.77 | 63 | 2 | 5 | 1.70 | 0.21 |
| 171 | isocitrate dehydrogenase | 25226 | 6.88 | Mytilus trossulus | gi|385268539 | 50918 | 6.77 | 63 | 2 | 5 | 2.21 | 0.18 |
| 136 | malate dehydrogenase, cytosolic | 30138 | 6.33 | Mytilus galloprovincialis | gi|73656337 | 36628 | 6.02 | 222 | 6 | 24 | 2.37 | 0.23 |
| 119 | malate dehydrogenase, mitochondrial | 34527 | 6.48 | Candida albicans | gi|68466091 | 34821 | 5.73 | 68 | 3 | 9 | 1.95 | 0.08 |
| 121 | malate dehydrogenase, mitochondrial | 34527 | 6.80 | Crassostrea gigas | gi|405963427 | 30046 | 8.20 | 64 | 2 | 7 | 0.62 | 0.06 |
| 118 | malate deshydrogenase, cytosolic | 35390 | 6.07 | Mytilus galloprovincialis | gi|73656337 | 36628 | 6.02 | 869 | 16 | 49 | 2.44 | 0.18 |
| 113 | pyruvate dehydrogenase E1 component subunit beta, mitochondrial | 32652 | 5.39 | Ascaris suum | gi|129066 | 39681 | 5.84 | 108 | 3 | 8 | 1.62 | 0.16 |
| Metabolism; Carbohydrate metabolism; Pentose phosphate pathway | ||||||||||||
| 36 | transketolase | 62469 | 6.90 | Strongylocentrotus purpuratus | gi|336455050 | 67029 | 5.96 | 118 | 3 | 5 | 0.61 | 0.08 |
| Metabolism; Energy metabolism; Transferring phosphorus-containing groups | ||||||||||||
| 107 | arginine kinase | 36605 | 7.29 | Macrobiotus occidentalis | gi|308199061 | 40207 | 6.91 | 89 | 2 | 8 | 2.44 | 0.17 |
| 108 | arginine kinase | 36605 | 7.68 | Conus novaehollandiae | gi|301341836 | 39664 | 6.34 | 123 | 2 | 3 | 3.95 | 0.43 |
| Metabolism; Energy metabolism; Oxidative phosphorylation | ||||||||||||
| 64 | ATP synthase alpha subunit mitochondrial | 51140 | 6.84 | Crassostrea gigas | gi|405974703 | 60000 | 8.48 | 505 | 11 | 18 | 1.56 | 0.12 |
| 65 | ATP synthase alpha subunit mitochondrial | 50843 | 6.92 | Litopenaeus vannamei | gi|288816877 | 59416 | 8.97 | 284 | 8 | 12 | 2.50 | 0.35 |
| 66 | ATP synthase alpha subunit mitochondrial | 50549 | 7.11 | Pinctada fucata | gi|116008297 | 59814 | 8.92 | 764 | 14 | 23 | 3.29 | 0.31 |
| 87 | ATP synthase beta subunit | 46535 | 4.90 | Mytilus edulis | gi|46909261 | 46288 | 4.97 | 885 | 16 | 53 | 1.94 | 0.14 |
| 152 | ETF beta-like | 27504 | 6.02 | Nasonia vitripennis | gi|156543370 | 27498 | 7.66 | 236 | 6 | 19 | 1.11 | 0.07 |
| 67 | NADH dehydrogenase (ubiquinone) flavoprotein 1, mitochondrial | 48850 | 8.37 | Crassostrea gigas | gi|405967555 | 51955 | 8.39 | 171 | 5 | 11 | 1.01 | 0.17 |
| 164 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 8, mitochondrial-like | 24770 | 4.83 | Metaseiulus occidentalis | gi|391342248 | 24721 | 5.42 | 62 | 2 | 10 | 0.86 | 0.05 |
| 14 | NADH dehydrogenase subunit, hypothetical protein DAPPUDRAFT_192333 | 75534 | 5.38 | Daphnia pulex | gi|321476647 | 80103 | 6.00 | 162 | 3 | 4 | 0.55 | 0.05 |
| 15 | NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial | 75534 | 5.33 | Crassostrea gigas | gi|405977043 | 81477 | 5.84 | 180 | 5 | 6 | 0.28 | 0.03 |
| 188 | nucleoside diphosphate kinase | 17020 | 7.99 | Ostrea edulis | gi|388571212 | 18860 | 6.82 | 65 | 2 | 12 | 4.98 | 0.89 |
| 22 | succinate dehydrogenase (ubiquinone) flavoprotein subunit | 67990 | 5.59 | Clonorchis sinensis | gi|358254399 | 72276 | 7.09 | 185 | 3 | 5 | 0.97 | 0.10 |
| 83 | succinate-semialdehyde dehydrogenase, mitochondrial | 45998 | 6.92 | Mytilus californianus | GE753097 | 29091 | 8.82 | 84 | 2 | 8 | 1.41 | 0.48 |
| 167 | ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 | 24770 | 5.37 | Mytilus galloprovincialis | FL489022 | 22838 | 9.08 | 283 | 5 | 30 | 1.53 | 0.15 |
| 149 | voltage-dependent anion selective channel protein 2, probable | 28679 | 8.24 | Mytilus californianus | GE752193 | 23286 | 5.38 | 164 | 3 | 17 | 6.18 | 0.61 |
| Metabolism; Lipid metabolism | ||||||||||||
| 154 | enoyl-CoA hydratase, mitochondrial-like | 27314 | 6.57 | Amphimedon queenslandica | gi|340375594 | 31912 | 5.82 | 74 | 2 | 10 | 1.27 | 0.07 |
| 155 | enoyl-CoA hydratase, mitochondrial-like | 27314 | 6.68 | Amphimedon queenslandica | gi|340375594 | 31912 | 5.82 | 100 | 2 | 10 | 1.54 | 0.08 |
| 165 | fatty acid-binding protein, provisional | 24021 | 5.06 | Mytilus galloprovincialis | FL498602 | 21271 | 8.51 | 171 | 4 | 33 | 3.69 | 0.25 |
| 111 | inorganic pyrophosphatase-like | 33434 | 5.17 | Mytilus californianus | ES407080 | 41244 | 8.71 | 152 | 3 | 7 | 1.63 | 0.11 |
| 97 | long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor | 40566 | 6.17 | Homo sapiens | gi|4501857 | 48024 | 7.68 | 90 | 2 | 6 | 0.70 | 0.08 |
| Metabolism; Amino acid metabolism | ||||||||||||
| 3 | glycine dehydrogenase | 100445 | 6.12 | Mytilus galloprovincialis | FL490887 | 29626 | 8.23 | 84 | 2 | 10 | 0.47 | 0.05 |
| 30 | delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial | 63776 | 5.27 | Crassostrea gigas | gi|405978465 | 64148 | 8.35 | 74 | 2 | 3 | 0.54 | 0.03 |
| 57 | amine oxidase, predicted | 54915 | 7.44 | Nematostella vectensis | gi|156382450 | 58581 | 6.54 | 54 | 2 | 4 | 0.76 | 0.09 |
| 60 | procollagen-proline dioxygenase beta subunit | 55950 | 4.62 | Mytilus galloprovincialis | gi|390979785 | 55402 | 4.53 | 449 | 13 | 25 | 7.01 | 0.42 |
| 95 | glutamine synthetase | 42166 | 5.98 | Tegillarca granosa | gi|306489668 | 41952 | 5.63 | 203 | 4 | 12 | 1.43 | 0.08 |
| 106 | cystathionine gamma-lyase | 39809 | 6.80 | Capitella teleta | gi|443685366 | 43775 | 6.14 | 78 | 2 | 4 | 1.23 | 0.08 |
| 123 | 3-hydroxyanthranilate 3,4-dioxygenase | 32399 | 6.70 | Suberites domuncula | gi|18076468 | 32433 | 5.57 | 70 | 2 | 5 | 0.77 | 0.07 |
| Metabolism; Metabolism of other amino-acids | ||||||||||||
| 135 | S-formylglutathione hydrolase | 29472 | 6.25 | Acromyrmex echinatior | gi|332027837 | 18955 | 6.58 | 138 | 2 | 9 | 1.03 | 0.08 |
| Metabolism; Glycan biosynthesis and metabolism | ||||||||||||
| 112 | short chain collagen C4, putative | 32148 | 5.20 | Mytilus galloprovincialis | EH 663252 | 32880 | 8.72 | 373 | 7 | 34 | 1.15 | 0.12 |
| Metabolism; Metabolism of cofactors and vitamins; Ubiquinone and other terpenoid-quinone biosynthesis | ||||||||||||
| 47 | ubiquinone biosynthesis monooxygenase COQ6 | 58506 | 6.53 | Harpegnathos saltator | gi|307192550 | 52851 | 8.79 | 66 | 2 | 2 | 0.68 | 0.07 |
| 193 | ubiquinone biosynthesis monooxygenase COQ6 | 15255 | 7.27 | Harpegnathos saltator | gi|307192550 | 52851 | 8.79 | 59 | 2 | 2 | 4.38 | 1.35 |
| Genetic Information Processing; Transcription | ||||||||||||
| 114 | transcriptional activator protein pur-alpha | 33170 | 5.45 | Crassostrea gigas | gi|405974727 | 27930 | 6.78 | 265 | 6 | 23 | 1.13 | 0.06 |
| 115 | pur-alpha, putative | 33170 | 5.55 | Ixodes scapularis | gi|242046488 | 26667 | 9.41 | 111 | 2 | 11 | 1.56 | 0.11 |
| Genetic Information Processing; Translation | ||||||||||||
| 86 | 40S ribosomal prot SA (p 40) (34/67 kDa laminin receptor) | 43011 | 4.69 | Pinctada fucata | gi|229891605 | 33727 | 5.24 | 185 | 4 | 12 | 1.73 | 0.13 |
| 183 | eIF5A like | 17469 | 5.35 | Mytilus galloprovincialis | AJ516752 | 19880 | 5.23 | 226 | 5 | 38 | 6.48 | 0.79 |
| 51 | elongation factor 1 alpha | 56303 | 5.51 | Mytilus edulis | gi|299474235 | 50827 | 9.12 | 174 | 5 | 14 | 0.56 | 0.07 |
| 71 | elongation factor 1 alpha 1 | 49971 | 5.53 | Saccoglossus kowalevskii | gi|296317283 | 50711 | 9.34 | 134 | 3 | 7 | 1.75 | 0.24 |
| 160 | Hadh2-prov protein isoform 1, similar | 26477 | 6.91 | Strongylocentrotus purpuratus | gi|72006882 | 27479 | 6.32 | 87 | 2 | 10 | 4.13 | 0.15 |
| 25 | phenylalanyl-tRNA synthetase beta chain, probable | 67990 | 5.92 | Mytilus galloprovincialis | FL494288 | 25820 | 5.63 | 138 | 4 | 17 | 0.84 | 0.07 |
| 26 | phenylalanyl-tRNA synthetase beta chain, probable | 67990 | 6.15 | Mytilus galloprovincialis | FL494288 | 25820 | 5.63 | 99 | 2 | 9 | 0.94 | 0.12 |
| 46 | PRP19/PSO4 pre-mRNA processing factor 19 homolog, predicted | 57755 | 6.43 | Saccoglossus kowalevskii | gi|291228334 | 56436 | 6.60 | 78 | 2 | 6 | 0.49 | 0.04 |
| 199 | ribosomal protein rps12 | 13157 | 5.94 | Lineus viridis | gi|166952363 | 13852 | 8.13 | 119 | 5 | 29 | 3.39 | 0.60 |
| 185 | ribosomal protein rps13 | 16142 | 5.11 | Arenicola marina | gi|158187708 | 17169 | 10.59 | 74 | 2 | 17 | 1.74 | 0.16 |
| 198 | ribosomal protein S12 | 13258 | 5.62 | Periplaneta americana | gi|21217441 | 15585 | 5.95 | 106 | 3 | 15 | 4.24 | 0.37 |
| 137 | ribosomal protein S2 | 30138 | 6.48 | Chlamys farreri | gi|22203717 | 27078 | 10.49 | 147 | 5 | 26 | 0.65 | 0.07 |
| Genetic Information Processing; Folding, sorting and degradation; Folding and sorting | ||||||||||||
| 13 | 78kDa glucose regulated protein | 75534 | 4.87 | Crassostrea gigas | gi|46359618 | 73088 | 5.02 | 567 | 11 | 16 | 3.93 | 0.36 |
| 68 | calreticulin, predicted | 50750 | 4.76 | Mytilus galloprovincialis | FL593839 | 27230 | 5.24 | 564 | 12 | 44 | 8.28 | 0.36 |
| 85 | calumenin precursor, putative | 44572 | 4.76 | Pediculus humanus corporis | gi|242005220 | 37885 | 4.61 | 65 | 2 | 3 | 3.60 | 0.20 |
| 38 | chaperonin | 56660 | 5.18 | Paracentrotus lividus | gi|5912574 | 62195 | 5.12 | 203 | 4 | 11 | 3.67 | 0.47 |
| 146 | endoplasmic reticulum protein ERp29 | 28479 | 5.85 | Crassostrea gigas | gi|405975720 | 28444 | 5.19 | 141 | 3 | 8 | 1.09 | 0.13 |
| 4 | glucose-regulated protein 94 | 95115 | 4.88 | Crassostrea gigas | gi|148717303 | 91795 | 4.83 | 384 | 8 | 10 | 1.60 | 0.18 |
| 186 | glucose-regulated protein 94 (fragment) | 16505 | 5.19 | Crassostrea gigas | gi|148717303 | 91795 | 4.83 | 101 | 2 | 3 | 1.25 | 0.08 |
| 20 | heat shock cognate 71 | 68990 | 5.25 | Mytilus galloprovincialis | gi|76780612 | 71508 | 5.29 | 1515 | 28 | 46 | 4.93 | 0.23 |
| 39 | heat shock protein 60 | 60059 | 5.23 | Biomphalaria glabrata | gi|218683627 | 31076 | 5.41 | 400 | 8 | 12 | 1.02 | 0.17 |
| 23 | heat shock protein 70 | 67013 | 5.64 | Mytilus galloprovincialis | gi|62989584 | 68848 | 5.35 | 90 | 3 | 6 | 0.75 | 0.09 |
| 24 | heat shock protein 70 | 67013 | 5.71 | Mytilus galloprovincialis | gi|62989584 | 69848 | 5.35 | 238 | 5 | 8 | 0.82 | 0.12 |
| 12 | heat shock protein 90 | 81772 | 5.54 | Mytilus galloprovincialis | gi|205362524 | 83358 | 4.85 | 179 | 4 | 7 | 0.45 | 0.07 |
| 75 | NFX1-type containing zinc finge, similar | 51140 | 5.99 | Hydra magnipapillata | gi|221116469 | 395486 | 8.08 | 59 | 3 | 0 | 3.20 | 1.15 |
| 143 | prohibitin | 27504 | 5.38 | Trichinella spiralis | gi|339249751 | 60213 | 6.90 | 129 | 4 | 6 | 2.40 | 0.17 |
| 70 | protein disulfide-isomerase, like | 50843 | 5.42 | Mytilus californianus | GE750884 | 30856 | 5.07 | 198 | 4 | 19 | 1.54 | 0.09 |
| 55 | protein disulfide-isomerase, predicted | 52991 | 6.64 | Trichoplax adhaerens | gi|196002337 | 52300 | 8.18 | 76 | 2 | 5 | 1.14 | 0.08 |
| 166 | putative small 22kd heat shock protein | 24770 | 5.35 | Mytilus californianus | ES737901 | 25707 | 5.94 | 80 | 2 | 11 | 1.39 | 0.11 |
| 168 | small 22kd heat shock protein, putative | 24518 | 5.49 | Mytilus californianus | ES737901 | 25707 | 5.94 | 80 | 2 | 11 | 1.98 | 0.13 |
| 131 | small heat shock protein 24.1 | 29692 | 5.4 | Mytilus galloprovincialis | gi|347545633 | 28691 | 5.61 | 163 | 3 | 12 | 1.42 | 0.12 |
| 132 | small heat shock protein 24.1 | 29582 | 5.54 | Mytilus galloprovincialis | gi|347545633 | 28691 | 5.61 | 200 | 4 | 15 | 2.34 | 0.27 |
| 133 | small heat shock protein 24.1 | 29582 | 5.73 | Mytilus galloprovincialis | gi|347545633 | 28691 | 5.61 | 534 | 12 | 48 | 2.57 | 0.20 |
| 144 | Small heat shock protein 24.1 | 28479 | 5.52 | Mytilus galloprovincialis | gi|347545633 | 28691 | 5.61 | 75 | 2 | 9 | 3.71 | 0.34 |
| 145 | small heat shock protein 24.1 | 28881 | 5.64 | Mytilus galloprovincialis | gi|347545633 | 28691 | 5.61 | 84 | 2 | 8 | 2.18 | 0.45 |
| 148 | small heat shock protein 24.1 | 28280 | 6.17 | Mytilus galloprovincialis | gi|347545633 | 28691 | 5.61 | 96 | 2 | 8 | 0.93 | 0.07 |
| 19 | stress-70 protein, mitochondrial, predicted mortaline-like | 67499 | 5.21 | Strongylocentrotus purpuratus | gi|72014569 | 76579 | 5.51 | 264 | 6 | 8 | 1.06 | 0.15 |
| 56 | TCP1 subunit epsilon like, hypothetical protein SINV_10604 | 54915 | 7.29 | Solenopsis invicta | gi|322800807 | 59845 | 5.80 | 172 | 4 | 9 | 0.77 | 0.08 |
| 48 | TCP1 subunit zeta | 57755 | 6.77 | Haliotis discus hannai | gi|379318220 | 58706 | 6.53 | 186 | 4 | 12 | 0.66 | 0.06 |
| 45 | TCP1, hypothetical protein | 58506 | 6.37 | Amblyomma maculatum | gi|346470969 | 59522 | 5.96 | 333 | 8 | 16 | 0.84 | 0.13 |
| 33 | TCP1, subunit beta-like | 61272 | 5.69 | Saccoglossus kowalevskii | gi|291227173 | 150827 | 8.07 | 148 | 4 | 3 | 1.05 | 0.07 |
| 44 | TCP1, subunit gamma isoform 1 | 60059 | 6.29 | Strongylocentrotus purpuratus | gi|115711990 | 60965 | 7.85 | 120 | 4 | 7 | 0.58 | 0.06 |
| 43 | TCP1, subunit eta-like isoform 1 | 55950 | 6.29 | Bombus terrestris | gi|340715736 | 60400 | 6.22 | 193 | 3 | 7 | 0.56 | 0.05 |
| 40 | TCP1, subunit theta | 58128 | 5.59 | Crassostrea gigas | gi|405961548 | 83831 | 5.67 | 175 | 4 | 6 | 0.70 | 0.06 |
| 180 | translationally controlled tumour protein | 20172 | 5.28 | Mytilus californianus | gi|359359687 | 19635 | 4.76 | 71 | 2 | 15 | 1.43 | 0.24 |
| 187 | tubulin-specific chaperone a-like | 16150 | 5.65 | Mytilus californianus | ES738008 | 26274 | 6.17 | 147 | 4 | 19 | 2.87 | 0.22 |
| 5 | valosin-containing protein-like | 93484 | 5.14 | Saccoglossus kowalevskii | gi|291242207 | 90395 | 5.18 | 296 | 6 | 10 | 0.87 | 0.10 |
| Genetic Information Processing; Folding, sorting and degradation; Proteasome | ||||||||||||
| 76 | 26S protease regulatory subunit 6a RPT5 | 48040 | 5.09 | Crassostrea gigas | gi|405957859 | 48206 | 5.08 | 303 | 5 | 14 | 0.86 | 0.17 |
| 77 | 26S protease regulatory subunit 6a RPT5 | 47776 | 5.18 | Aedes aegypti | gi|157129681 | 47953 | 5.20 | 269 | 6 | 17 | 1.71 | 0.17 |
| 96 | 26S proteasome regulatory complex ATPase RPT4 | 42584 | 6.12 | Daphnia pulex | gi|321461635 | 44199 | 6.10 | 291 | 6 | 22 | 1.04 | 0.09 |
| 69 | 26S proteasome regulatory subunit T3 | 49405 | 5.21 | Schistosoma japonicum | gi|226471414 | 46930 | 5.29 | 563 | 13 | 32 | 0.85 | 0.10 |
| 17 | E3 ubiquitin-protein ligase TRIM33 | 75534 | 6.05 | Mytilus galloprovincialis | AJ625521 | 20697 | 7.07 | 174 | 4 | 24 | 0.26 | 0.04 |
| 120 | proteasome 26S subunit, non-ATPase 14-like, predicted | 33836 | 6.69 | Saccoglossus kowalevskii | gi|291239801 | 34852 | 6.07 | 89 | 3 | 9 | 0.82 | 0.09 |
| 142 | proteasome alpha 5 subunit-like | 27889 | 4.88 | Saccoglossus kowalevskii | gi|291243435 | 26525 | 4.74 | 268 | 4 | 22 | 1.24 | 0.07 |
| 169 | proteasome alpha type 2 | 24435 | 5.79 | Haliotis discus discus | gi|126697376 | 26249 | 5.73 | 173 | 3 | 18 | 1.35 | 0.10 |
| 177 | proteasome beta type-6 subunit | 22320 | 6.33 | Mytilus californianus | ES387982 | 30469 | 7.13 | 421 | 9 | 43 | 1.15 | 0.12 |
| 147 | proteasome subunit alpha type-4 | 28280 | 6.04 | Crassostrea gigas | gi|405964515 | 21464 | 5.69 | 70 | 2 | 13 | 1.26 | 0.17 |
| 153 | proteasome subunit alpha type-6 | 27600 | 6.12 | Crassostrea gigas | gi|405975869 | 25429 | 7.57 | 182 | 4 | 18 | 1.13 | 0.10 |
| 176 | ubiquination linked effector, hypothetical protein CRE_31518 | 22248 | 6.26 | Caenorhabditis remanei | gi|308460407 | 37338 | 8.82 | 57 | 1 | 2 | 0.97 | 0.08 |
| Genetic Information Processing; Replication and repair | ||||||||||||
| 197 | histone H2B | 13942 | 5.23 | Mytilus edulis | gi|23304756 | 13781 | 10.69 | 91 | 2 | 19 | 2.35 | 0.22 |
| 202 | histone H4 | 12350 | 5.76 | Diprion pini | gi|1883030 | 11141 | 11.51 | 79 | 3 | 32 | 1.17 | 0.08 |
| 41 | meiosis-specific nuclear structural protein 1-like | 57021 | 5.91 | Saccoglossus kowalevskii | gi|291241736 | 61112 | 5.52 | 107 | 3 | 3 | 0.76 | 0.09 |
| Environmental Information Processing; Signal transduction | ||||||||||||
| 128 | 14-3-3 epsilon protein | 29364 | 4.71 | Bombyx mori | gi|148298752 | 29767 | 4.66 | 267 | 7 | 24 | 1.55 | 0.14 |
| 130 | 14-3-3 epsilon protein | 29364 | 4.95 | Lepeophtheirus salmonis | gi|155966250 | 28466 | 4.67 | 102 | 3 | 8 | 3.44 | 0.27 |
| 129 | 14-3-3 epsilon protein | 28980 | 4.69 | Bombyx mori | gi|148298752 | 29767 | 4.66 | 214 | 7 | 24 | 2.07 | 0.28 |
| 174 | calcyphosin-like protein | 24505 | 5.61 | Mytilus galloprovincialis | FL489968 | 22644 | 7.00 | 141 | 2 | 10 | 1.64 | 0.28 |
| 18 | EF-hand domain-containing protein 1 | 72142 | 6.66 | Crassostrea gigas | gi|405964721 | 74735 | 6.23 | 126 | 4 | 5 | 1.10 | 0.08 |
| Environmental Information Processing; Signaling molecules and interaction | ||||||||||||
| 182 | cyclophilin-type peptidyl-prolyl cis-trans isomerase-15 | 21545 | 5.95 | Mytilus galloprovincialis | FL494508 | 21737 | 5.77 | 305 | 6 | 28 | 1.22 | 0.26 |
| 32 | dedicator of cytokinesis protein 8, partial, predicted | 61686 | 5.57 | Amphimedon queenslandica | gi|340379755 | 236465 | 6.29 | 53 | 2 | 2 | 0.68 | 0.06 |
| 116 | G protein subunit beta-1 | 34110 | 5.65 | Loligo forbesii | gi|121014 | 37983 | 5.76 | 403 | 9 | 34 | 1.12 | 0.08 |
| 162 | GTP-binding nuclear protein Ran, provisional | 25139 | 7.99 | Crassostrea gigas | gi|405971745 | 24274 | 6.96 | 72 | 2 | 10 | 3.01 | 0.32 |
| 195 | peptidyl prolyl cis-trans isomerase A (II) | 14887 | 8.55 | Conus novaehollandiae | gi|289064183 | 17759 | 7.68 | 178 | 4 | 20 | 6.17 | 0.75 |
| 138 | receptor of Activated Kinase C 1 | 30251 | 6.96 | Mya arenaria | gi|115501910 | 35534 | 6.74 | 233 | 7 | 30 | 0.88 | 0.06 |
| 139 | receptor of Activated Kinase C 1 | 30251 | 7.29 | Mya arenaria | gi|115501910 | 35534 | 6.74 | 662 | 15 | 58 | 2.24 | 0.10 |
| 140 | receptor of Activated Kinase C 1 | 30251 | 7.41 | Mya arenaria | gi|115501910 | 35534 | 6.74 | 510 | 12 | 41 | 1.03 | 0.07 |
| 80 | RIB43A-like with coiled-coils protein 2 | 46744 | 6.04 | Crassostrea gigas | gi|405963849 | 45583 | 6.09 | 124 | 4 | 5 | 1.64 | 0.34 |
| 31 | serine/threonine-protein kinase pelle-like | 63776 | 5.44 | Bombus impatiens | gi|350396247 | 58945 | 8.87 | 54 | 1 | 1 | 1.01 | 0.10 |
| 102 | SET protein | 37354 | 4.49 | Crassostrea gigas | gi|405963180 | 28144 | 4.34 | 197 | 4 | 12 | 2.76 | 0.25 |
| 124 | sirtuin-5 | 32399 | 7.05 | Aplysia californica | gi|325197143 | 39468 | 9.03 | 117 | 2 | 6 | 0.82 | 0.07 |
| Cellular Processes; Transport and catabolism | ||||||||||||
| 181 | C1q domain containing protein MgC1q64, putative | 20473 | 5.85 | Mytilus galloprovincialis | gi|325504427 | 24551 | 8.32 | 65 | 2 | 20 | 0.85 | 0.09 |
| 59 | catalase | 53914 | 7.99 | Mytilus californianus | gi|46909299 | 30345 | 6.01 | 235 | 7 | 34 | 1.31 | 0.28 |
| 141 | cathepsin L-like, predicted | 27219 | 4.55 | Strongylocentrotus purpuratus | gi|115715524 | 37335 | 5.14 | 64 | 2 | 3 | 2.23 | 0.23 |
| 16 | dipeptidyl peptidase family member 6 | 72142 | 5.68 | Crassostrea gigas | gi|405969597 | 74497 | 5.66 | 60 | 1 | 1 | 0.46 | 0.03 |
| 134 | dyp-type peroxidase like | 30364 | 6.00 | Trichoplax adhaerens | gi|195996389 | 33144 | 6.21 | 59 | 2 | 6 | 1.03 | 0.11 |
| 173 | glutathione S-transferase sigma 3 | 22964 | 5.56 | Mytilus galloprovincialis | gi|402227995 | 22940 | 5.44 | 121 | 3 | 18 | 0.65 | 0.06 |
| 158 | glutathione S-transferase, Class Beta | 25489 | 6.57 | Mytilus californianus | ES392983 | 38159 | 5.76 | 74 | 2 | 8 | 1.96 | 0.15 |
| 110 | heavy metal-binding protein HIP | 34810 | 4.92 | Mytilus edulis | gi|46395578 | 24388 | 5.09 | 165 | 6 | 45 | 2.43 | 0.38 |
| 105 | kin 17-mid super family, hypothetical protein AND_04962 | 38721 | 6.53 | Anopheles darlingi | gi|312382372 | 48048 | 9.44 | 55 | 2 | 5 | 0.57 | 0.04 |
| 58 | leucine aminopeptidase, predictive | 54915 | 8.03 | Mytilus californianus | ES400183 | 36649 | 7.01 | 332 | 8 | 40 | 2.32 | 0.25 |
| 6 | major vault protein | 91110 | 5.48 | Mytilus edulis | gi|5714749 | 31855 | 5.45 | 343 | 9 | 46 | 0.50 | 0.07 |
| 8 | major vault protein | 91892 | 5.55 | Mytilus edulis | gi|5714749 | 31855 | 5.45 | 718 | 16 | 56 | 1.27 | 0.12 |
| 7 | major vault protein | 91892 | 5.53 | Crassostrea gigas | gi|405974681 | 96651 | 5.58 | 73 | 2 | 2 | 0.79 | 0.08 |
| 9 | major vault protein | 90338 | 5.61 | Mytilus edulis | gi|5714749 | 31855 | 5.45 | 276 | 8 | 35 | 1.07 | 0.10 |
| 170 | peroxiredoxin | 24186 | 6.7 | Pinctada fucata | gi|306451460 | 22530 | 7.63 | 99 | 2 | 9 | 1.81 | 0.23 |
| 184 | peroxiredoxin V | 17924 | 6.38 | Chlamys farreri | gi|149688674 | 20431 | 8.20 | 69 | 1 | 5 | 4.55 | 0.24 |
| 81 | Rab GDP dissociation inhibitor alpha | 45998 | 6.21 | Schistosoma japonicum | gi|226484726 | 50623 | 6.41 | 60 | 2 | 4 | 1.32 | 0.51 |
| 190 | superoxide dismutase | 14887 | 5.77 | Mytilus chilensis | gi|332356353 | 15925 | 5.84 | 173 | 4 | 30 | 2.23 | 0.18 |
| 191 | superoxide dismutase (Cu/Zn-SOD) | 14673 | 6.11 | Mytilus edulis | gi|34481600 | 16046 | 5.84 | 289 | 4 | 31 | 3.40 | 0.22 |
| 175 | superoxide dismutase, mitochondrial (Mn-SOD) | 22327 | 6.00 | Mytilus galloprovincialis | gi|402122769 | 25412 | 6.44 | 124 | 2 | 9 | 1.67 | 0.15 |
| 203 | thioredoxin 1 | 12520 | 4.69 | Mytilus galloprovincialis | gi|391358072 | 11667 | 4.47 | 244 | 4 | 33 | 7.72 | 0.72 |
| 178 | thioredoxin peroxidase | 23774 | 6.70 | Cristaria plicata | gi|306451460 | 22143 | 5.95 | 75 | 2 | 10 | 1.63 | 0.16 |
| 21 | V-type proton ATPase catalytic subunit A | 72142 | 5.45 | Crassostrea gigas | gi|405950221 | 71148 | 5.21 | 314 | 7 | 11 | 0.69 | 0.06 |
| Cellular Processes; Cell motility; Cytoskeleton proteins | ||||||||||||
| 92 | actin | 43011 | 5.60 | Mytilus sp. | gi|120564812 | 35392 | 5.26 | 93 | 3 | 14 | 0.90 | 0.10 |
| 189 | actin | 15106 | 5.15 | Schistosoma japonicum | gi|257215973 | 10215 | 5.40 | 273 | 6 | 55 | 3.99 | 0.45 |
| 196 | actin | 14330 | 5.05 | Hydroides elegans | gi|73532714 | 41520 | 5.39 | 357 | 9 | 19 | 2.90 | 0.28 |
| 88 | actin 2 = cytoplasmic actin = beta actin | 42200 | 5.22 | Crassostrea gigas | gi|18565104 | 42002 | 5.30 | 669 | 15 | 47 | 5.83 | 0.44 |
| 89 | actin 2 = cytoplasmic actin = beta actin | 42200 | 5.31 | Aedes aegypti | gi|67782283 | 42194 | 5.30 | 648 | 14 | 49 | 15.31 | 0.97 |
| 91 | actin 2 = cytoplasmic actin = beta actin | 42200 | 5.4 | Mytilus sp. | gi|120564812 | 35392 | 5.26 | 454 | 11 | 51 | 1.86 | 0.13 |
| 151 | actin 2 = cytoplasmic actin = beta actin | 26568 | 5.49 | Crassostrea gigas | gi|18565104 | 42002 | 5.30 | 444 | 11 | 32 | 3.25 | 0.23 |
| 94 | actin 5 | 40566 | 5.73 | Aedes aegypti | gi|67782283 | 42194 | 5.3 | 404 | 10 | 35 | 1.06 | 0.10 |
| 150 | actin-87E isoform 1, similar | 26845 | 5.28 | Tribolium castaneum | gi|91078486 | 42158 | 5.29 | 419 | 10 | 36 | 5.20 | 0.59 |
| 2 | catchin protein | 113783 | 5.32 | Mytilus galloprovincialis | gi|6682323 | 112777 | 5.22 | 701 | 16 | 21 | 0.40 | 0.02 |
| 179 | centrin-3 | 19785 | 4.66 | Crassostrea gigas | gi|405964350 | 20761 | 4.58 | 139 | 4 | 22 | 2.93 | 0.21 |
| 192 | destrin, partial | 15330 | 6.38 | Macaca mulatta | gi|73696362 | 12274 | 8.64 | 64 | 2 | 7 | 7.79 | 0.54 |
| 52 | fascin | 53914 | 6.01 | Crassostrea gigas | gi|405961655 | 56081 | 6.21 | 99 | 3 | 5 | 1.29 | 0.08 |
| 53 | fascin-like domain protein | 53914 | 6.15 | Tetraodon nigroviridis | gi|47209051 | 106026 | 8.68 | 85 | 2 | 2 | 0.74 | 0.08 |
| 78 | gelsolin | 46245 | 5.61 | Suberites domuncula | gi|27528508 | 42414 | 5.23 | 115 | 2 | 7 | 2.17 | 0.09 |
| 194 | hypothetical protein KGM_09271 with pleckstrin homology-like domain | 14603 | 7.58 | Danaus plexippus | gi|357623784 | 110881 | 9.64 | 72 | 2 | 2 | 4.85 | 0.72 |
| 29 | Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | 63335 | 4.83 | Mytilus galloprovincialis | FL501152 | 22127 | 4.93 | 347 | 6 | 42 | 1.39 | 0.37 |
| 62 | non-neuronal cytoplasmic intermediate filament protein | 56303 | 5.27 | Mytilus californianus | GE750313 | 31541 | 7.63 | 410 | 9 | 29 | 2.03 | 0.27 |
| 200 | profilin like | 12468 | 6.68 | Mytilus galloprovincialis | FL496207 | 20580 | 8.33 | 243 | 6 | 37 | 5.42 | 0.37 |
| 117 | radial spoke head protein 9, like | 32909 | 5.82 | Crassostrea gigas | gi|405959092 | 31220 | 5.20 | 118 | 3 | 8 | 1.21 | 0.22 |
| 1 | spectrin alpha chain | 105775 | 4.83 | Crassostrea gigas | gi|405973516 | 287684 | 4.88 | 143 | 5 | 2 | 0.44 | 0.04 |
| 90 | tektin 1 | 45755 | 5.36 | Crassostrea gigas | gi|405975636 | 48654 | 6.12 | 55 | 2 | 3 | 1.86 | 0.12 |
| 72 | tektin-2 | 49971 | 5.58 | Crassostrea gigas | gi|405950079 | 48059 | 5.71 | 172 | 6 | 18 | 2.60 | 0.14 |
| 73 | tektin-4 | 48307 | 5.64 | Crassostrea gigas | gi|405967050 | 52952 | 5.53 | 172 | 7 | 12 | 2.72 | 0.18 |
| 109 | tropomyosin | 35098 | 4.65 | Mytilus galloprovincialis | gi|6647862 | 32807 | 4.62 | 559 | 12 | 36 | 5.25 | 0.23 |
| 126 | tropomyosin | 30478 | 4.69 | Mytilus edulis | gi|6647862 | 32836 | 4.64 | 312 | 6 | 12 | 1.81 | 0.13 |
| 127 | tropomyosin | 30593 | 4.77 | Mytilus galloprovincialis | gi|6647862 | 32807 | 4.62 | 190 | 4 | 8 | 2.17 | 0.08 |
| 49 | tubulin alpha-1 chain | 54915 | 5.09 | Schistosoma mansoni | gi|256087763 | 50660 | 4.97 | 780 | 18 | 47 | 11.21 | 0.75 |
| 61 | tubulin beta chain | 51744 | 4.93 | Crassostrea gigas | gi|56603670 | 50371 | 4.79 | 705 | 15 | 37 | 31.68 | 1.30 |
| 63 | tubulin, beta 2C-like, predicted | 56303 | 5.40 | Saccoglossus kowalevskii | gi|291243365 | 50516 | 4.74 | 266 | 6 | 16 | 3.06 | 0.32 |
| Unknown function | ||||||||||||
| 35 | CCDC 151 like, coiled-coil domain containing 151 | 62469 | 6.68 | Crassostrea gigas | gi|405957528 | 63895 | 6.65 | 68 | 2 | 2 | 1.21 | 0.12 |
| 50 | selenium-binding protein 1, partial | 57755 | 5.48 | Crassostrea gigas | gi|405971621 | 54060 | 6.11 | 56 | 2 | 2 | 0.67 | 0.08 |
| 201 | hypothetical protein AND_08398 | 12519 | 7.95 | Anopheles darlingi | gi|312379666 | 38819 | 8.84 | 53 | 2 | 0 | 4.58 | 0.55 |
| N° | Proteoforme | cqv | ||
|---|---|---|---|---|
| 99 | isocitrate dehydrogenase | 5.9 | <10% | |
| 165 | fatty acid-binding protein, provisional | 6.7 | ||
| 72 | tektin-2 | 6.8 | ||
| 192 | destrin, partial | 7.4 | ||
| 78 | gelsolin | 7.7 | ||
| 73 | tektin-4 | 7.8 | ||
| 157 | triosephosphate isomerase, partial | 8.0 | ||
| 139 | receptor of Activated Kinase C 1 | 8.2 | ||
| 160 | Hadh2-prov protein isoform 1, similar | 8.3 | ||
| 23 | heat shock protein 70 | 8.4 | ||
| 60 | procollagen-proline dioxygenase beta subunit | 8.4 | ||
| 61 | tubulin beta chain | 8.9 | ||
| 109 | tropomyosin | 9.0 | ||
| 175 | superoxide dismutase, mitochondrial (Mn-SOD) | 9.1 | ||
| 68 | calreticulin, predicted | 9.1 | ||
| 119 | malate dehydrogenase, mitochondrial | 9.2 | ||
| 74 | enolase | 9.5 | ||
| 127 | tropomyosin | 9.5 | ||
| 156 | glyceraldehyde-3-phosphate dehydrogenase A (EC 1.2.1.12) | 9.6 | ||
| 104 | fructose-bisphosphate aldolase | 9.7 | ||
| 144 | Small heat shock protein 24.1 | 9.9 | ||
| 150 | actin-87E isoform 1, similar | 10.2 | <15% | |
| 105 | kin 17-mid super family, hypothetical protein AND_04962 | 10.2 | ||
| 115 | pur-alpha, putative | 10.3 | ||
| 93 | fructose-bisphosphate aldolase | 10.5 | ||
| 155 | enoyl-CoA hydratase, mitochondrial-like | 10.6 | ||
| 86 | 40S ribosomal prot SA (p 40) (34/67 kDa laminin receptor) | 10.7 | ||
| 143 | prohibitin | 10.9 | ||
| 142 | proteasome alpha 5 subunit-like | 11.0 | ||
| 16 | dipeptidyl peptidase family member 6 | 11.0 | ||
| 88 | actin 2 = cytoplasmic actin= beta actin | 11.0 | ||
| 95 | glutamine synthetase | 11.0 | ||
| 106 | cystathionine gamma-lyase | 11.0 | ||
| 118 | malate deshydrogenase, cytosolic | 11.0 | ||
| 138 | receptor of Activated Kinase C 1 | 11.0 | ||
| 122 | glyceraldehyde-3-phosphate dehydrogenase | 11.1 | ||
| 20 | heat shock cognate 71 | 11.1 | ||
| 136 | malate dehydrogenase, cytosolic | 11.5 | ||
| 89 | actin 2 = cytoplasmic actin = beta actin | 11.6 | ||
| 202 | histone H4 | 11.8 | ||
| 66 | ATP synthase alpha subunit mitochondrial | 11.9 | ||
| 168 | small 22kd heat shock protein, putative | 12.0 | ||
| 30 | delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial | 12.0 | ||
| 96 | 26S proteasome regulatory complex ATPase RPT4 | 12.0 | ||
| 185 | ribosomal protein rps13 | 12.2 | ||
| 70 | protein disulfide-isomerase, like | 12.2 | ||
| 82 | citrate synthase, mitochondrial, predicted | 12.4 | ||
| 52 | fascin | 12.4 | ||
| 35 | CCDC 151 like, coiled-coil domain containing 151 | 12.4 | ||
| 200 | profilin like | 12.5 | ||
| 152 | ETF beta-like | 12.5 | ||
| 184 | peroxiredoxin V | 12.6 | ||
| 135 | S-formylglutathione hydrolase | 12.7 | ||
| 116 | G protein subunit beta-1 | 12.8 | ||
| 179 | centrin-3 | 12.8 | ||
| 55 | protein disulfide-isomerase, predicted | 12.9 | ||
| 108 | arginine kinase | 13.0 | ||
| 128 | 14-3-3 epsilon protein | 13.0 | ||
| 154 | enoyl-CoA hydratase, mitochondrial-like | 13.1 | ||
| 9 | major vault protein | 13.3 | ||
| 18 | EF-hand domain-containing protein 1 | 13.3 | ||
| 113 | pyruvate dehydrogenase E1 component subunit beta, mitochondrial | 13.3 | ||
| 203 | thioredoxin 1 | 13.3 | ||
| 186 | glucose-regulated protein 94 (fragment) | 13.4 | ||
| 141 | cathepsin L-like, predicted | 13.4 | ||
| 33 | TCP1, subunit beta-like | 13.4 | ||
| 85 | calumenin precursor, putative | 13.4 | ||
| 111 | inorganic pyrophosphatase-like | 13.6 | ||
| 147 | proteasome subunit alpha type-4 | 13.6 | ||
| 114 | transcriptional activator protein pur-alpha | 13.6 | ||
| 53 | fascin-like domain protein | 13.7 | ||
| 191 | superoxide dismutase (Cu/Zn-SOD) | 13.7 | ||
| 32 | dedicator of cytokinesis protein 8, partial, predicted | 14.0 | ||
| 71 | elongation factor 1 alpha 1 | 14.0 | ||
| 129 | 14-3-3 epsilon ptotein | 14.2 | ||
| 183 | eIF5A like | 14.2 | ||
| 40 | TCP1, subunit theta | 14.3 | ||
| 13 | 78kDa glucose regulated protein | 14.4 | ||
| 126 | tropomyosin | 14.4 | ||
| 84 | phosphoglycerate kinase | 14.7 | ||
| 90 | tektin 1 | 15.0 | <20% | |
| 43 | TCP1, subunit eta-like isoform 1 | 15.1 | ||
| 130 | 14-3-3 epsilon protein | 15.1 | ||
| 91 | actin 2 = cytoplasmic actin = beta actin | 15.2 | ||
| 25 | phenylalanyl-tRNA synthetase beta chain, probable | 15.2 | ||
| 190 | superoxide dismutase | 15.3 | ||
| 37 | NADPH-dependent aldehyde reductase, putative | 15.4 | ||
| 2 | catchin protein | 15.4 | ||
| 159 | triosephosphate isomerase | 15.5 | ||
| 197 | histone H2B | 15.7 | ||
| 76 | 26S protease regulatory subunit 6a RPT5 | 15.9 | ||
| 194 | hypothetical protein KGM_09271 with pleckstrin homology-like domain | 15.9 | ||
| 164 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 8, mitochondrial-like | 15.9 | ||
| 158 | glutathione S-transferase, Class Beta | 16.0 | ||
| 67 | NADH dehydrogenase (ubiquinone) flavoprotein 1, mitochondrial | 16.0 | ||
| 140 | receptor of Activated Kinase C 1 | 16.0 | ||
| 62 | non-neuronal cytoplasmic intermediate filament protein | 16.1 | ||
| 188 | nucleoside diphosphate kinase | 16.1 | ||
| 87 | ATP synthase beta subunit | 16.2 | ||
| 151 | actin 2 = cytoplasmic actin = beta actin | 16.5 | ||
| 132 | small heat shock protein 24.1 | 16.6 | ||
| 107 | arginine kinase | 16.9 | ||
| 94 | actin 5 | 16.9 | ||
| 8 | major vault protein | 16.9 | ||
| 195 | peptidyl prolyl cis-trans isomerase A (II) | 16.9 | ||
| 4 | glucose-regulated protein 94 | 17.0 | ||
| 46 | PRP19/PSO4 pre-mRNA processing factor 19 homolog, predicted | 17.0 | ||
| 10 | aconitase 2, mitochondrial isoform 2, similar | 17.0 | ||
| 7 | major vault protein | 17.1 | ||
| 121 | malate dehydrogenase, mitochondrial | 17.2 | ||
| 173 | glutathione S-transferase sigma 3 | 17.3 | ||
| 187 | tubulin-specific chaperone a-like | 17.4 | ||
| 65 | ATP synthase alpha subunit mitochondrial | 17.4 | ||
| 14 | NADH dehydrogenase subunit, hypothetical protein DAPPUDRAFT_192333 | 17.5 | ||
| 79 | fumarylacetoacetate hydrolase, similar | 17.5 | ||
| 64 | ATP synthase alpha subunit mitochondrial | 17.9 | ||
| 123 | 3-hydroxyanthranilate 3,4-dioxygenase | 17.9 | ||
| 117 | radial spoke head protein 9, like | 17.9 | ||
| 153 | proteasome subunit alpha type-6 | 18.0 | ||
| 59 | catalase | 18.1 | ||
| 80 | RIB43A-like with coiled-coils protein 2 | 18.2 | ||
| 189 | actin | 18.2 | ||
| 44 | TCP1, subunit gamma isoform 1 | 18.4 | ||
| 100 | isocitrate dehydrogenase | 18.4 | ||
| 48 | TCP1 subunit zeta | 18.4 | ||
| 171 | isocitrate dehydrogenase | 18.5 | ||
| 3 | glycine dehydrogenase | 18.5 | ||
| 177 | proteasome beta type-6 subunit | 18.5 | ||
| 178 | thioredoxin peroxidase | 18.5 | ||
| 45 | TCP1, hypothetical protein | 18.7 | ||
| 131 | small heat shock protein 24.1 | 19.2 | ||
| 69 | 26S proteasome regulatory subunit T3 | 19.3 | ||
| 21 | V-type proton ATPase catalytic subunit A | 19.3 | ||
| 133 | small heat shock protein 24.1 | 19.3 | ||
| 149 | voltage-dependent anion selective channel protein 2, probable | 19.6 | ||
| 102 | SET protein | 19.6 | ||
| 12 | heat shock protein 90 | 19.6 | ||
| 41 | meiosis-specific nuclear structural protein 1-like | 19.7 | ||
| 148 | small heat shock protein 24.1 | 19.8 | ||
| 174 | calcyphosin-like protein | 19.8 | ||
| 198 | ribosomal protein S12 | 19.9 | ||
| 176 | ubiquination linked effector, hypothetical protein CRE_31518 | 20.0 | <25% | |
| 49 | tubulin alpha-1 chain | 20.2 | ||
| 17 | E3 ubiquitin-protein ligase TRIM33 | 20.3 | ||
| 103 | UDP-glucose 4-epimerase | 20.3 | ||
| 77 | 26S protease regulatory subunit 6a RPT5 | 20.4 | ||
| 19 | stress-70 protein, mitochondrial, predicted mortaline-like | 20.6 | ||
| 22 | succinate dehydrogenase (ubiquinone) flavoprotein subunit | 20.6 | ||
| 172 | glucosamine phosphate isomerase | 20.6 | ||
| 27 | phosphoenolpyruvate carboxykinase | 20.6 | ||
| 124 | sirtuin-5 | 20.6 | ||
| 15 | NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial | 20.8 | ||
| 169 | proteasome alpha type 2 | 20.8 | ||
| 199 | ribosomal protein rps12 | 20.9 | ||
| 134 | dyp-type peroxidase like | 21.0 | ||
| 98 | isocitrate dehydrogenase | 21.0 | ||
| 47 | ubiquinone biosynthesis monooxygenase COQ6 | 21.0 | ||
| 29 | Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | 21.1 | ||
| 170 | peroxiredoxin | 21.1 | ||
| 196 | actin | 21.1 | ||
| 166 | putative small 22kd heat shock protein | 21.2 | ||
| 181 | C1q domain containing protein MgC1q64, putative | 21.2 | ||
| 162 | GTP-binding nuclear protein Ran, provisional | 21.5 | ||
| 57 | amine oxidase, predicted | 21.5 | ||
| 39 | heat shock protein 60 | 21.7 | ||
| 31 | serine/threonine-protein kinase pelle-like | 21.8 | ||
| 56 | TCP1 subunit epsilon like, hypothetical protein SINV_10604 | 22.1 | ||
| 36 | transketolase | 22.1 | ||
| 97 | long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor | 22.4 | ||
| 63 | tubulin, beta 2C-like, predicted | 22.5 | ||
| 182 | cyclophilin-type peptidyl-prolyl cis-trans isomerase-15 | 22.5 | ||
| 112 | short chain collagen C4, putative | 22.6 | ||
| 1 | spectrin alpha chain | 22.8 | ||
| 42 | UDP-N-acetylglucosamine pyrophosphorylase, provisional | 23.1 | ||
| 34 | UDP-N-acetylglucosamine pyrophosphorylase, provisional | 23.2 | ||
| 146 | endoplasmic reticulum protein ERp29 | 23.4 | ||
| 193 | ubiquinone biosynthesis monooxygenase COQ6 | 23.5 | ||
| 28 | phosphoenolpyruvate carboxykinase | 23.8 | ||
| 38 | chaperonin | 24.1 | ||
| 167 | ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 | 24.1 | ||
| 58 | leucine aminopeptidase, predictive | 24.6 | ||
| 50 | selenium-binding protein 1, partial | 24.7 | ||
| 5 | valosin-containing protein-like | 24.8 | ||
| 51 | elongation factor 1 alpha | 25.9 | <30% | |
| 137 | ribosomal protein S2 | 25.9 | ||
| 163 | phosphoglycerate mutase 1 | 25.9 | ||
| 92 | actin | 26.4 | ||
| 26 | phenylalanyl-tRNA synthetase beta chain, probable | 26.9 | ||
| 54 | dihydrolipoamide dehydrogenase | 27.1 | ||
| 101 | phosphoglycerate kinase | 27.6 | ||
| 201 | hypothetical protein AND_08398 | 27.7 | ||
| 125 | GDP-L-fucose synthetase | 28.0 | ||
| 145 | small heat shock protein 24.1 | 31.4 | >30% | |
| 24 | heat shock protein 70 | 33.5 | ||
| 81 | Rab GDP dissociation inhibitor alpha | 37.1 | ||
| 11 | aconitase 2, mitochondrial isoform 2, similar | 37.4 | ||
| 180 | translationally controlled tumour protein | 40.8 | ||
| 83 | succinate-semialdehyde dehydrogenase, mitochondrial | 43.8 | ||
| 75 | NFX1-type containing zinc finge, similar | 47.0 | ||
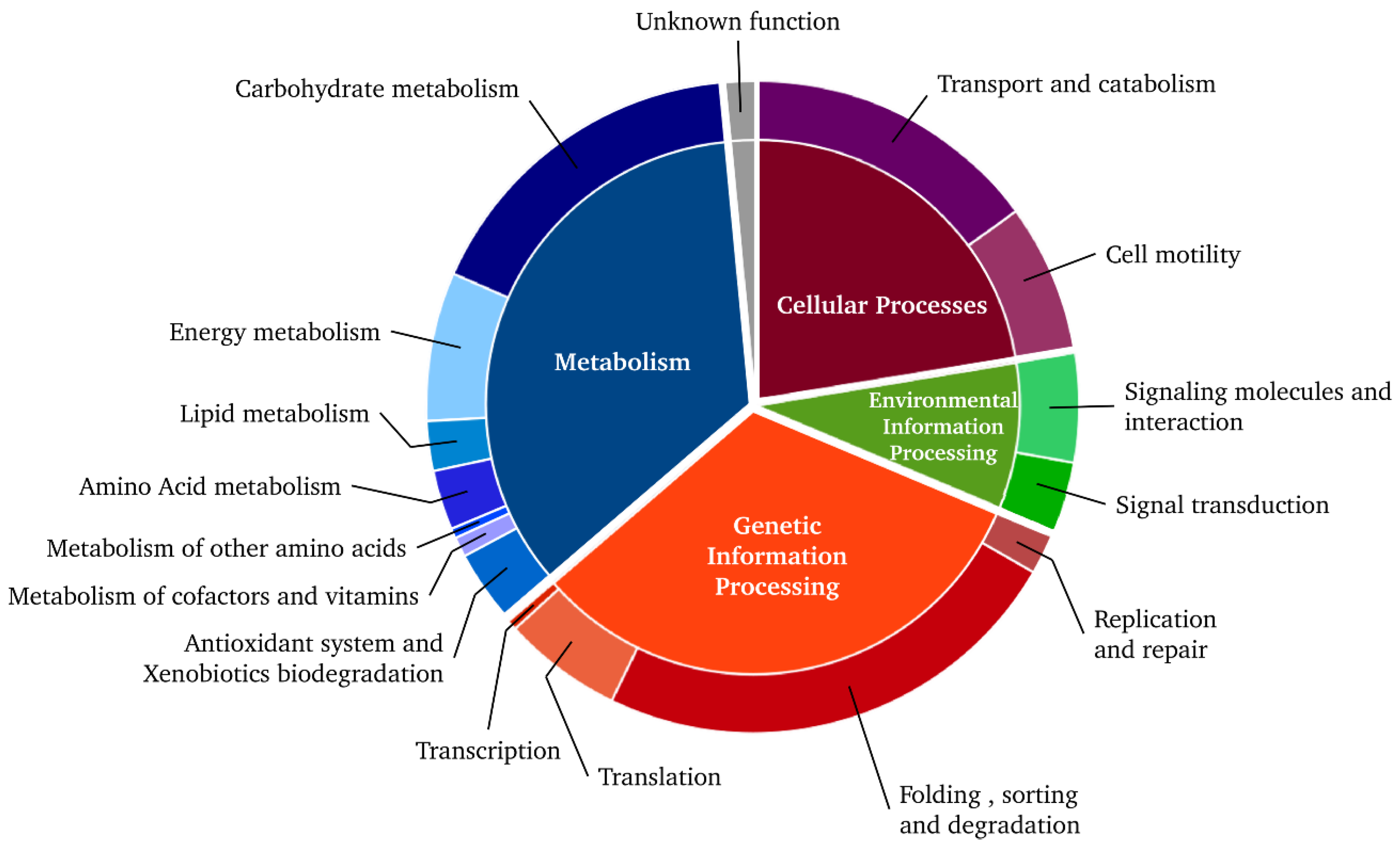
3.1. Transcriptional and Translational Actors
3.2. Cytoskeleton
3.3. Energetic, Carbohydrate and Amino Acid Metabolisms
3.4. Antioxidant and Defence Systems
3.5. Protein Stabilisation, Folding and Sequestration
3.6. Intracellular Protein Trafficking
3.7. Ubiquitin Proteasome System
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Odronitz, F.; Hellkamp, M.; Kollmar, M. diArk—A resource for eukaryotic genome research. BMC Genomics 2007, 8, e103. [Google Scholar] [CrossRef]
- Zhang, G.; Fang, X.; Guo, X.; Li, L.; Luo, R.; Xu, F.; Yang, P.; Zhang, L.; Wang, X.; Qi, H.; et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 2012, 490, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Knigge, T.; Monsinjon, T.; Andersen, O.K. Surface-enhanced laser desorption/ionization-time of flight-mass spectrometry approach to biomarker discovery in blue mussels (Mytilus edulis) exposed to polyaromatic hydrocarbons and heavy metals under field conditions. Proteomics 2004, 4, 2722–2727. [Google Scholar] [CrossRef] [PubMed]
- Manduzio, H.; Cosette, P.; Gricourt, L.; Jouenne, T.; Lenz, C.; Andersen, O.K.; Leboulenger, F.; Rocher, B. Proteome modifications of blue mussel (Mytilus edulis L.) gills as an effect of water pollution. Proteomics 2005, 5, 4958–4963. [Google Scholar] [CrossRef] [PubMed]
- Venier, P.; de Pitta, C.; Bernante, F.; Varotto, L.; de Nardi, B.; Bovo, G.; Roch, P.; Novoa, B.; Figueras, A.; Pallavicini, A.; et al. MytiBase: A knowledgebase of mussel (M. galloprovincialis) transcribed sequences. BMC Genomics 2009, 10, e72. [Google Scholar] [CrossRef]
- Goldberg, E.D.; Bowen, V.T.; Farrington, J.W.; Harvey, G.; Martin, J.H.; Parker, P.L.; Risebrough, R.W.; Robertson, W.; Schneider, E.; Gamble, E. The Mussel Watch. Environ. Conserv. 1978, 5, 101–125. [Google Scholar] [CrossRef]
- Connor, K.M.; Gracey, A.Y. Circadian cycles are the dominant transcriptional rhythm in the intertidal mussel Mytilus californianus. Proc. Natl. Acad. Sci. USA 2011, 108, 16110–16115. [Google Scholar] [CrossRef] [PubMed]
- Place, S.P.; Menge, B.A.; Hofmann, G.E. Transcriptome profiles link environmental variation and physiological response of Mytilus californianus between Pacific tides. Funct. Ecol. 2012, 26, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Schneider, K.R. Heat stress in the intertidal: Comparing survival and growth of an invasive and native mussel under a variety of thermal conditions. Biol. Bull. 2008, 215, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Letendre, J.; Chouquet, B.; Rocher, B.; Manduzio, H.; Leboulenger, F.; Durand, F. Differential pattern of Cu/Zn superoxide dismutase isoforms in relation to tidal spatio-temporal changes in the blue mussel Mytilus edulis. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2008, 148, 211–216. [Google Scholar] [CrossRef]
- Letendre, J.; Dupont-Rouzeyrol, M.; Hanquet, A.C.; Durand, F.; Budzinski, H.; Chan, P.; Vaudry, D.; Rocher, B. Impact of toxicant exposure on the proteomic response to intertidal condition in Mytilus edulis. Comp. Biochem. Physiol. Part D Genomics Proteomics 2011, 6, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Pantzartzi, C.; Drosopoulou, E.; Yiangou, M.; Drozdov, I.; Tsoka, S.; Ouzounis, C.A.; Scouras, Z.G. Promoter complexity and tissue-specific expression of stress response components in Mytilus galloprovincialis, a sessile marine invertebrate species. PLoS Comput. Biol. 2010, 6, e1000847. [Google Scholar] [CrossRef] [PubMed]
- Olsson, B.; Bradley, B.P.; Gilek, M.; Reimer, O.; Shepard, J.L.; Tedengren, M. Physiological and proteomic responses in Mytilus edulis exposed to PCBs and PAHs extracted from Baltic Sea sediments. Hydrobiologia 2004, 514, 15–27. [Google Scholar] [CrossRef]
- Tomanek, L.; Zuzow, M.J. The proteomic response of the mussel congeners Mytilus galloprovincialis and M. trossulus to acute heat stress: Implications for thermal tolerance limits and metabolic costs of thermal stress. J. Exp. Biol. 2010, 213, 3559–3574. [Google Scholar] [CrossRef] [PubMed]
- Dowd, W.W.; Somero, G.N. Behavior and survival of Mytilus congeners following episodes of elevated body temperature in air and seawater. J. Exp. Biol. 2013, 216, 502–514. [Google Scholar] [CrossRef] [PubMed]
- Dutton, J.M.; Hofmann, G.E. Spatial and temporal variation in distribution and protein ubiquitination for Mytilus congeners in the California hybrid zone. Arine Biol. 2008, 154, 1067–1075. [Google Scholar]
- Tomanek, L. Proteomics to study adaptations in marine organisms to environmental stress. J. Proteomics 2014, 105C, 92–106. [Google Scholar] [CrossRef] [PubMed]
- Suchanek, T.H.; Geller, G.B.; Kreiser, B.R.; Mitton, J.B. Zoogeographic Distributions of the Sibling Species Mytilus galloprovincialis and M. trossulus (Bivalvia: Mytilidae) and Their Hybrids in the North Pacific. Biol. Bull. 1997, 193, 187–194. [Google Scholar] [CrossRef]
- Rawson, P.D.; Hilbish, T.J. Asymmetric introgression of mtDNA among European populations of blue mussels (Mytilus spp.). Evolution 1998, 52, 100–108. [Google Scholar] [CrossRef]
- Westfall, K.M.; Gardner, J.P. Genetic diversity of Southern hemisphere blue mussels (Bivalvia: Mytilidae) and the identification of non-indigenous taxa. Biol. J. Linn. Soc. 2010, 101, 898–909. [Google Scholar] [CrossRef]
- Lopez, J.L.; Marina, A.; Alvarez, G.; Vazquez, J. Application of proteomics for fast identification of species-specific peptides from marine species. Proteomics 2002, 2, 1658–1665. [Google Scholar] [CrossRef] [PubMed]
- Lopez, J.L.; Abalde, S.L.; Fuentes, J. Proteomic approach to probe for larval proteins of the mussel Mytilus galloprovincialis. Mar. Biotechnol. (N.Y.) 2005, 7, 396–404. [Google Scholar] [CrossRef]
- Tomanek, L. Environmental proteomics of the mussel Mytilus: Implications for tolerance to stress and change in limits of biogeographic ranges in response to climate change. Integr. Comp. Biol. 2012, 52, 648–664. [Google Scholar] [CrossRef] [PubMed]
- Diz, A.P.; Dudley, E.; Cogswell, A.; MacDonald, B.W.; Kenchington, E.L.; Zouros, E.; Skibinski, D.O. Proteomic analysis of eggs from Mytilus edulis females differing in mitochondrial DNA transmission mode. Mol. Cell Proteomics 2013, 12, 3068–3080. [Google Scholar] [CrossRef] [PubMed]
- Diz, A.P.; Skibinski, D.O. Evolution of 2-DE protein patterns in a mussel hybrid zone. Proteomics 2007, 7, 2111–2120. [Google Scholar] [CrossRef] [PubMed]
- Suarez-Ulloa, V.; Fernandez-Tajes, J.; Manfrin, C.; Gerdol, M.; Venier, P.; Eirin-Lopez, J.M. Bivalve omics: State of the art and potential applications for the biomonitoring of harmful marine compounds. Mar. Drugs 2013, 11, 4370–4389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monsinjon, T.; Knigge, T. Proteomic applications in ecotoxicology. Proteomics 2007, 7, 2997–3009. [Google Scholar] [CrossRef] [PubMed]
- Campos, A.; Puerto, M.; Prieto, A.; Camean, A.; Almeida, A.M.; Coelho, A.V.; Vasconcelos, V. Protein extraction and two-dimensional gel electrophoresis of proteins in the marine mussel Mytilus galloprovincialis: An important tool for protein expression studies, food quality and safety assessment. J. Sci. Food Agric. 2013, 93, 1779–1787. [Google Scholar] [CrossRef] [PubMed]
- Rabilloud, T.; Chevallet, M.; Luche, S.; Lelong, C. Two-dimensional gel electrophoresis in proteomics: Past, present and future. J. Proteomics 2010, 73, 2064–2077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cannuel, R.; Beninger, P.G.; McCombie, H.; Boudry, P. Gill Development and its functional and evolutionary implications in the blue mussel Mytilus edulis (Bivalvia: Mytilidae). Biol. Bull. 2009, 217, 173–188. [Google Scholar] [PubMed]
- Barker Jorgensen, C. A Hydromechanical Principle for Particle Retention in Mytilus edulis and Other Ciliary Suspension Feeders. Mar. Biol. 1981, 61, 277–282. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
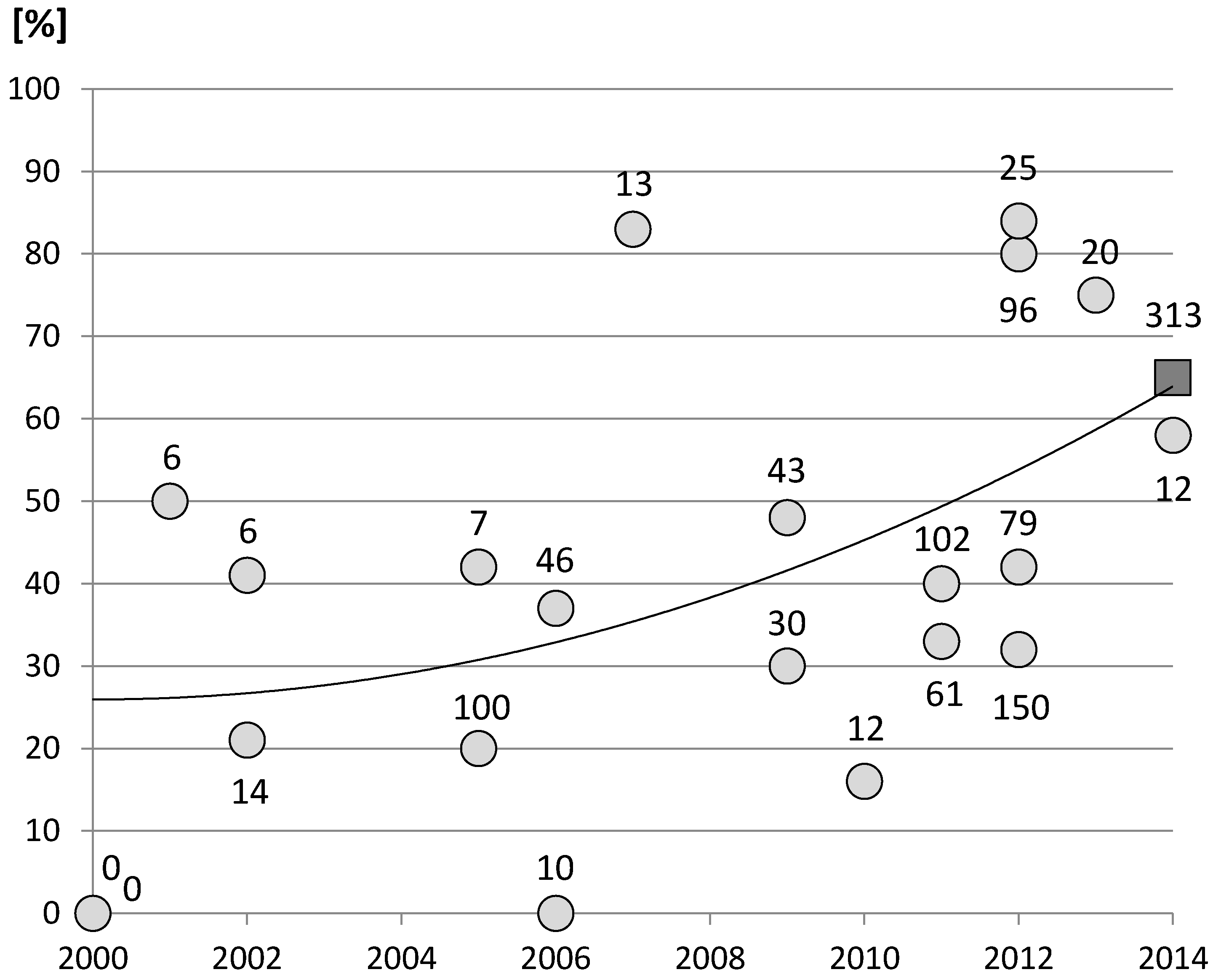
- Bonett, D.G. Confidence interval for a coefficient of quartile variation. Comput. Stat. Data Anal. 2006, 50, 2953–2957. [Google Scholar] [CrossRef]
- Dejda, A.; Chan, P.; Seaborn, T.; Coquet, L.; Jouenne, T.; Fournier, A.; Vaudry, H.; Vaudry, D. Involvment of Stathmin 1 in the neurotrophic effects of PACAP in PC12 cells. J. Neurochem. 2010, 114, 1498–1510. [Google Scholar] [PubMed]
- Shepard, J.L.; Olsson, B.; Tedengren, M.; Bradley, B.P. Protein expression signatures identified in Mytilus edulis exposed to PCBs, copper and salinity stress. Mar. Environ. Res. 2000, 50, 337–340. [Google Scholar] [CrossRef] [PubMed]
- Fields, P.A.; Zuzow, M.J.; Tomanek, L. Proteomic responses of blue mussel (Mytilus) congeners to temperature acclimation. J. Exp. Biol. 2012, 215, 1106–1116. [Google Scholar] [CrossRef] [PubMed]
- Dondero, F.; Negri, A.; Boatti, L.; Marsano, F.; Mignone, F.; Viarengo, A. Transcriptomic and proteomic effects of a neonicotinoid insecticide mixture in the marine mussel (Mytilus galloprovincialis, Lam.). Sci. Total Environ. 2010, 408, 3775–3786. [Google Scholar] [CrossRef] [PubMed]
- Leung, P.T.; Wang, Y.; Mak, S.S.; Ng, W.C.; Leung, K.M. Differential proteomic responses in hepatopancreas and adductor muscles of the green-lipped mussel Perna viridis to stresses induced by cadmium and hydrogen peroxide. Aquat. Toxicol. 2011, 105, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Puerto, M.; Campos, A.; Prieto, A.; Camean, A.; de Almeida, A.M.; Coelho, A.V.; Vasconcelos, V. Differential protein expression in two bivalve species; Mytilus galloprovincialis and Corbicula fluminea; exposed to Cylindrospermopsis raciborskii cells. Aquat. Toxicol. 2011, 101, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Gomes, T.; Araujo, O.; Pereira, R.; Almeida, A.C.; Cravo, A.; Bebianno, M.J. Genotoxicity of copper oxide and silver nanoparticles in the mussel Mytilus galloprovincialis. Mar. Environ. Res. 2013, 84, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Ji, C.; Wei, L.; Zhao, J.; Lu, H. Proteomic and metabolomic responses in hepatopancreas of Mytilus galloprovincialis challenged by Micrococcus luteus and Vibrio anguillarum. J. Proteomics 2013, 94, 54–67. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Ji, C.; Wei, L.; Zhao, J. Evaluation of protein extraction protocols for 2DE in marine ecotoxicoproteomics. Proteomics 2013, 13, 3205–3210. [Google Scholar] [CrossRef] [PubMed]
- Kolli, S.; Zito, C.I.; Mossink, M.H.; Wiemer, E.A.; Bennett, A.M. The major vault protein is a novel substrate for the tyrosine phosphatase SHP-2 and scaffold protein in epidermal growth factor signaling. J. Biol. Chem. 2004, 279, 29374–29385. [Google Scholar] [CrossRef] [PubMed]
- Lin, G.; Brownsey, R.W.; MacLeod, K.M. Regulation of mitochondrial aconitase by phosphorylation in diabetic rat heart. Cell. Mol. Life Sci. 2009, 66, 919–932. [Google Scholar] [CrossRef] [PubMed]
- Muller, P.; Ruckova, E.; Halada, P.; Coates, P.J.; Hrstka, R.; Lane, D.P.; Vojtesek, B. C-Terminal phosphorylation of Hsp70 and Hsp90 regulates alternate binding to co-chaperones CHIP and HOP to determine cellular protein folding/degradation balances. Oncogene 2013, 32, 3101–3110. [Google Scholar] [CrossRef] [PubMed]
- Walker, R.P.; Leegood, R.C. Phosphorylation of phosphoenolpyruvate carboxykinase in plants: Studies in plants with C4 photosynthesis and Crassulacean acid metabolism and in germinating seeds. Biochem. J. 1996, 317, 653–658. [Google Scholar] [PubMed]
- Petrak, J.; Ivanek, R.; Toman, O.; Cmejla, R.; Cmejlova, J.; Vyoral, D.; Zivny, J.; Vulpe, C.D. Deja vu in proteomics. A hit parade of repeatedly identified differentially expressed proteins. Proteomics 2008, 8, 1744–1749. [Google Scholar] [CrossRef] [PubMed]
- Evans, T.G.; Somero, G.N. Phosphorylation events catalyzed by major cell signaling proteins differ in response to thermal and osmotic stress among native (Mytilus californianus and Mytilus trossulus) and invasive (Mytilus galloprovincialis) species of mussels. Physiol. Biochem. Zool. 2010, 83, 984–996. [Google Scholar] [CrossRef] [PubMed]
- Storey, K.B.; Storey, J.M. Heat schock proteins and hypometabolism: Adaptive strategy for protein preservation. Res. Rep. Biol. 2011, 2, 57–68. [Google Scholar] [CrossRef]
- McDonagh, B.; Tyther, R.; Sheehan, D. Carbonylation and glutathionylation of proteins in the blue mussel Mytilus edulis detected by proteomic analysis and Western blotting: Actin as a target for oxidative stress. Aquat. Toxicol. 2005, 73, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Apraiz, I.; Mi, J.; Cristobal, S. Identification of proteomic signatures of exposure to marine pollutants in mussels (Mytilus edulis). Mol. Cell Proteomics 2006, 5, 1274–1285. [Google Scholar] [CrossRef] [PubMed]
- Company, R.; Torreblanca, A.; Cajaraville, M.; Bebianno, M.J.; Sheehan, D. Comparison of thiol subproteome of the vent mussel Bathymodiolus azoricus from different Mid-Atlantic Ridge vent sites. Sci. Total Environ. 2012, 437, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Culloty, S.; Darmody, G.; Lynch, S.; Davenport, J.; Ramirez-Garcia, S.; Dawson, K.A.; Lynch, I.; Blasco, J.; Sheehan, D. Toxicity of copper oxide nanoparticles in the blue mussel, Mytilus edulis: A redox proteomic investigation. Chemosphere 2014, 108, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Schluter, H.; Apweiler, R.; Holzhutter, H.G.; Jungblut, P.R. Finding one’s way in proteomics: A protein species nomenclature. Chem. Cent. J. 2009, 3, e11. [Google Scholar] [CrossRef]
- Lisitsa, A.; Moshkovskii, S.; Chernobrovkin, A.; Ponomarenko, E.; Archakov, A. Profiling proteoforms: Promising follow-up of proteomics for biomarker discovery. Expert Rev. Proteomics 2014, 11, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Shepard, J.L.; Bradley, B.P. Protein expression signatures and lysosomal stability in Mytilus edulis exposed to graded copper concentrations. Mar. Environ. Res. 2000, 50, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Lopez, J.L.; Mosquera, E.; Fuentes, J.; Marina, A.; Vlazquez, J.; Alvarez, G. Two-dimensional gel electrophoresis of Mytilus galloprovincialis: Differences in protein expression of intertidal and cultured mussels. Mar. Ecol. Progr. Ser. 2001, 224, 149–156. [Google Scholar] [CrossRef]
- Fuentes, J.; López, J.L.; Mosquera, E.; Vázquez, J.; Villalba, A.; Álvarez, G. Growth, mortality, pathological conditions and protein expression of Mytilus edulis and M. galloprovincialis crosses cultured in the Rı́a de Arousa (NW of Spain). Aquaculture 2002, 213, 233–251. [Google Scholar] [CrossRef]
- Mi, J.; Orbea, A.; Syme, N.; Ahmed, M.; Cajaraville, M.P.; Cristobal, S. Peroxisomal proteomics, a new tool for risk assessment of peroxisome proliferating pollutants in the marine environment. Proteomics 2005, 5, 3954–3965. [Google Scholar] [CrossRef] [PubMed]
- Amelina, H.; Apraiz, I.; Sun, W.; Cristobal, S. Proteomics-based method for the assessment of marine pollution using liquid chromatography coupled with two-dimensional electrophoresis. J. Proteome Res. 2007, 6, 2094–2104. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, H.; Schiedek, D.; Grøsvik, B.E.; Goksøyr, A. Protein responses in blue mussels (Mytilus edulis) exposed to organic pollutants: A combined CYP-antibody/proteomic approach. Aquat. Toxicol. 2006, 78, S49–S56. [Google Scholar] [CrossRef] [PubMed]
- Apraiz, I.; Cajaraville, M.P.; Cristobal, S. Peroxisomal proteomics: Biomonitoring in mussels after the Prestige’s oil spill. Mar. Pollut. Bull. 2009, 58, 1815–1826. [Google Scholar] [CrossRef] [PubMed]
- Diz, A.P.; Dudley, E.; MacDonald, B.W.; Pina, B.; Kenchington, E.L.; Zouros, E.; Skibinski, D.O. Genetic variation underlying protein expression in eggs of the marine mussel Mytilus edulis. Mol. Cell Proteomics 2009, 8, 132–144. [Google Scholar] [CrossRef] [PubMed]
- Diz, A.P.; Dudley, E.; Skibinski, D.O. Identification and characterization of highly expressed proteins in sperm cells of the marine mussel Mytilus edulis. Proteomics 2012, 12, 1949–1956. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, W.; Rainville, L.C.; McEneff, G.; Sheehan, D.; Quinn, B. A proteomic evaluation of the effects of the pharmaceuticals diclofenac and gemfibrozil on marine mussels (Mytilus spp.): Evidence for chronic sublethal effects on stress-response proteins. Drug Test. Anal. 2014, 6, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Schwelberger, H.G.; Kang, H.A.; Hershey, J.W. Translation initiation factor eIF-5A expressed from either of two yeast genes or from human cDNA. Functional identity under aerobic and anaerobic conditions. J. Biol. Chem. 1993, 268, 14018–14025. [Google Scholar] [PubMed]
- Borradaile, N.M.; Buhman, K.K.; Listenberger, L.L.; Magee, C.J.; Morimoto, E.T.; Ory, D.S.; Schaffer, J.E. A critical role for eukaryotic elongation factor 1A-1 in lipotoxic cell death. Mol. Biol. Cell 2006, 17, 770–778. [Google Scholar] [CrossRef] [PubMed]
- Merrick, W.C. Mechanism and regulation of eukaryotic protein synthesis. Microbiol. Rev. 1992, 56, 291–315. [Google Scholar] [PubMed]
- Gonen, H.; Smith, C.E.; Siegel, N.R.; Kahana, C.; Merrick, W.C.; Chakraburtty, K.; Schwartz, A.L.; Ciechanover, A. Protein synthesis elongation factor EF-1 alpha is essential for ubiquitin-dependent degradation of certain N alpha-acetylated proteins and may be substituted for by the bacterial elongation factor EF-Tu. Proc. Natl. Acad. Sci. USA 1994, 91, 7648–7652. [Google Scholar] [CrossRef] [PubMed]
- Darbinian, N.; Gallia, G.L.; Kundu, M.; Shcherbik, N.; Tretiakova, A.; Giordano, A.; Khalili, K. Association of Pur alpha and E2F-1 suppresses transcriptional activity of E2F-1. Oncogene 1999, 18, 6398–6402. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Matsumura, I.; Ezoe, S.; Satoh, Y.; Sakamaki, T.; Albanese, C.; Machii, T.; Pestell, R.G.; Kanakura, Y. E2F1 and c-Myc potentiate apoptosis through inhibition of NF-kappaB activity that facilitates MnSOD-mediated ROS elimination. Mol. Cell 2002, 9, 1017–1029. [Google Scholar] [CrossRef] [PubMed]
- Skorzewski, R.; Sliwinska, M.; Borys, D.; Sobieszek, A.; Moraczewska, J. Effect of actin C-terminal modification on tropomyosin isoforms binding and thin filament regulation. Biochim. Biophys. Acta 2009, 1794, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Norrander, J.M.; deCathelineau, A.M.; Brown, J.A.; Porter, M.E.; Linck, R.W. The Rib43a protein is associated with forming the specialized protofilament ribbons of flagellar microtubules in Chlamydomonas. Mol. Biol. Cell 2000, 11, 201–215. [Google Scholar] [CrossRef] [PubMed]
- Sandoz, D.; Chailley, B.; Boisvieux-Ulrich, E.; Lemullois, M.; Laine, M.C.; Bautista-Harris, G. Organization and functions of cytoskeleton in metazoan ciliated cells. Biol. Cell 1988, 63, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Brett, C.L.; Donowitz, M.; Rao, R. Evolutionary origins of eukaryotic sodium/proton exchangers. Am. J. Physiol. Cell Physiol. 2005, 288, C223–C239. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Brautigan, D.L. Phosphatase inhibitor 2 promotes acetylation of tubulin in the primary cilium of human retinal epithelial cells. BMC Cell Biol. 2008, 9, e62. [Google Scholar] [CrossRef]
- Blond, D.M.; Whittam, R. The regulation of kidney respiration by sodium and potassium ions. Biochem. J. 1964, 92, 158–167. [Google Scholar] [PubMed]
- Balaban, R.S.; Mandel, L.J.; Soltoff, S.P.; Storey, J.M. Coupling of active ion transport and aerobic respiratory rate in isolated renal tubules. Proc. Natl. Acad. Sci. USA 1980, 77, 447–451. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A.; Heller, H.; Elias, S.; Haas, A.L.; Hershko, A. ATP-dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proc. Natl. Acad. Sci. USA 1980, 77, 1365–1368. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Maes, E.G.; Taylor, A.B.; Wang, L.; Hinck, A.P.; Lafer, E.M.; Sousa, R. Structural basis of J cochaperone binding and regulation of Hsp70. Mol. Cell 2007, 28, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Livingstone, D.R. Origins and evolution of pathways of anaerobic metabolism in the animal kingdom. Amer. Zool. 1991, 31, 522–534. [Google Scholar]
- Pereira, C.A.; Alonso, G.D.; Paveto, M.C.; Iribarren, A.; Cabanas, M.L.; Torres, H.N.; Flawia, M.M. Trypanosoma cruzi arginine kinase characterization and cloning. A novel energetic pathway in protozoan parasites. J. Biol. Chem. 2000, 275, 1495–1501. [Google Scholar] [CrossRef] [PubMed]
- Reddy, S.R.; Houmeida, A.; Benyamin, Y.; Roustan, C. Interaction in vitro of scallop muscle arginine kinase with filamentous actin. Eur. J. Biochem. 1992, 206, 251–257. [Google Scholar]
- Bishop, S.H.; Ellis, L.L.; Burcham, J.M. Amino Acid Metabolism in Molluscs. In Metabolic Biochemistry and Molecular Biomechanics; Hochachka, P.W., Ed.; Academic Press: Waltham, MA, USA, 1983; pp. 243–327. [Google Scholar]
- De Zwaan, A. Carbohydrate Catabolism in Bivalves. In Metabolic Biochemistry and Molecular Biomechanics, 1983 ed.; Hochachka, P.W., Ed.; Academic Press: Waltham, MA, USA, 1983; pp. 137–175. [Google Scholar]
- Behrens, J.W.; Elias, J.P.; Taylor, H.H.; Weber, R.E. The archaeogastropod mollusc Haliotis iris: Tissue and blood metabolites and allosteric regulation of haemocyanin function. J. Exp. Biol. 2002, 205, 253–263. [Google Scholar]
- Bradshaw, J.C.; Kumai, Y.; Perry, S.F. The effects of gill remodeling on transepithelial sodium fluxes and the distribution of presumptive sodium-transporting ionocytes in goldfish (Carassius auratus). J. Comp. Physiol. B 2012, 182, 351–366. [Google Scholar]
- Wang, L.; Zhang, H.; Zhou, Z.; Siva, V.S.; Song, L. A C1q domain containing protein from scallop Chlamys farreri serving as pattern recognition receptor with heat-aggregated IgG binding activity. PLOS ONE 2012, 7, e43289. [Google Scholar]
- Liu, H.H.; Xiang, L.X.; Shao, J.Z. A novel C1q-domain-containing (C1qDC) protein from Mytilus coruscus with the transcriptional analysis against marine pathogens and heavy metals. Dev. Comp. Immunol. 2014, 44, 70–75. [Google Scholar]
- Moreira, R.; Balseiro, P.; Romero, A.; Dios, S.; Posada, D.; Novoa, B.; Figueras, A. Gene expression analysis of clams Ruditapes philippinarum and Ruditapes decussatus following bacterial infection yields molecular insights into pathogen resistance and immunity. Dev. Comp. Immunol. 2012, 36, 140–149. [Google Scholar]
- Gestal, C.; Costa, M.; Figueras, A.; Novoa, B. Analysis of differentially expressed genes in response to bacterial stimulation in hemocytes of the carpet-shell clam Ruditapes decussatus: Identification of new antimicrobial peptides. Gene 2007, 406, 134–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livingstone, D.R. Cytochrome P-450 and oxidative metabolism in invertebrates. Biochem. Soc. Trans. 1990, 18, 15–19. [Google Scholar] [PubMed]
- Ercal, N.; Gurer-Orhan, H.; Aykin-Burns, N. Toxic metals and oxidative stress part I: Mechanisms involved in metal-induced oxidative damage. Curr. Top. Med. Chem. 2001, 1, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Allen, R.G.; Tresini, M. Oxidative stress and gene regulation. Free Radic. Biol. Med. 2000, 28, 463–499. [Google Scholar] [CrossRef] [PubMed]
- Khessiba, A.; Romeo, M.; Aissa, P. Effects of some environmental parameters on catalase activity measured in the mussel (Mytilus galloprovincialis) exposed to lindane. Environ. Pollut. 2005, 133, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Regoli, F.; Gorbi, S.; Fattorini, D.; Tedesco, S.; Notti, A.; Machella, N.; Bocchetti, R.; Benedetti, M.; Piva, F. Use of the land snail Helix aspersa as sentinel organism for monitoring ecotoxicologic effects of urban pollution: An integrated approach. Environ. Health Perspect. 2006, 114, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Letendre, J.; Chouquet, B.; Manduzio, H.; Marin, M.; Bultelle, F.; Leboulenger, F.; Durand, F. Tidal height influences the levels of enzymatic antioxidant defences in Mytilus edulis. Mar. Environ. Res. 2009, 67, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Ross, S.J.; Findlay, V.J.; Malakasi, P.; Morgan, B.A. Thioredoxin peroxidase is required for the transcriptional response to oxidative stress in budding yeast. Mol. Biol. Cell 2000, 11, 2631–2642. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, P.J.; Sheehan, D. Separation of multiple forms of glutathione S-transferase from the blue mussel, Mytilus edulis. Xenobiotica 1993, 23, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Sheehan, D.; Meade, G.; Foley, V.M.; Dowd, C.A. Structure, function and evolution of glutathione transferases: Implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem. J. 2001, 360, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Young, J.C.; Agashe, V.R.; Siegers, K.; Hartl, F.U. Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 2004, 5, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.P.; Bukau, B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell. Mol. Life Sci. 2005, 62, 670–684. [Google Scholar] [CrossRef] [PubMed]
- Richter, K.; Haslbeck, M.; Buchner, J. The heat shock response: Life on the verge of death. Mol. Cell 2010, 40, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Hilton, G.R.; Lioe, H.; Stengel, F.; Baldwin, A.J.; Benesch, J.L. Small heat-shock proteins: Paramedics of the cell. Top. Curr. Chem. 2013, 328, 69–98. [Google Scholar] [PubMed]
- Garrido, C.; Paul, C.; Seigneuric, R.; Kampinga, H.H. The small heat shock proteins family: The long forgotten chaperones. Int. J. Biochem. Cell Biol. 2012, 44, 1588–1592. [Google Scholar] [CrossRef] [PubMed]
- Mogk, A.; Deuerling, E.; Vorderwulbecke, S.; Vierling, E.; Bukau, B. Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol. Microbiol. 2003, 50, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Tissieres, A.; Mitchell, H.K.; Tracy, U.M. Protein synthesis in salivary glands of Drosophila melanogaster: Relation to chromosome puffs. J. Mol. Biol. 1974, 84, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Young, J.T.; Heikkila, J.J. Proteasome inhibition induces hsp30 and hsp70 gene expression as well as the acquisition of thermotolerance in Xenopus laevis A6 cells. Cell Stress Chaperones 2010, 15, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Feder, M.E.; Hofmann, G.E. Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annu. Rev. Physiol. 1999, 61, 243–282. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, J.G.; Kristensen, T.N.; Loeschcke, V. The evolutionary and ecological role of heat shock proteins. Ecol. Lett. 2003, 6, 1025–1037. [Google Scholar] [CrossRef]
- Clark, M.S.; Peck, L.S. Triggers of the HSP70 stress response: Environmental responses and laboratory manipulation in an Antarctic marine invertebrate (Nacella concinna). Cell Stress Chaperones 2009, 14, 649–660. [Google Scholar] [CrossRef] [PubMed]
- Cottin, D.; Shillito, B.; Chertemps, T.; Tanguy, A.; Leger, N.; Ravaux, J. Identification of differentially expressed genes in the hydrothermal vent shrimp Rimicaris exoculata exposed to heat stress. Mar. Genomics 2010, 3, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Knigge, T.; Bachmann, L.; Kohler, H.R. An intron-containing, heat-inducible stress-70 gene in the millipede Tachypodoiulus niger (Julidae, Diplopoda). Cell Stress Chaperones 2014, 19, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Carpentier, S.; KU Leuven, Division of Biosystems: Leuven, Belgium. Personal communication, 2014.
- Meiri, D.; Breiman, A. Arabidopsis ROF1 (FKBP62) modulates thermotolerance by interacting with HSP90.1 and affecting the accumulation of HsfA2-regulated sHSPs. Plant J. 2009, 59, 387–399. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Metzler, B.; Jahangiri, M.; Mandal, K. Molecular chaperones and heat shock proteins in atherosclerosis. Am. J. Physiol. Heart Circ. Physiol. 2012, 302, H506–H514. [Google Scholar] [CrossRef] [PubMed]
- Kohler, H.R.; Lazzara, R.; Dittbrenner, N.; Capowiez, Y.; Mazzia, C.; Triebskorn, R. Snail phenotypic variation and stress proteins: Do different heat response strategies contribute to Waddington’s widget in field populations? J. Exp. Zool. B Mol. Dev. Evol. 2009, 312, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Leach, M.D.; Budge, S.; Walker, L.; Munro, C.; Cowen, L.E.; Brown, A.J. Hsp90 orchestrates transcriptional regulation by Hsf1 and cell wall remodelling by MAPK signalling during thermal adaptation in a pathogenic yeast. PLoS Pathog. 2012, 8, e1003069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inglis, P.N.; Boroevich, K.A.; Leroux, M.R. Piecing together a ciliome. Trends Genet. 2006, 22, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Hershko, A.; Ciechanover, A.; Heller, H.; Haas, A.L.; Rose, I.A. Proposed role of ATP in protein breakdown: Conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proc. Natl. Acad. Sci. USA 1980, 77, 1783–1786. [Google Scholar] [CrossRef] [PubMed]
- Gotze, S.; Bose, A.; Abele, D.; Sokolova, I.M.; Saborowski, R. Pitfalls in invertebrate proteasome assays. J. Exp. Biol. 2013, 216, 1351–1354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varotto, L.; Domeneghetti, S.; Rosani, U.; Manfrin, C.; Cajaraville, M.P.; Raccanelli, S.; Pallavicini, A.; Venier, P. DNA damage and transcriptional changes in the gills of mytilus galloprovincialis exposed to nanomolar doses of combined metal salts (Cd, Cu, Hg). PLoS One 2013, 8, e54602. [Google Scholar] [CrossRef] [PubMed]
- Tanguy, M.; McKenna, P.; Gauthier-Clerc, S.; Pellerin, J.; Danger, J.M.; Siah, A. Functional and molecular responses in Mytilus edulis hemocytes exposed to bacteria, Vibrio splendidus. Dev. Comp. Immunol. 2013, 39, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Dupont, S.; Mamidi, A.; Cordenonsi, M.; Montagner, M.; Zacchigna, L.; Adorno, M.; Martello, G.; Stinchfield, M.J.; Soligo, S.; Morsut, L.; et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell 2009, 136, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, A.; Oza, J.; Yao, M.; Sohail, H.; Ginjala, V.; Tomas-Loba, A.; Horejsi, Z.; Tan, A.R.; Boulton, S.J.; Ganesan, S. Tripartite Motif-containing 33 (TRIM33) protein functions in the poly(ADP-ribose) polymerase (PARP)-dependent DNA damage response through interaction with Amplified in Liver Cancer 1 (ALC1) protein. J. Biol. Chem. 2013, 288, 32357–32369. [Google Scholar] [CrossRef] [PubMed]
- Chondrogianni, N.; Petropoulos, I.; Grimm, S.; Georgila, K.; Catalgol, B.; Friguet, B.; Grune, T.; Gonos, E.S. Protein damage, repair and proteolysis. Mol. Aspects Med. 2014, 35, 1–71. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K. The proteasome: From basic mechanisms to emerging roles. Keio J. Med. 2013, 62, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Groll, M.; Bajorek, M.; Kohler, A.; Moroder, L.; Rubin, D.M.; Huber, R.; Glickman, M.H.; Finley, D. A gated channel into the proteasome core particle. Nat. Struct. Biol. 2000, 7, 1062–1067. [Google Scholar] [CrossRef] [PubMed]
- Jager, S.; Groll, M.; Huber, R.; Wolf, D.H.; Heinemeyer, W. Proteasome beta-type subunits: Unequal roles of propeptides in core particle maturation and a hierarchy of active site function. J. Mol. Biol. 1999, 291, 997–1013. [Google Scholar] [CrossRef] [PubMed]
- Kunjappu, M.J.; Hochstrasser, M. Assembly of the 20S proteasome. Biochim. Biophys. Acta 2014, 1843, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.J.; Lee, B.H.; Hanna, J.; King, R.W.; Finley, D. Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Mol. Cell Proteomics 2011, 10, R110.003871. [Google Scholar] [PubMed]
- Butler, L.R.; Densham, R.M.; Jia, J.; Garvin, A.J.; Stone, H.R.; Shah, V.; Weekes, D.; Festy, F.; Beesley, J.; Morris, J.R. The proteasomal de-ubiquitinating enzyme POH1 promotes the double-strand DNA break response. EMBO J. 2012, 31, 3918–3934. [Google Scholar] [CrossRef] [PubMed]
- Lam, Y.A.; Lawson, T.G.; Velayutham, M.; Zweier, J.L.; Pickart, C.M. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature 2002, 416, 763–767. [Google Scholar] [CrossRef] [PubMed]
- Shanmuganathan, A.; Avery, S.V.; Willetts, S.A.; Houghton, J.E. Copper-induced oxidative stress in Saccharomyces cerevisiae targets enzymes of the glycolytic pathway. FEBS Lett. 2004, 556, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Santos, P.M.; Benndorf, D.; Sa-Correia, I. Insights into Pseudomonas putida KT2440 response to phenol-induced stress by quantitative proteomics. Proteomics 2004, 4, 2640–2652. [Google Scholar] [CrossRef] [PubMed]
- Kultz, D. Evolution of the cellular stress proteome: From monophyletic origin to ubiquitous function. J. Exp. Biol. 2003, 206, 3119–3124. [Google Scholar] [CrossRef] [PubMed]
- McDonagh, B.; Sheehan, D. Redox proteomics in the blue mussel Mytilus edulis: Carbonylation is not a prerequisitefor ubiquination in acute free radical-mediated oxidative stress. Aquat. Toxicol. 2006, 79, 325–333. [Google Scholar] [CrossRef] [PubMed]
- McDonagh, B.; Sheehan, D. Effect of oxidative stress on protein thiols in the blue mussel Mytilus edulis: Proteomic identification of target proteins. Proteomics 2007, 7, 3395–3403. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rocher, B.; Bultelle, F.; Chan, P.; Foll, F.L.; Letendre, J.; Monsinjon, T.; Olivier, S.; Péden, R.; Poret, A.; Vaudry, D.; et al. 2-DE Mapping of the Blue Mussel Gill Proteome: The Usual Suspects Revisited. Proteomes 2015, 3, 3-41. https://doi.org/10.3390/proteomes3010003
Rocher B, Bultelle F, Chan P, Foll FL, Letendre J, Monsinjon T, Olivier S, Péden R, Poret A, Vaudry D, et al. 2-DE Mapping of the Blue Mussel Gill Proteome: The Usual Suspects Revisited. Proteomes. 2015; 3(1):3-41. https://doi.org/10.3390/proteomes3010003
Chicago/Turabian StyleRocher, Béatrice, Florence Bultelle, Philippe Chan, Frank Le Foll, Julie Letendre, Tiphaine Monsinjon, Stéphanie Olivier, Romain Péden, Agnès Poret, David Vaudry, and et al. 2015. "2-DE Mapping of the Blue Mussel Gill Proteome: The Usual Suspects Revisited" Proteomes 3, no. 1: 3-41. https://doi.org/10.3390/proteomes3010003
APA StyleRocher, B., Bultelle, F., Chan, P., Foll, F. L., Letendre, J., Monsinjon, T., Olivier, S., Péden, R., Poret, A., Vaudry, D., & Knigge, T. (2015). 2-DE Mapping of the Blue Mussel Gill Proteome: The Usual Suspects Revisited. Proteomes, 3(1), 3-41. https://doi.org/10.3390/proteomes3010003






