Proteomic Profiling of Sugar Beet (Beta vulgaris) Leaves during Rhizomania Compatible Interactions
Abstract
:1. Introduction
2. Experimental
2.1. Plant Propagation and Inoculation
2.2. Protein Digestion and Mass Spectrometry
2.3. Protein Identification and Data Analysis
2.4. Statistical Analyses for Differentially Expressed Proteins
3. Results and Discussion
3.1. Protein Characterization

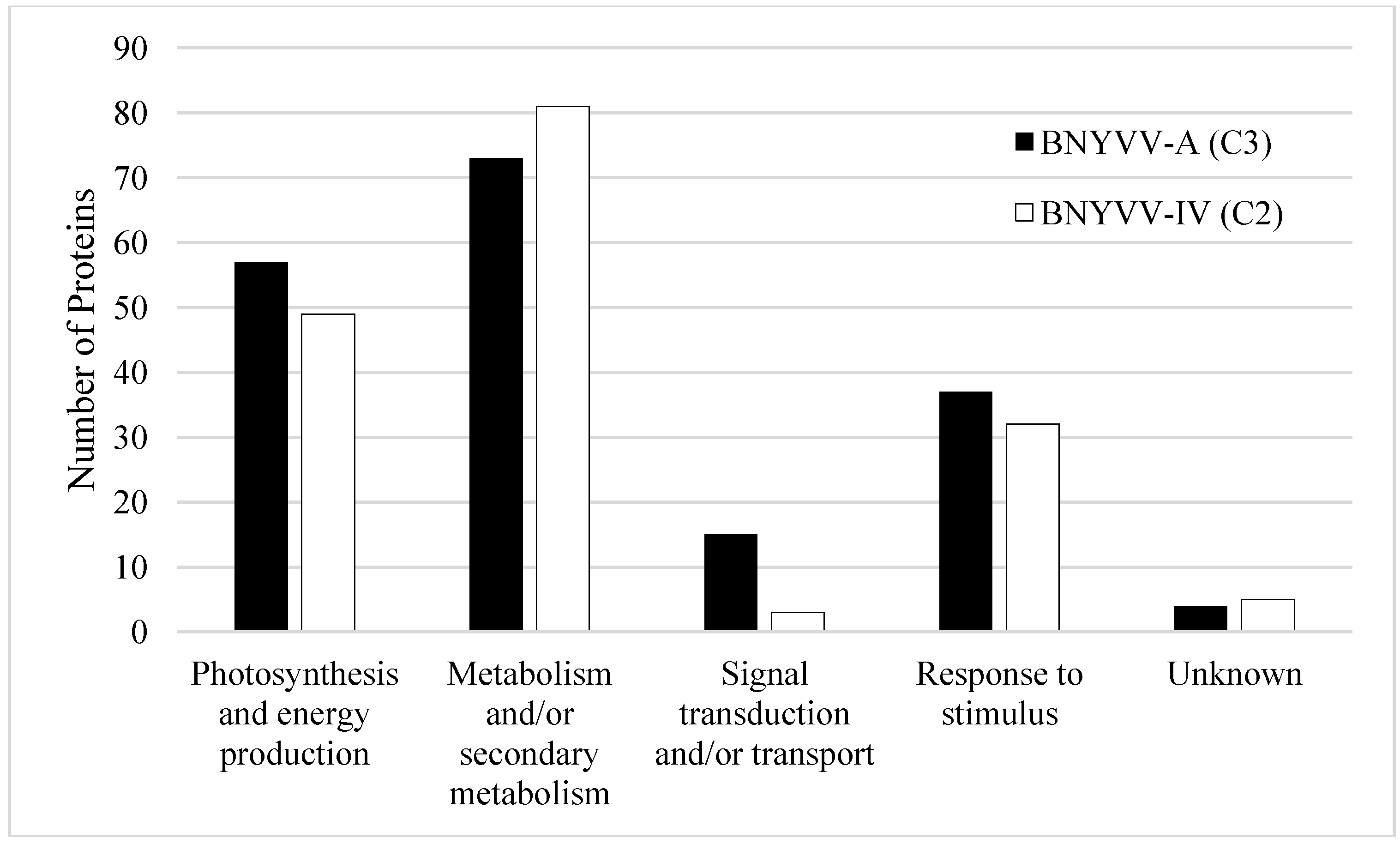
3.2. Response to Stimulus (Including Plant Defense Response) Proteins
3.3. Differential Expression of Proteins during Compatible BNYVV Interactions
| Protein Name | Accession Number | Spectral Counting p-value (p < 0.1) 1 | Average TIC p-value (p < 0.1) 2 |
|---|---|---|---|
| Photosystem II CP43 chlorophyll apoprotein | PSBC_SPIOL | 0.025 | 0.001 |
| Cytosolic heat shock 70 protein | Q41374_SPIOL | 0.053 | |
| Superoxide dismutase [Cu-Zn] | H9BQP7_SUASA | 0.100 | |
| Glyceraldehyde-3-phosphate dehydrogenase B, chloroplastic | G3PB_SPIOL | 0.010 | |
| Choline monooxygenase, chloroplastic | CHMO_BETVU | 0.032 | |
| Fructose-bisphosphate aldolase, chloroplastic | ALFC_SPIOL | 0.033 | |
| Dehydroascorbate reductase | Q9FVE4_SPIOL | 0.061 | |
| Formate—tetrahydrofolate ligase | FTHS_SPIOL | 0.061 |
4. Conclusions
Supplementary Materials
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gidner, S.; Lennefors, B.L.; Nilsson, N.O.; Bensefelt, J.; Johansson, E.; Gyllenspetz, U.; Kraft, T. QTL mapping of BNYVV resistance from the WB41 source in sugar beet. Genome 2005, 48, 279–285. [Google Scholar] [CrossRef]
- Rush, C.M.; Liu, H.Y.; Lewellen, R.T.; Acosta-Leal, R. The continuing saga of rhizomania of sugar beets in the United States. Plant Dis. 2006, 90, 4–15. [Google Scholar] [CrossRef]
- Pavli, O.I.; Stevanato, P.; Biancardi, E.; Skaracis, G.N. Achievements and prospects in breeding for rhizomania resistance in sugar beet. Field Crops Res. 2011, 122, 165–172. [Google Scholar] [CrossRef]
- Abe, H.; Tamada, T. Association of Beet necrotic yellow vein virus with isolates of Polymyxa betae Keskin. Ann. Phytopathol. Soc. Jpn. 1986, 52, 235–247. [Google Scholar] [CrossRef]
- Liu, H.; Sears, J.L.; Lewellen, R.T. Occurrence of resistance breaking Beet necrotic yellow vein virus of sugar beet. Plant Dis. 2005, 89, 464–468. [Google Scholar] [CrossRef]
- Fujisawa, I.; Sugimoto, T. Transmission of Beet necrotic yellow vein virus by Polymyxa betae. Ann. Phytopathol. Soc. Jpn. 1976, 43, 583–586. [Google Scholar] [CrossRef]
- Tamada, T.; Baba, T. Beet necrotic yellow vein virus from rhizomania-affected sugar beet in Japan. Ann. Phytopathol. Soc. Jpn. 1973, 39, 325–332. [Google Scholar] [CrossRef]
- Lewellen, R.T.; Skoyen, I.O.; Erichsen, A.W. Breeding sugar beet for resistance to rhizomania: Evaluation of host-plant reactions and selection for and inheritance of resistance. In Proceeings of the 50th Congress of the IIRB, Brussels, Belgium, 11–12 February 1987; pp. 139–156.
- Lewellen, R.T. Selection for resistance to rhizomania in sugar beet. In Proceeings of the 5th International Congress Plant Pathology, Kyoto, Japan, 20–27 August 1988; p. 455.
- Scholten, O.E.; Lange, W. Breeding for resistance to rhizomania in sugar beet: A review. Euphytica 2000, 112, 219–231. [Google Scholar] [CrossRef]
- Biancardi, E.; Lewellen, R.T.; de Biaggi, M.; Erichsen, A.W.; Stevanato, P. The origin of rhizomania resistance to sugar beet. Euphytica 2002, 127, 383–397. [Google Scholar] [CrossRef]
- Scholten, O.E.; Klein-Lankhorst, R.M.; Esselink, D.G.; de Bock, T.S.; Lange, W. Identification and mapping of random amplified polymorphic DNA (RAPD) markers linked to resistance against Beet necrotic yellow vein virus (BNYVV) in Beta accessions. Theor. Appl. Genet. 1997, 94, 123–130. [Google Scholar] [CrossRef]
- Scholten, O.E.; de Bock, T.S.; Klein-Lankhorst, R.M.; Lange, W. Inheritance of resistance to Beet necrotic yellow vein virus in Beta vulgaris conferred by a second gene for resistance. Theor. Appl. Genet. 1999, 99, 740–746. [Google Scholar] [CrossRef]
- Koenig, R.; Luddecke, P.; Kaeberle, A.M. Detection of Beet necrotic yellow vein virus strains, variants, and mixed infections by examining single-strand conformation polymorphisms of immunocapture RT-PCR products. J. Gen. Virol. 1995, 76, 2051–2055. [Google Scholar] [CrossRef]
- Koenig, R.; Lennefors, B.L. Molecular analyses of European A, B, and P type sources of Beet necrotic yellow vein virus and detection of the rare P type in Kazakhstan. Arch. Virol. 2000, 145, 1561–1570. [Google Scholar] [CrossRef]
- Kruse, M.; Koenig, R.; Hoffmann, A.; Kaufmann, A.; Commandeur, U.; Solovyev, A.G.; Savenkov, I.; Burgermeister, W. Restriction fragment length polymorphism analysis of reverse transcription PCR products reveals the existence of two mojor strain groups of Beet necrotic yellow vein virus. J. Gen. Virol. 1994, 75, 1835–1842. [Google Scholar] [CrossRef]
- Schirmer, A.; Link, D.; Cognat, V.; Moury, B.; Bouve, M.; Meunier, A.; Bragard, C.; Gilmer, D.; Lemaire, O. Phylogenetic analysis of isolates of Beet necrotic yellow vein virus collected worldwide. J. Gen. Virol. 2005, 86, 2897–2911. [Google Scholar] [CrossRef]
- McGrann, G.R.D.; Grimmer, M.K.; Mutasa-Göttgens, E.S.; Stevens, M. Progress towards the understanding and control of sugarbeet rhizomania disease. Mol. Plant Pathol. 2009, 10, 129–141. [Google Scholar] [CrossRef]
- Miyanishi, M.; Kusume, T.; Saito, M.; Tamada, T. Evidence for three groups of sequence variants of been necrotic yellow vein virus RNA5. Arch. Virol. 1999, 144, 879–892. [Google Scholar] [CrossRef]
- Tamada, T.; Kusume, T.; Uchino, H.; Kiguchi, T.; Saito, M. Evidence that Beet necrotic yellow vein virus RNA-5 is involved in symptom development of sugarbeet roots. In Proceedings of the Third Symposium of the International Working Group on Plant Viruses with Fungal Vectors, Dundee, UK, 6–7 August 1996; 1996; pp. 49–52. [Google Scholar]
- Koenig, R.; Haeberle, A.M.; Commandeur, U. Detection and characterization of resistance-breaking isolates of Beet necrotic yellow vein virus in the United States. Eur. Arch. Virol. 1997, 142, 1499–1504. [Google Scholar] [CrossRef]
- Ward, L.; Koenig, R.; Budge, G.; Garrido, C.; McGrath, C.; Stubbley, H.; Boonham, N. Occurrence of two different types of RNA-5 containing Beet necrotic yellow vein virus in the UK. Arch. Virol. 2007, 152, 59–73. [Google Scholar] [CrossRef]
- Mehrvar, M.; Valizadeh, J.; Koenig, R.; Bragard, C. Iranian Beet necrotic yellow vein virus (BNYVV): Pronounced diversity of the p25 coding region in A-type BNYVV and identification of P-type BNYVV lacking a fifth RNA species. Arch. Virol. 2009, 154, 501–506. [Google Scholar] [CrossRef]
- Liu, H.Y.; Lewellen, R.T. Distribution and molecular characterization of resistance-breaking isolates of Beet necrotic yellow vein virus in the United States. Plant Dis. 2007, 91, 847–851. [Google Scholar] [CrossRef]
- Hajeidari, M.; Abdollahian-Noghabi, M.; Askari, H.; Heidari, M.; Sadeghian, S.Y.; Ober, E.S.; Salekdeh, G.H. Proteome analysis of sugar beet leaves under drought stress. Proteomics 2005, 5, 950–960. [Google Scholar] [CrossRef]
- Wakeel, A.; Asif, A.R.; Pitann, B.; Schubert, S. Proteome analysis of sugar beet (Beta vulgaris L.) elucidates constitutive adaptation during the first phase of salt stress. J. Plant Physiol. 2011, 168, 519–526. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, Y.; Zhu, N.; Koh, J.; Ma, C.; Pan, Y.; Yu, B.; Chen, S.; Li, H. Proteomic analysis of salt tolerance in sugar beet monosomic addition line M14. J. Proteome Res. 2013, 12, 4931–4920. [Google Scholar] [CrossRef]
- Rellan-Alvarez, R.; Andaluz, S.; Rodriguez-Celma, J.; Wohlgemuth, G.; Zocchi, G.; Alvarez-Fernandez, A.; Fiehn, O.; Lopez-Millan, A.F.; Abadia, J. Changes in the proteomic and metabolomic profiles of Beta vulgaris root tips in response to iron deficiency and resupply. BMC Plant Biol. 2010, 10, e120. [Google Scholar] [CrossRef] [Green Version]
- Catusse, J.; Strub, J.; Job, C.; van Dorsselaer, A.; Job, D. Proteome-wide characterization of sugarbeet seed vigor and its tissue specific expression. Proc. Natl. Acad. Sci. USA 2008, 105, 10262–10267. [Google Scholar] [CrossRef]
- Catusse, J.; Meinhard, J.; Job, C.; Strub, J.; Fischer, U.; Pestsova, E.; Westhoff, P.; van Dorsselaer, A.; Job, D. Proteomics reveals potential biomarkers of seed vigor in sugarbeet. Proteomics 2011, 11, 1569–1580. [Google Scholar] [CrossRef]
- Larson, R.L.; Hill, A.L.; Nunez, A. Characterization of protein changes associated with sugar beet (Beta vulgaris) resistance and susceptibility to Fusarium oxysporum. J. Agric. Food Chem. 2007, 55, 7905–7915. [Google Scholar] [CrossRef]
- Larson, R.L.; Wintermantel, W.M.; Hill, A.L.; Fortis, L.; Nunez, A. Proteome changes in sugar beet in response to Beet necrotic yellow vein virus. Physiol. Mol. Plant Pathol. 2008, 72, 62–72. [Google Scholar] [CrossRef]
- Wisler, G.C.; Lewellen, R.T.; Sears, J.L.; Liu, H.; Duffus, J.E. Specificity of TAS-ELISA for Beet necrotic yellow vein virus and its application for determining rhizomania resistance in field-grown sugar beets. Plant Dis. 1999, 83, 864–870. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Freund, D.M.; Prenni, J.E.; Curthoys, N.P. Response of the mitochondrial proteome of rat renal proximal convoluted tubules to chronic metabolic acidosis. Am. J. Physiol. 2013, 304, F145–F155. [Google Scholar]
- Uniprot Amaranthaceae database. Available online: http://www.uniprot.org/taxonomy/3563 (accessed on 6 March 2013).
- Uniprot Mus musculus database. Available online: http://www.uniprot.org/taxonomy/10090 (accessed on 6 March 2013).
- Giri, P.K.; Kruh, N.A.; Dobos, K.M.; Schorey, J.S. Proteomic analysis identifies highly antigenic proteins in exosomes from M. tuberculosis-infected and culture filtrate protein-treated macrophages. Proteomics 2010, 10, 3190–3202. [Google Scholar] [CrossRef]
- The Sugarbeet EST database. Available online: http://genomics.msu.edu/cgi-bin/sugarbeet/est_search.cgi (accessed on 5 April 2014).
- Beta vulgaris Gene Index (BvGI). Available online: http://compbio.dfci.harvard.edu/cgi-bin/tgi/tc_ann.pl?gudb=beet (accessed on 5 April 2014).
- Keller, A.; Nesvizhskii, A.I.; Kolker, E.; Aebersold, R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002, 74, 5383–5392. [Google Scholar] [CrossRef]
- Nesvizhskii, A.I.; Keller, A.; Kolker, E.; Aebersold, R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003, 75, 4646–4658. [Google Scholar] [CrossRef]
- Elias, J.E.; Gygi, S.P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 2007, 4, 207–214. [Google Scholar] [CrossRef]
- ProteomeXchange consortium. Available online: http://proteomecentral.proteomexchange.org (accessed on 6 March 2013).
- Vizcaino, J.A.; Cote, R.G.; Csordas, A.; Dianes, J.A.; Fabregat, A.; Foster, J.M.; Griss, J.; Alpi, E.; Birim, M.; Contell, J.; et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: Status in 2013. Nucleic Acids Res. 2013, 4, D1063–D1069. [Google Scholar]
- Paoletti, A.C.; Parmely, T.J.; Tomomori-Sato, C.; Sato, S.; Zhu, D.; Conaway, R.C.; Conaway, J.W.; Florens, L.; Washburn, M.P. Quantitative proteomic analysis of distinct mamalian mediator complexes using normalized spectral abundance factors. Proc. Natl. Acad. Sci. USA 2006, 103, 18928–18933. [Google Scholar] [CrossRef]
- Carli, M.D.; Benvenuto, E.; Donini, M. Recent insights into plant-virus interactions through proteomic analysis. J. Proteome Res. 2012, 11, 4765–4780. [Google Scholar] [CrossRef]
- Ranty, B.; Alson, D.; Galaud, J. Plant calmodulins and calmodulin-related proteins. Plant Signal. Behav. 2006, 1, 96–104. [Google Scholar] [CrossRef]
- Beffa, R.S.; Hofer, R.M.; Thomas, M.; Meins, F., Jr. Decreased susceptibility to viral disease of [beta]-1,3-glucanase-deficient plants generated by antisense transformation. Plant Cell 1996, 8, 1001–1011. [Google Scholar]
- Bell, A.A. Biochemical mechanisms of disease resitance. Annu. Rev. Plant Physiol. 1981, 32, 21–81. [Google Scholar] [CrossRef]
- Stone, B.A.; Clarke, A.E. Chemistry and Biology of (1-3)-beta-glucans; La Trobe University Press: Melbourne, Austrailia, 1992. [Google Scholar]
- Aist, J.R. Papillae and related wounds plugs of plant cells. Annu. Rev. Phytopathol. 1976, 14, 145–163. [Google Scholar] [CrossRef]
- Allison, A.V.; Shalla, T.A. The ultrastructure of local lesions induced by potato virus X: A sequence of cytological events in the course of infection. Phytopathology 1974, 64, 784–793. [Google Scholar] [CrossRef]
- Iglesias, V.A.; Meins, F., Jr. Movement of plant viruses is delayed in a beta-1,3-glucanase deficient mutant showing a reduced plasmodesmatal size exclusion limit and enhanced callose deposition. Plant J. 2000, 21, 157–166. [Google Scholar] [CrossRef]
- Roberts, M.R.; Salinas, J.; Collinge, D.B. 14-3-3 proteins and the response to abiotic and biotic stress. Plant Mol. Biol. 2002, 1031, 1031–1039. [Google Scholar]
- Denison, F.C.; Paul, A.L.; Zupanska, A.K.; Ferl, R.J. 14-3-3 proteins in plant physiology. Semin. Cell Dev. Biol. 2011, 22, 720–727. [Google Scholar] [CrossRef]
- Van Loon, L.C.; van Strien, E.A. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 1999, 55, 85–97. [Google Scholar] [CrossRef]
- Chittoor, J.M.; Leach, J.E.; White, F.F. Induction of peroxidase during defense against pathogens. In Pathogenesis-Related Proteins in Plants; Datta, S.K., Muthukrishnan, S., Eds.; CRC Press: Boca Raton, FL, USA, 1999; pp. 171–193. [Google Scholar]
- Kawano, T. Roles of the reactive oxygen species-generating peroxidase reactions in plant defese and growth induction. Plant Cell Rep. 2003, 21, 829–837. [Google Scholar]
- Caverzan, A.; Passaia, G.; Barcellos Rosa, S.; Werner Ribeiro, C.; Lazzarotto, F.; Margis-Pinheiro, M. Plant resonses to stresses: Role of ascorbate peroxidase in the antioxidant protection. Genet. Mol. Biol. 2012, 35, 1011–1019. [Google Scholar] [CrossRef]
- Gupta, A.S.; Heinen, J.L.; Holaday, A.S.; Burke, J.J.; Allen, R.D. Increased resistance to oxidative stress in transgenic plants that overexpress chloroplastic Cu/Zn superoxide dismutase. Proc. Natl. Acad. Sci. USA 1993, 90, 1629–1633. [Google Scholar] [CrossRef]
- Tsang, E.; Bowler, C.; Herouart, D.; van Camp, W.; Villarroel, R.; Genetello, C.; van Montagu, M.; Inze, D. Differential regulation of superoxide dismutases in plants exposed to environmental stress. Plant Cell 1991, 3, 783–792. [Google Scholar]
- Crow, J.F. Basic Concepts in Population, Quantitative, and Evolutionary Genetics; Academic Press: New York, NY, USA, 1986; p. 273. [Google Scholar]
- Antonovics, J.; Alexander, H.M. The concept of fitness in plant-fungal pathogen systems. In Plant Disease Epidemiology; Leonard, K.J., Fry, W.E., Eds.; McGraw-Hill: New York, NY, USA, 1989; pp. 185–214. [Google Scholar]
- Leach, J.E.; Vera Cruz, C.M.; Bai, J.; Leung, H. Pathogen fitness penalty as a predictor of durability of disease resistance genes. Annu. Rev. Phytopathol. 2001, 39, 187–224. [Google Scholar] [CrossRef]
- Eujayl, I.; Strausbaugh, C. Beta vulgaris subsp. Vulgaris. Available online: http://www.ncbi.nlm.nih.gov/Traces/wgs/?val=ARYA01 (accessed on 5 April 2014).
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Webb, K.M.; Broccardo, C.J.; Prenni, J.E.; Wintermantel, W.M. Proteomic Profiling of Sugar Beet (Beta vulgaris) Leaves during Rhizomania Compatible Interactions. Proteomes 2014, 2, 208-223. https://doi.org/10.3390/proteomes2020208
Webb KM, Broccardo CJ, Prenni JE, Wintermantel WM. Proteomic Profiling of Sugar Beet (Beta vulgaris) Leaves during Rhizomania Compatible Interactions. Proteomes. 2014; 2(2):208-223. https://doi.org/10.3390/proteomes2020208
Chicago/Turabian StyleWebb, Kimberly M., Carolyn J. Broccardo, Jessica E. Prenni, and William M. Wintermantel. 2014. "Proteomic Profiling of Sugar Beet (Beta vulgaris) Leaves during Rhizomania Compatible Interactions" Proteomes 2, no. 2: 208-223. https://doi.org/10.3390/proteomes2020208




